Artificial Intelligence in Food Safety: A Decade Review and Bibliometric Analysis
Abstract
1. Introduction
- We selected 1855 articles as research samples to explore which AI technologies were applied for a sustainable food system from farm to fork;
- Our review elaborated on the development trend and current research hotspots on AI technologies in food safety and predicted the future research direction;
- This review should be helpful for researchers and practitioners to comprehensively understand the application status of AI technologies in the food sector;
- We have elaborated on the countries, institutions, and journals that have contributed much to the research on AI technologies in food safety.
2. Theoretical Background
3. Method and Data
3.1. Method and Software
3.2. Sample
3.3. Analyses
4. Analysis and Results
4.1. Current Status of Al Research
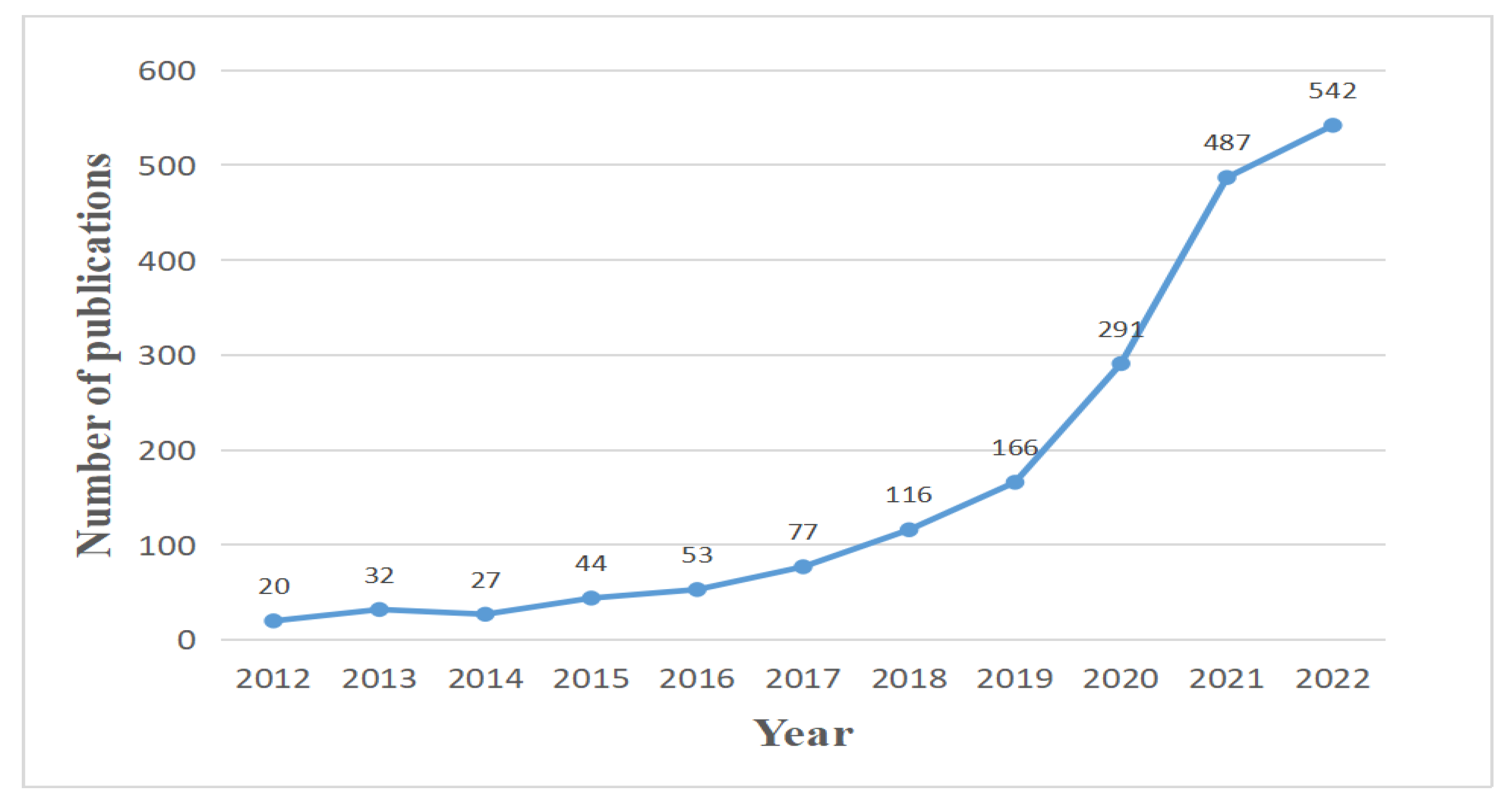
4.1.1. Annual Trends
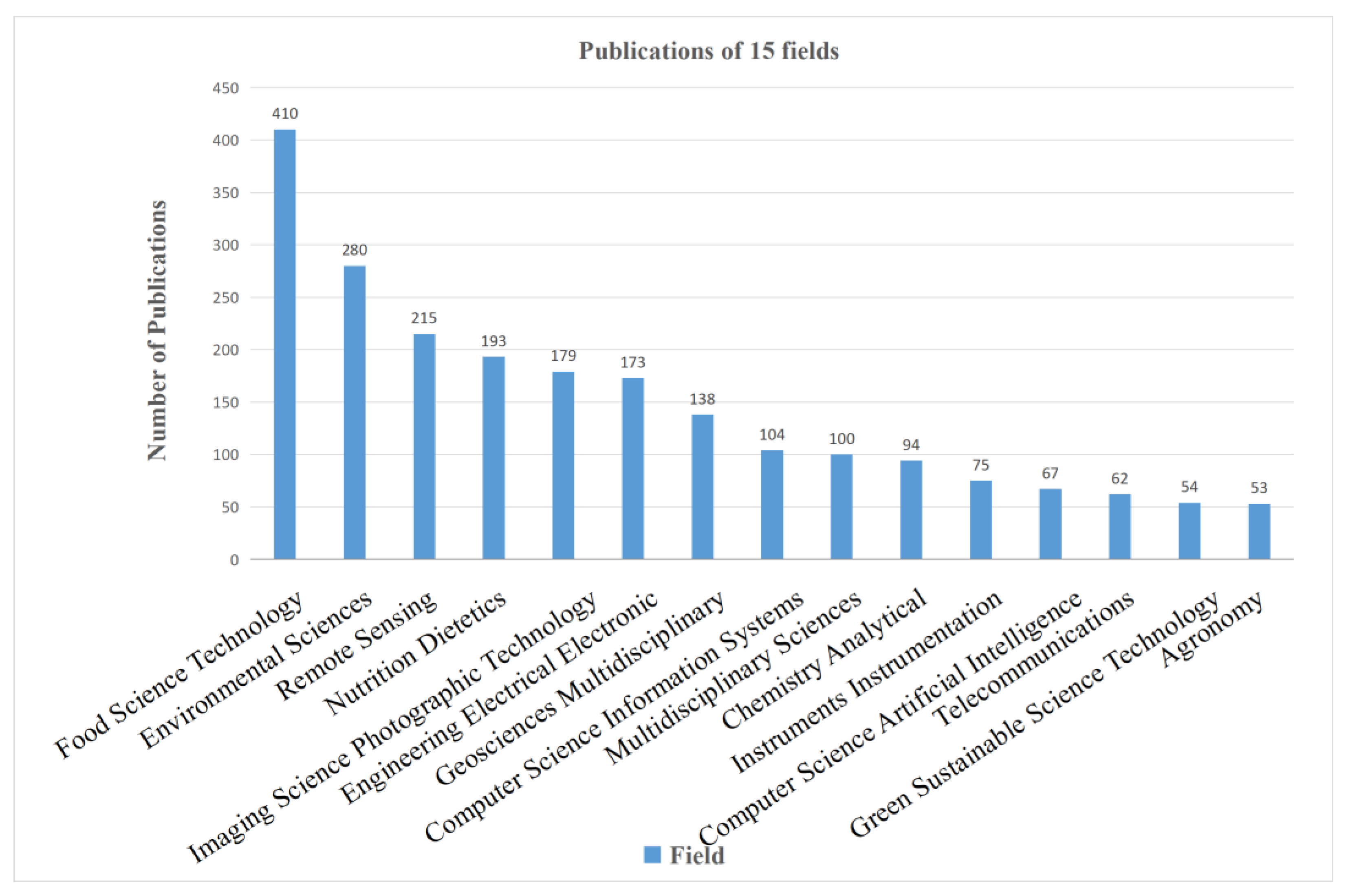
4.1.2. Distribution of Publications
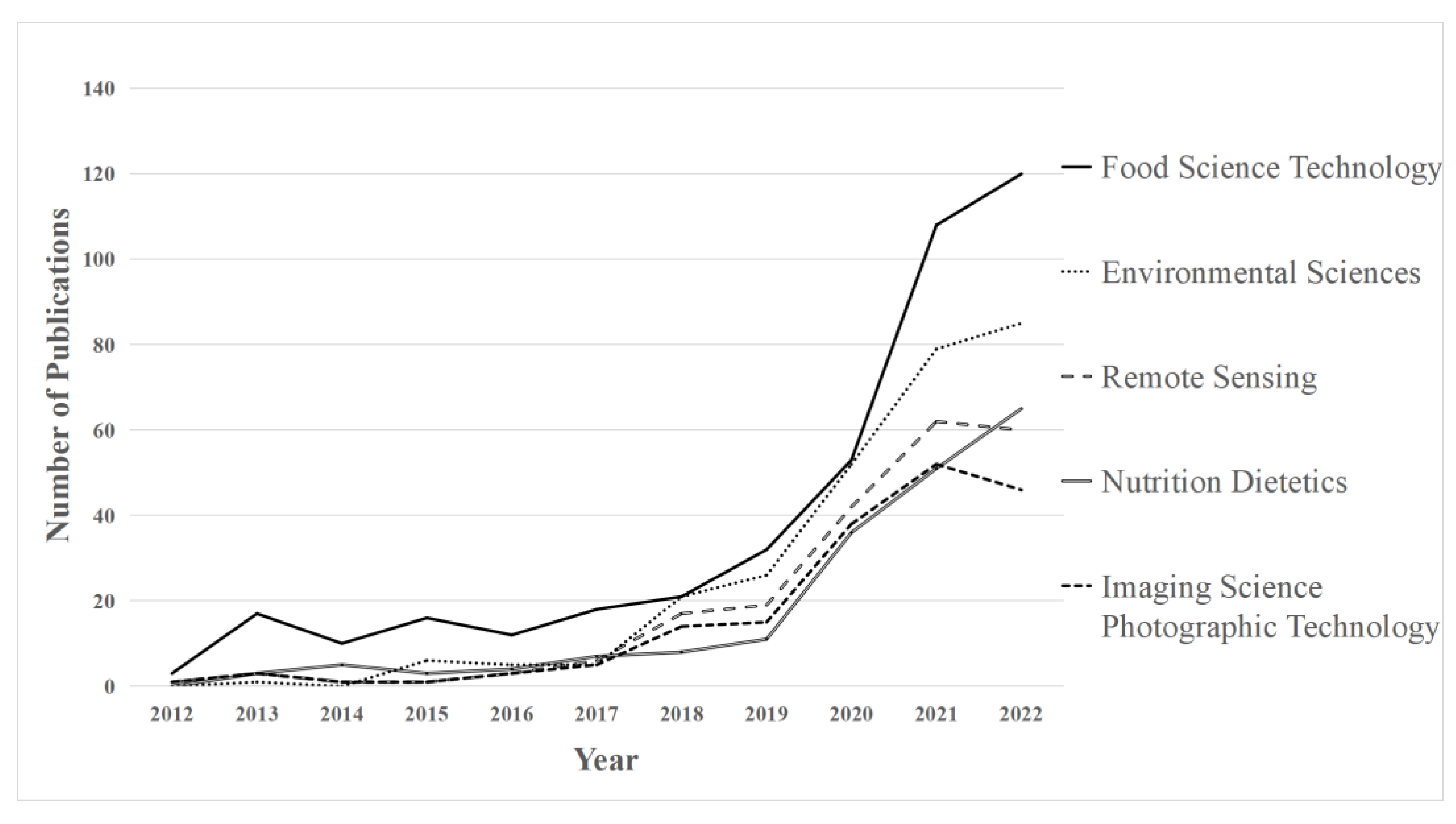
4.1.3. Publication Timeline
4.2. Co-Citation Analyses
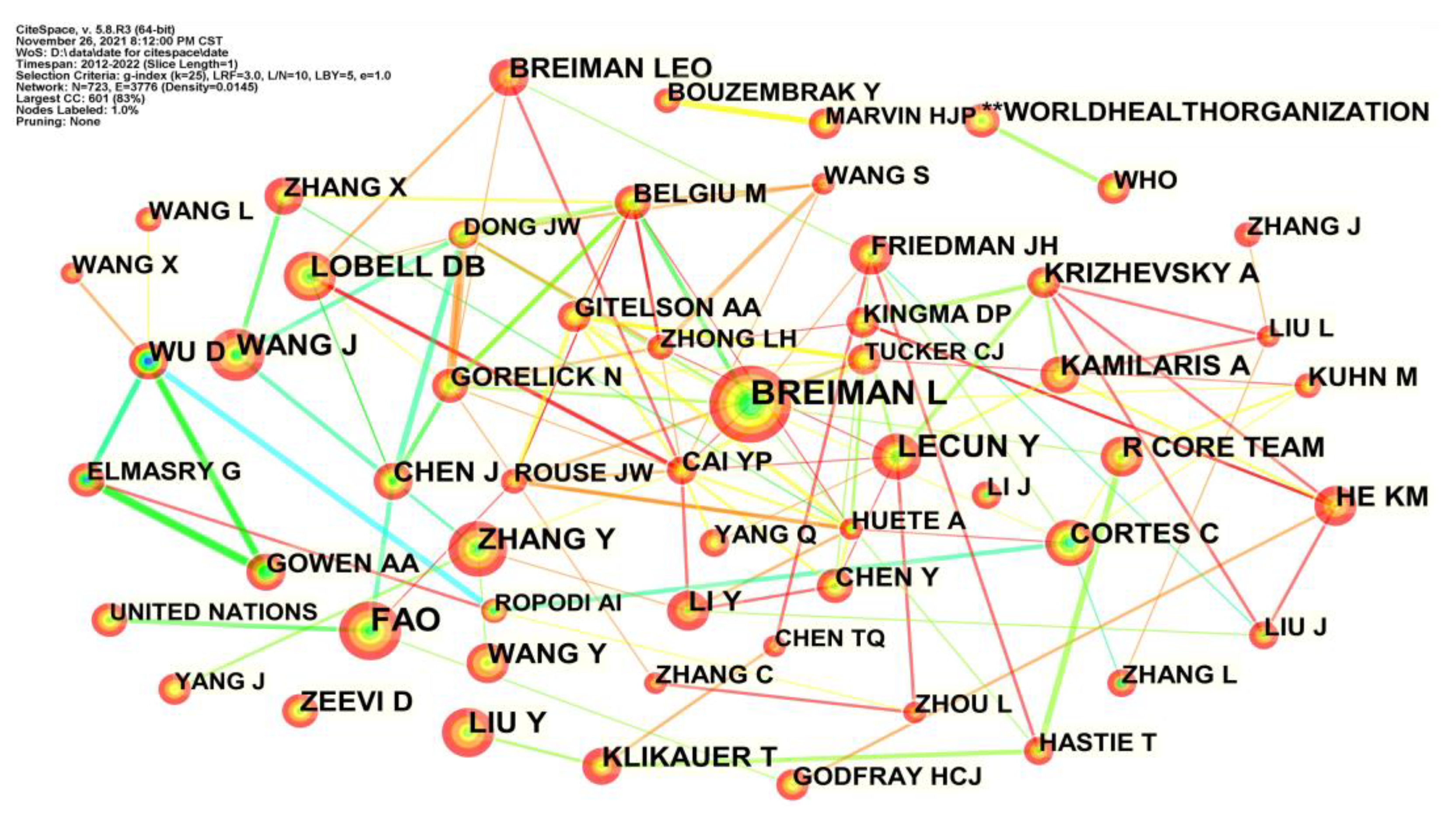
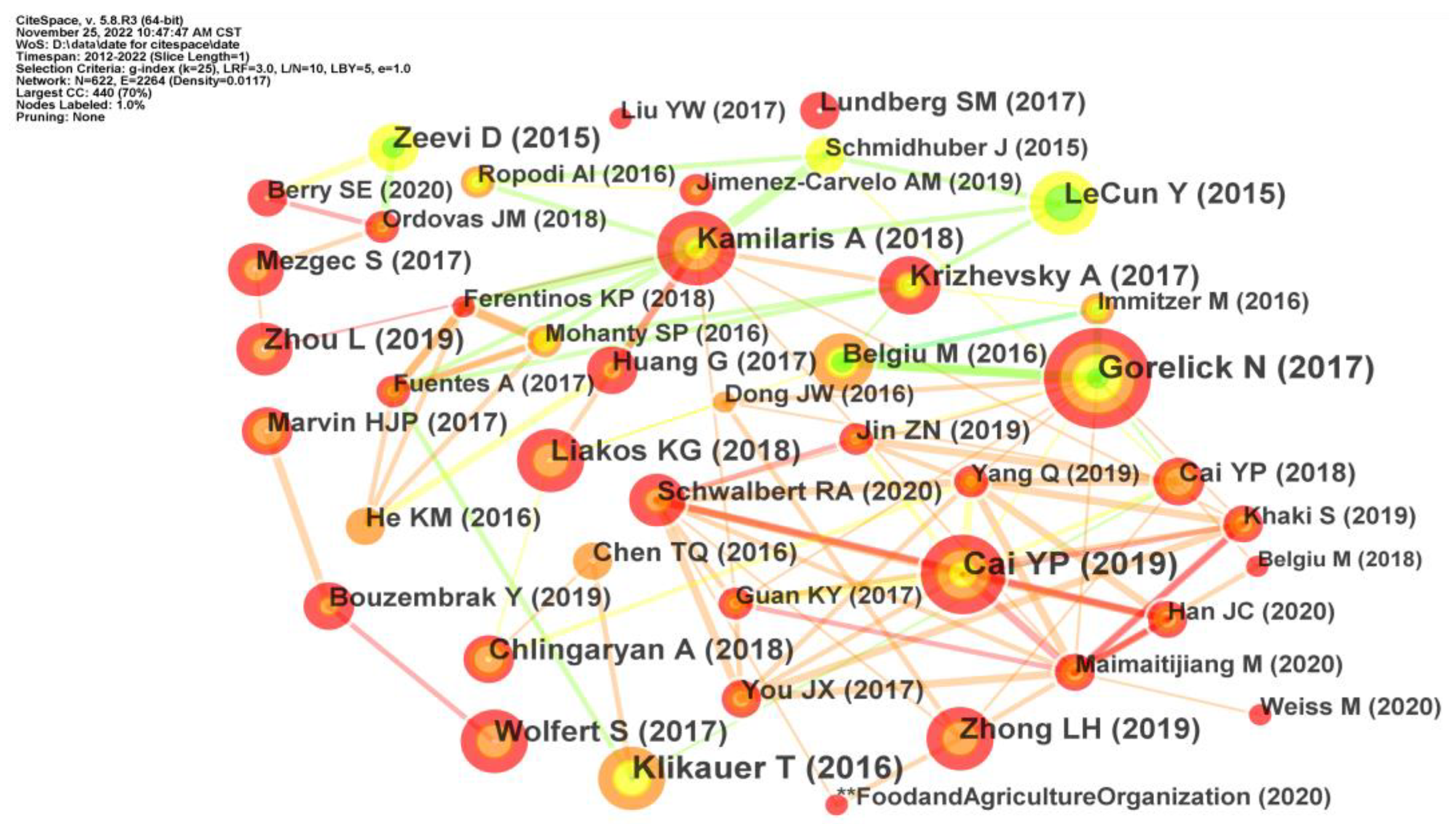
4.2.1. Author Co-Citation Analysis
4.2.2. Reference Co-Citation Analysis
4.2.3. Citation Burst Analysis
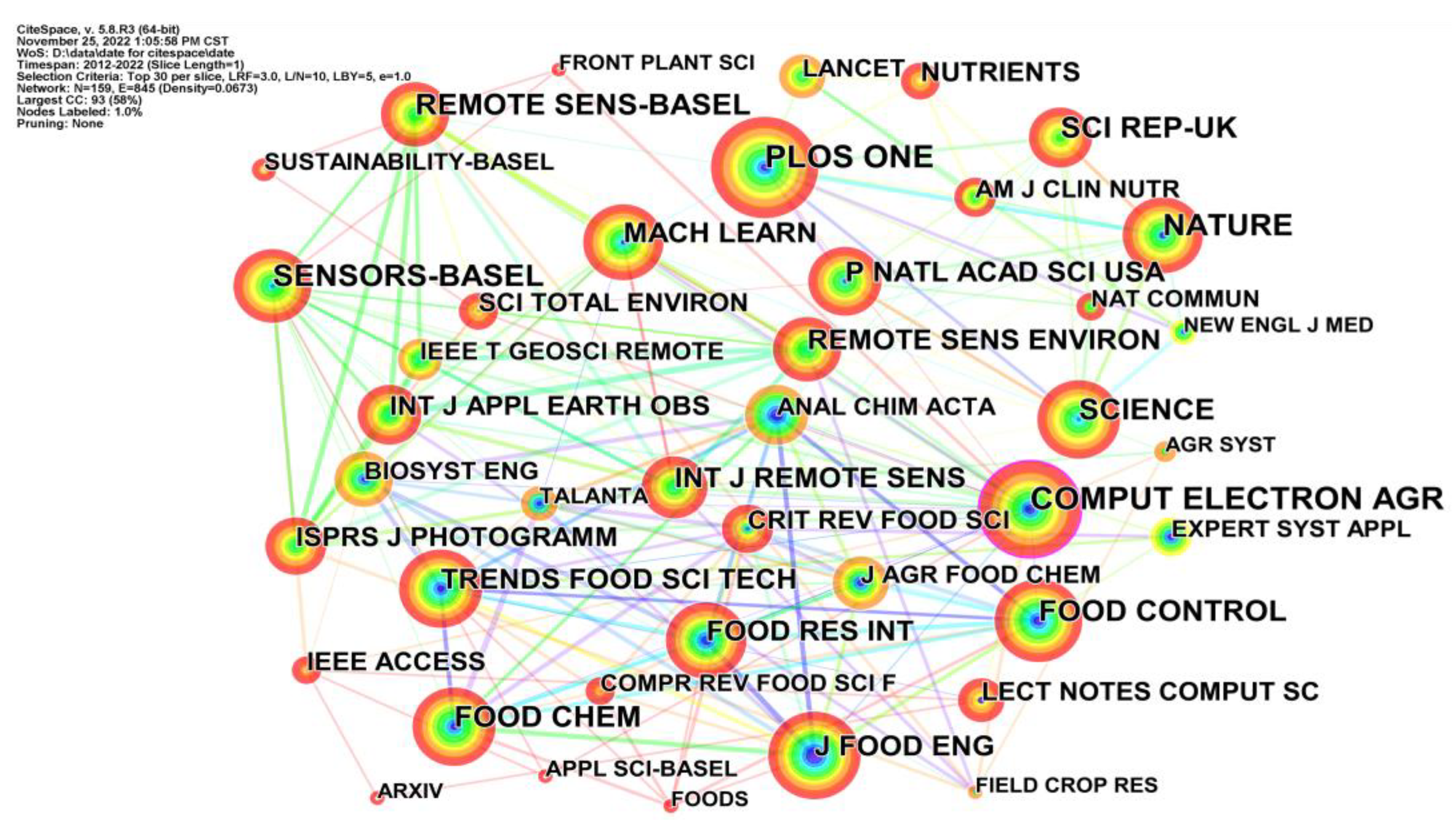
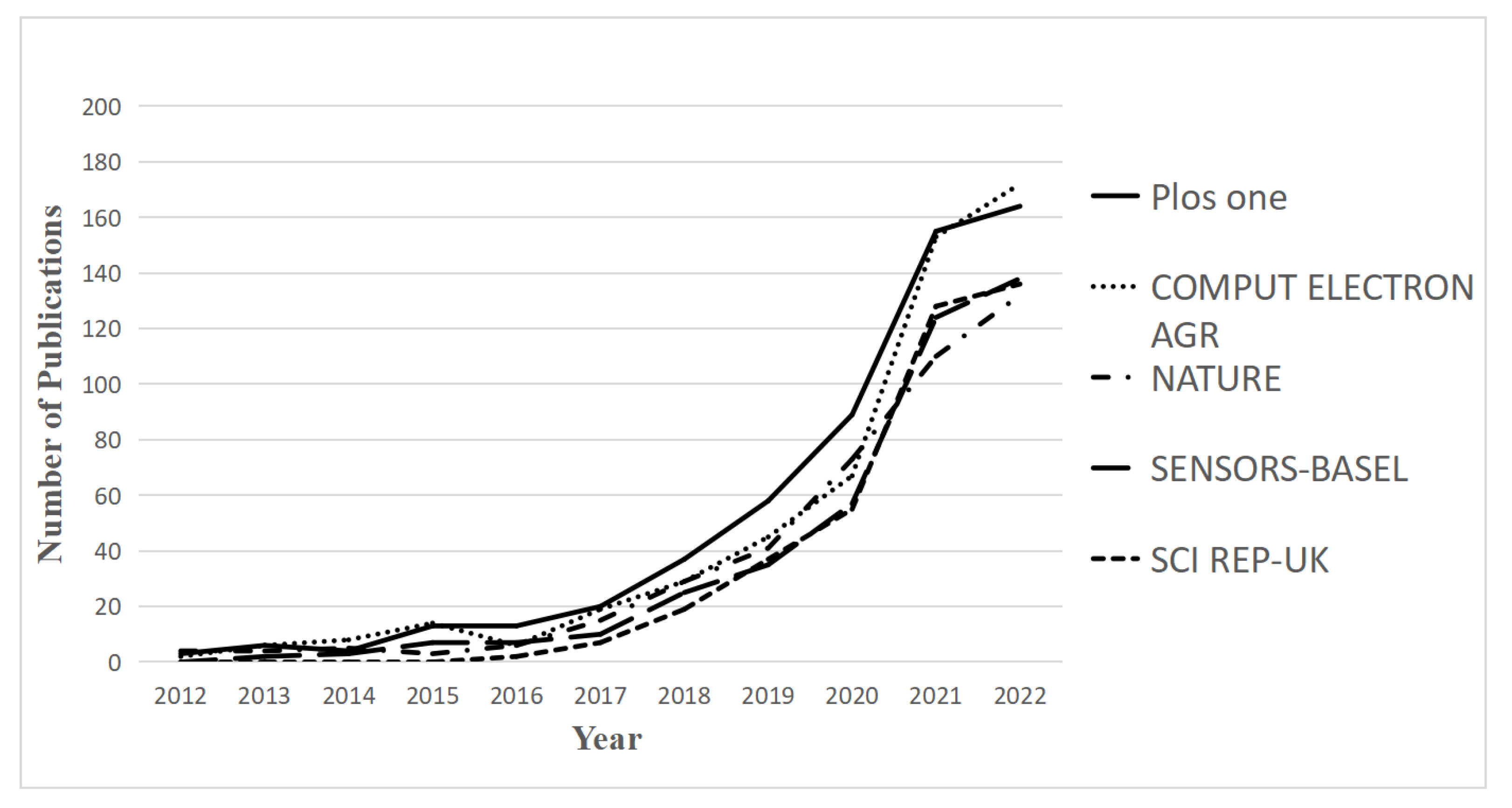
4.2.4. Journal Co-Citation Analysis
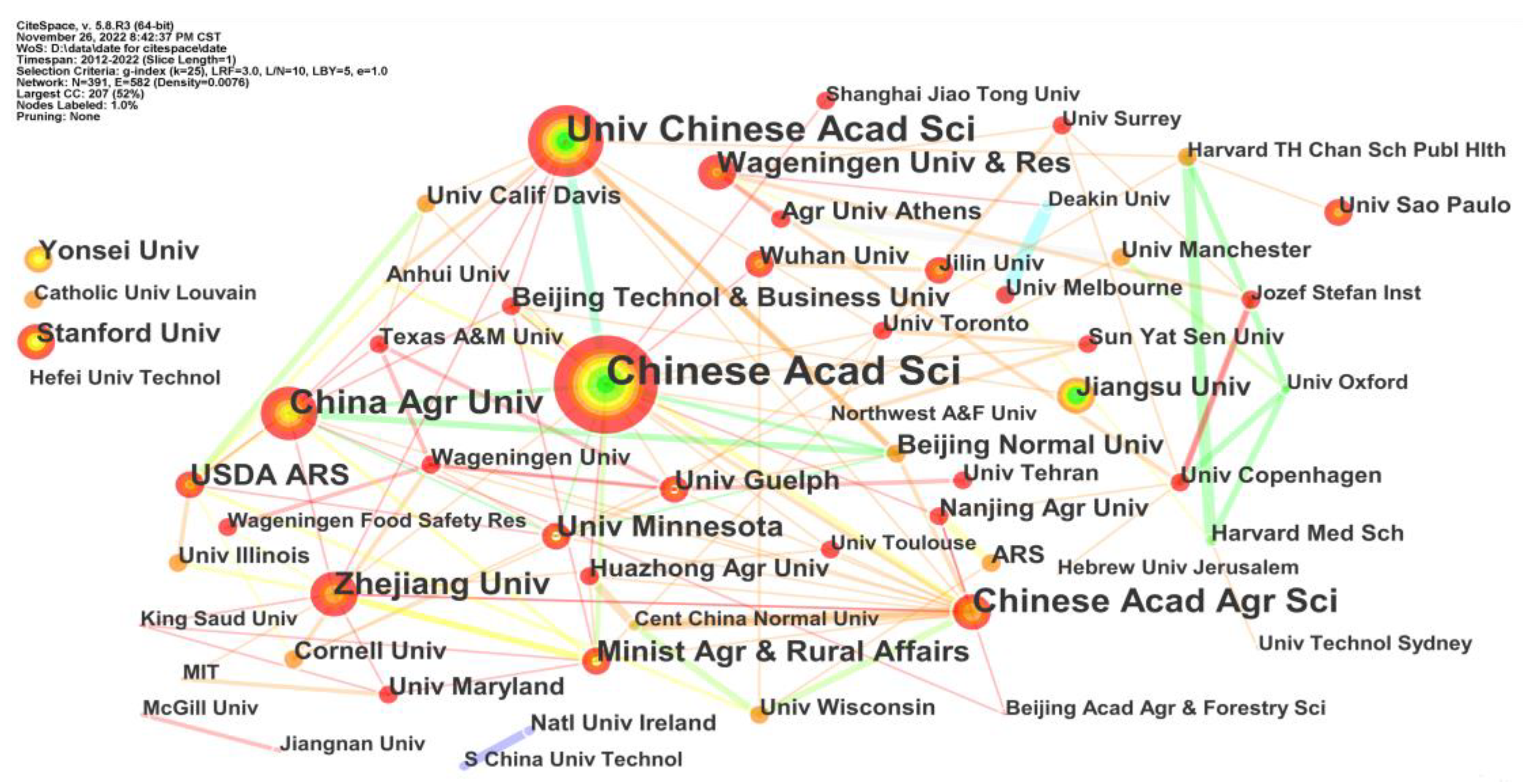
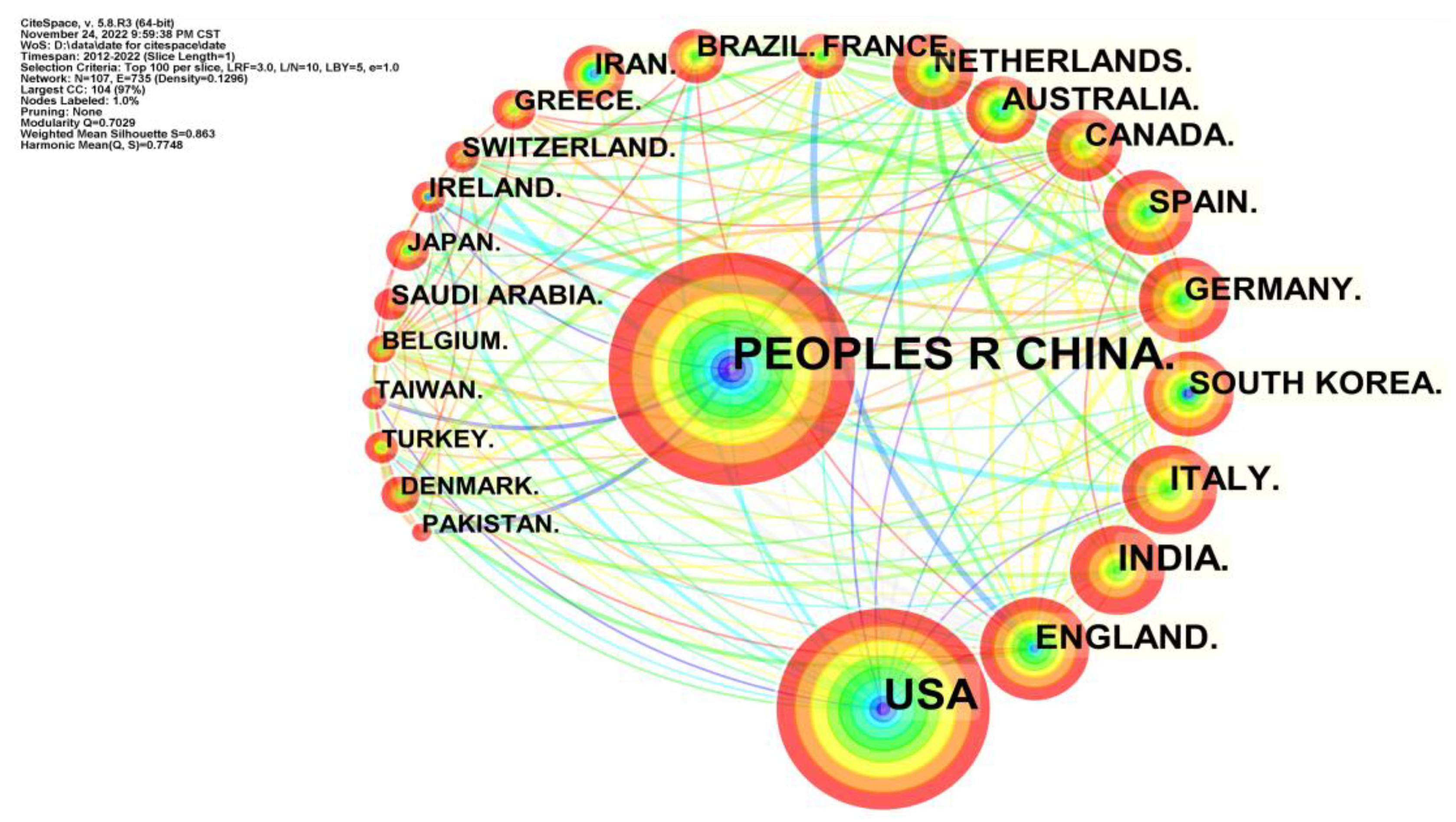
4.3. Co-authorship Analysis
5. Keywords and Hot Spots
5.1. Keyword Co-Occurrence Analysis
5.2. Research Hotspots
5.2.1. Cluster #0—Remote Sensing
5.2.2. Cluster #1—Food Quality
5.2.3. Cluster #2—Personalized Nutrition
5.2.4. Cluster #3—Big Data
5.2.5. Cluster #4—Food Safety
5.2.6. Cluster #5—Deep Learning
5.2.7. Cluster #6—Artificial Neural Network
6. Discussion
6.1. Theoretical Implications
6.1.1. AI Technologies in Molecular Breeding
6.1.2. AI Technologies in Agricultural Production
6.1.3. AI Technologies in Food Processing and Distribution
6.1.4. AI Technologies in Food Nutrition
6.2. Practical Implications
6.3. Limitations
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | adaptive boosting |
| ABC | artificial bee colony |
| AI | artificial intelligence |
| ANFIS | adaptive network-based fuzzy inference system |
| ANN | artificial neural network |
| BN | Bayesian network |
| BNN | Bayesian neural network |
| BNPK | Bayesian network model fusing prior knowledge |
| BP-ANN | back propagation artificial neural network |
| BPNN | back propagation neural network |
| BayesR | Bayesian mixture model |
| CGBN | conditional Gaussian Bayesian network |
| CNN | convolutional neural network |
| CS | cuckoo search |
| CVS | computer vision system |
| DAL_CL | deep attention layer-based convolutional learning |
| DCNN | deep convolutional neural network |
| DE | differential evolution |
| DNN | deep neural network |
| DNN-MCP | MCP regularization for sparse deep neural network |
| DR | dimensionality reduction |
| DS | data science |
| DT | decision tree |
| FFM | feature fusion module |
| ELM | extreme learning machine |
| GA | genetic algorithm |
| GBLUP | genomic best linear unbiased predictor |
| GP | Gaussian processes |
| GS | greed search |
| Hy-CNN | hybrid convolutional neural network |
| K2 | K2 algorithm for Bayesian network structure |
| KNN | k-nearest neighbours |
| LDA | linear discriminant analysis |
| LightGBM | light gradient boosting machine |
| LR | logistic regression |
| LP | linear programming |
| J48 | J48 decision tree |
| MCP | minmax concave penalty |
| ML | machine learning |
| MLP | multilayer perceptron |
| MLR | multiple linear regression |
| MOB | model-based recursive partitioning |
| MODAS | multi-omics data association studies |
| MODIS | moderate resolution imaging spectra radiometer |
| MOLO | multi-objective local optimization |
| NIR | near-infrared spectroscopy |
| NN | neural network |
| OLR | optimal linear regression |
| PC | Phenocentric |
| PLS | partial least squares regression |
| PLS-DA | partial least-squares discriminant analysis |
| PLSR | partial least squares regression |
| QDA | quadratic discriminant analysis |
| R2 | determination coefficient |
| RF | random forest |
| RF-r | random forest regression |
| RMSE | root-mean-square error |
| RNN | recurrent neural network |
| ResNet | residual network |
| SA | simulated annealing |
| SNP | single nucleotide polymorphism |
| ST | Swin Transformer |
| SVM | support vector machine |
| SVR | support vector regression |
| TL | transfer learning |
| XGBoost | extreme gradient boosting |
References
- Haenlein, M.; Kaplan, A. A brief history of artificial intelligence: On the past, present, and future of artificial intelligence. Calif. Manag. Rev. 2019, 61, 5–14. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kudashkina, K.; Corradini, M.G.; Thirunathan, P.; Yada, R.Y.; Fraser, E.D. Artificial Intelligence technology in food safety: A behavioral approach. Trends Food Sci. Technol. 2022, 123, 376–381. [Google Scholar] [CrossRef]
- Kim, S.S.; Kim, S. Impact and prospect of the fourth industrial revolution in food safety: Mini-review. Food Sci. Biotechnol. 2022, 31, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Tahir, G.A.; Loo, C.K. A comprehensive survey of image-based food recognition and volume estimation methods for dietary assessment. Healthcare 2021, 9, 1676. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Kumar, I.; Rawat, J.; Mohd, N.; Husain, S. Opportunities of artificial intelligence and machine learning in the food industry. J. Food Qual. 2021, 4535567. [Google Scholar] [CrossRef]
- Marvin, H.J.; Bouzembrak, Y.; Van der Fels-Klerx, H.J.; Kempenaar, C.; Veerkamp, R.; Chauhan, A.; Stroosnijder, S.; Top, J.; Simsek-Senel, G.; Tekinerdogan, B. Digitalisation and Artificial Intelligence for sustainable food systems. Trends Food Sci. Technol. 2022, 120, 344–348. [Google Scholar] [CrossRef]
- Deng, X.; Cao, S.; Horn, A.L. Emerging applications of machine learning in food safety. Annu. Rev. Food Sci. Technol. 2022, 12, 513–538. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Li, J.; Liu, G.; Yang, M.M.; Liu, S. A bibliometric review of a decade of research: Big data in business research–setting a research agenda. J. Bus. Res. 2021, 131, 374–390. [Google Scholar] [CrossRef]
- Smith, P.D. Hands-On Artificial Intelligence for Beginners: An Introduction to AI Concepts, Algorithms, and Their Implementation; Packt Publishing Ltd.: Birmingham, UK, 2018; pp. 6–7. [Google Scholar]
- Zhou, L.; Zhang, C.; Liu, F.; Qiu, Z.; He, Y. Application of deep learning in food: A review. Compr. Rev. Food Sci. Food Safety 2019, 18, 1793–1811. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q.V. Mnasnet: Platform-aware neural architecture search for mobile. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 2820–2828. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8697–8710. [Google Scholar]
- Shen, C.; Wei, M.; Sheng, Y. A bibliometric analysis of food safety governance research from 1999 to 2019. Food Sci. Nutr. 2021, 9, 2316–2334. [Google Scholar] [CrossRef]
- Jovanovic, M.; Mitrov, G.; Zdravevski, E.; Lameski, P.; Colantonio, S.; Kampel, M.; Tellioglu, H.; Florez-Revuelta, F. Ambient Assisted Living: Scoping Review of Artificial Intelligence Models, Domains, Technology, and Concerns. J. Med. Internet Res. 2022, 24, e36553. [Google Scholar] [CrossRef]
- Camaréna, S. Artificial intelligence in the design of the transitions to sustainable food systems. J. Clean. Prod. 2020, 271, 122574. [Google Scholar] [CrossRef]
- Pranckutė, R. Web of Science (WoS) and Scopus: The titans of bibliographic information in today’s academic world. Publ. 2021, 9, 12. [Google Scholar] [CrossRef]
- Donthu, N.; Kumar, S.; Mukherjee, D.; Pandey, N.; Lim, W.M. How to conduct a bibliometric analysis: An overview and guidelines. J. Bus. Res. 2021, 133, 285–296. [Google Scholar] [CrossRef]
- Moral-Muñoz, J.A.; Herrera-Viedma, E.; Santisteban-Espejo, A.; Cobo, M.J. Software tools for conducting bibliometric analysis in science: An up-to-date review. Prof. De La Inf. 2020, 29, e290103. [Google Scholar] [CrossRef]
- Chen, C. Science mapping: A systematic review of the literature. J. Data Inf. Sci. 2017, 2, 1–40. [Google Scholar] [CrossRef]
- White, H.D.; McCain, K.W. Visualizing a discipline: An author co-citation analysis of information science, 1972–1995. J. Am. Soc. Inf. Sci. 1998, 49, 327–355. [Google Scholar]
- Persson, O. Identifying research themes with weighted direct citation links. J. Informetr. 2010, 4, 415–422. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006, 57, 359–377. [Google Scholar] [CrossRef]
- Chen, C.; Dubin, R.; Kim, M.C. Emerging trends and new developments in regenerative medicine: A scientometric update (2000–2014). Expert Opin. Biol. Ther. 2014, 14, 1295–1317. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Determination of total viable count (TVC) in chicken breast fillets by near-infrared hyperspectral imaging and spectroscopic transforms. Talanta 2013, 105, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Barbin, D.F.; ElMasry, G.; Sun, D.W.; Allen, P. Predicting quality and sensory attributes of pork using near-infrared hyperspectral imaging. Anal. Chim. Acta. 2012, 719, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Sun, D.W.; He, Y. Application of long-wave near infrared hyperspectral imaging for measurement of color distribution in salmon fillet. Innov. Food Sci. Emerg. Technol. 2012, 16, 361–372. [Google Scholar] [CrossRef]
- ElMasry, G.; Sun, D.W.; Allen, P. Non-destructive determination of water-holding capacity in fresh beef by using NIR hyperspectral imaging. Food Res. Int. 2011, 44, 2624–2633. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Papadopoulou, O.; Carstensen, J.M.; Nychas, G.J.E. Potential of multispectral imaging technology for rapid and non-destructive determination of the microbiological quality of beef filets during aerobic storage. Int. J. Food Microbiol. 2014, 174, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Chao, K.; Kim, M.S.; Lu, R.; Burks, T.F. Hyperspectral and multispectral imaging for evaluating food safety and quality. J. Food Eng. 2013, 118, 157–171. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.J. Data mining derived from food analyses using non-invasive/non-destructive analytical techniques; determination of food authenticity, quality & safety in tandem with computer science disciplines. Trends Food Sci. Technol. 2016, 50, 11–25. [Google Scholar]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Segal, E. Personalized nutrition by prediction of glycemic responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Mosleh, M.K.; Hassan, Q.K.; Chowdhury, E.H. Application of remote sensors in mapping rice area and forecasting its production: A review. Sensors 2015, 15, 769–791. [Google Scholar] [CrossRef]
- Xiong, J.; Thenkabail, P.S.; Tilton, J.C.; Gumma, M.K.; Teluguntla, P.; Oliphant, A.; Congalton, R.G.; Yadav, K.; Gorelick, N. Nominal 30-m cropland extent map of continental Africa by integrating pixel-based and object-based algorithms using Sentinel-2 and Landsat-8 data on Google Earth Engine. Remote Sens. 2017, 9, 1065. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Zhao, X. A scientometric review of global BIM research: Analysis and visualization. Autom. Constr. 2017, 80, 37–47. [Google Scholar] [CrossRef]
- Li, H.; An, H.; Wang, Y.; Huang, J.; Gao, X. Evolutionary features of academic articles co-keyword network and keywords co-occurrence network: Based on two-mode affiliation network. Phys. A Stat. Mech. Its Appl. 2016, 450, 657–669. [Google Scholar] [CrossRef]
- Goyal, S. Artificial neural networks (ANNs) in food science–A review. Int. J. Sci. World. 2013, 1, 19–28. [Google Scholar] [CrossRef]
- Duan, C.; Chen, C.; Khan, M.N.; Liu, Y.; Zhang, R.; Lin, H.; Cao, L. Non-destructive determination of the total bacteria in flounder fillet by portable near infrared spectrometer. Food Control 2014, 42, 18–22. [Google Scholar] [CrossRef]
- Liu, C.; Cao, Y.; Luo, Y.; Chen, G.; Vokkarane, V.; Yunsheng, M.; Chen, S.; Hou, P. A new deep learning-based food recognition system for dietary assessment on an edge computing service infrastructure. IEEE Trans. Serv. Comput. 2017, 11, 249–261. [Google Scholar] [CrossRef]
- Johnson, M.D.; Hsieh, W.W.; Cannon, A.J.; Davidson, A.; Bédard, F. Crop yield forecasting on the Canadian Prairies by remotely sensed vegetation indices and machine learning methods. Agric. For. Meteorol. 2016, 218, 74–84. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Kong, J.L.; Jin, X.B.; Wang, X.Y.; Su, T.L.; Zuo, M. CropDeep: The crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors 2019, 19, 1058. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, K.M.; Ramos-Lopez, O.; Pérusse, L.; Kato, H.; Ordovas, J.M.; Martínez, J.A. Precision nutrition: A review of current approaches and future endeavors. Trends Food Sci. Technol. 2022, 128, 253–264. [Google Scholar] [CrossRef]
- Pradana-López, S.; Pérez-Calabuig, A.M.; Cancilla, J.C.; Lozano, M.Á.; Rodrigo, C.; Mena, M.L.; Torrecilla, J.S. Deep transfer learning to verify quality and safety of ground coffee. Food Control 2021, 122, 107801. [Google Scholar] [CrossRef]
- Chen, C.; Ibekwe-Sanjuan, F.; Hou, J. The structure and dynamics of co-citation clusters: A multiple-perspective co-citation analysis. J. Am. Soc. Inf. Sci. Technol. 2010, 61, 1386–1409. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Hao, P.; Zhan, Y.; Wang, L.; Niu, Z.; Shakir, M. Feature selection of time series MODIS data for early crop classification using random forest: A case study in Kansas, USA. Remote Sens. 2015, 7, 5347–5369. [Google Scholar] [CrossRef]
- Vreugdenhil, M.; Wagner, W.; Bauer-Marschallinger, B.; Pfeil, I.; Teubner, I.; Rüdiger, C.; Strauss, P. Sensitivity of Sentinel-1 backscatter to vegetation dynamics: An Austrian case study. Remote Sens. 2018, 10, 1396. [Google Scholar] [CrossRef]
- Han, J.; Zhang, Z.; Cao, J.; Luo, Y.; Zhang, L.; Li, Z.; Zhang, J. Prediction of winter wheat yield based on multi-source data and machine learning in China. Remote Sens. 2020, 12, 236. [Google Scholar] [CrossRef]
- Teluguntla, P.; Thenkabail, P.S.; Oliphant, A.; Xiong, J.; Gumma, M.K.; Congalton, R.G.; Yadav, K.; Huete, A. A 30-m landsat-derived cropland extent product of Australia and China using random forest machine learning algorithm on Google Earth Engine cloud computing platform. ISPRS J. Photogramm. Remote Sens. 2018, 144, 325–340. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Z.; Tao, F.; Zhang, L.; Luo, Y.; Han, J.; Li, Z. Identifying the contributions of multi-source data for winter wheat yield prediction in China. Remote Sens. 2020, 12, 750. [Google Scholar] [CrossRef]
- Duke, O.P.; Alabi, T.; Neeti, N.; Adewopo, J. Comparison of UAV and SAR performance for Crop type classification using machine learning algorithms: A case study of humid forest ecology experimental research site of West Africa. Int. J. Remote Sens. 2022, 43, 4259–4286. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Z.; Kang, Y.; Özdoğan, M. Corn yield prediction and uncertainty analysis based on remotely sensed variables using a Bayesian neural network approach. Remote Sens. Environ. 2021, 259, 112408. [Google Scholar] [CrossRef]
- Hu, Q.; Yin, H.; Friedl, M.A.; You, L.; Li, Z.; Tang, H.; Wu, W. Integrating coarse-resolution images and agricultural statistics to generate sub-pixel crop type maps and reconciled area estimates. Remote Sens. Environ. 2021, 258, 112365. [Google Scholar] [CrossRef]
- Tao, J.; Wu, W.; Xu, M. Using the Bayesian Network to map large-scale cropping intensity by fusing multi-source data. Remote Sens. 2019, 11, 168. [Google Scholar] [CrossRef]
- Saha, D.; Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. Curr. Res. Food Sci. 2021, 4, 28–44. [Google Scholar] [CrossRef]
- Jiménez-Carvelo, A.M.; González-Casado, A.; Bagur-González, M.G.; Cuadros-Rodríguez, L. Alternative data mining/machine learning methods for the analytical evaluation of food quality and authenticity–A review. Food Res. Int. 2019, 122, 25–39. [Google Scholar] [CrossRef]
- Pu, H.; Sun, D.W.; Ma, J.; Cheng, J.H. Classification of fresh and frozen-thawed pork muscles using visible and near infrared hyperspectral imaging and textural analysis. Meat Sci. 2015, 99, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Bansal, A. Fruits and vegetables quality evaluation using computer vision: A review. J. King Saud Univ. -Comput. Inf. Sci. 2021, 33, 243–257. [Google Scholar] [CrossRef]
- Lopes, J.F.; Ludwig, L.; Barbin, D.F.; Grossmann, M.V.E.; Barbon, S., Jr. Computer vision classification of barley flour based on spatial pyramid partition ensemble. Sensors 2019, 19, 2953. [Google Scholar] [CrossRef]
- Barbon, S.; Costa Barbon, A.P.A.D.; Mantovani, R.G.; Barbin, D.F. Machine learning applied to near-infrared spectra for chicken meat classification. J. Spectrosc. 2018, 8949741. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, E.H.; Lee, O.K.; Park, S.Y.; Lee, B.; Kim, S.H.; Park, I.; Chung, I.M. Variation and correlation analysis of phenolic compounds in mungbean (Vigna radiata L.) varieties. Food Chem. 2013, 141, 2988–2997. [Google Scholar] [CrossRef]
- Lin, Y.; Ma, J.; Wang, Q.; Sun, D.W. Applications of machine learning techniques for enhancing nondestructive food quality and safety detection. Crit. Rev. Food Sci. Nutr. 2022, 1–21. [Google Scholar] [CrossRef]
- Crusiol, L.G.T.; Sun, L.; Sibaldelli, R.N.R.; Junior, V.F.; Furlaneti, W.X.; Chen, R.; Sun, Z.; Wuyun, D.; Chen, Z.; Farias, J.R.B. Strategies for monitoring within-field soybean yield using Sentinel-2 Vis-NIR-SWIR spectral bands and machine learning regression methods. Precis. Agric. 2022, 23, 1093–1123. [Google Scholar] [CrossRef]
- Talukdar, S.; Naikoo, M.W.; Mallick, J.; Praveen, B.; Sharma, P.; Islam, A.R.M.T.; Pal, S.; Rahman, A. Coupling geographic information system integrated fuzzy logic-analytical hierarchy process with global and machine learning based sensitivity analysis for agricultural suitability mapping. Agric. Syst. 2022, 196, 103343. [Google Scholar] [CrossRef]
- Liao, D.; Niu, J.; Lu, N.; Shen, Q. Towards crop yield estimation at a finer spatial resolution using machine learning methods over agricultural regions. Theor. Appl. Climatol. 2021, 146, 1387–1401. [Google Scholar] [CrossRef]
- Rezapour, S.; Jooyandeh, E.; Ramezanzade, M.; Mostafaeipour, A.; Jahangiri, M.; Issakhov, A.; Chowdhury, S.; Techato, K. Forecasting rainfed agricultural production in arid and semi-arid lands using learning machine methods: A case study. Sustainability 2021, 13, 4607. [Google Scholar] [CrossRef]
- Guo, Y.; Fu, Y.; Hao, F.; Zhang, X.; Wu, W.; Jin, X.; Bryant, C.R.; Senthilnath, J. Integrated phenology and climate in rice yields prediction using machine learning methods. Ecol. Indic. 2021, 120, 106935. [Google Scholar] [CrossRef]
- Löw, F.; Biradar, C.; Dubovyk, O.; Fliemann, E.; Akramkhanov, A.; Narvaez Vallejo, A.; Waldner, F. Regional-scale monitoring of cropland intensity and productivity with multi-source satellite image time series. GIScience Remote Sens. 2018, 55, 539–567. [Google Scholar] [CrossRef]
- Cetin, N. Prediction of moisture ratio and drying rate of orange slices using machine learning approaches. J. Food Process Preserv. 2022, 46, e17011. [Google Scholar] [CrossRef]
- Meenu, M.; Kurade, C.; Neelapu, B.C.; Kalra, S.; Ramaswamy, H.S.; Yu, Y. A concise review on food quality assessment using digital image processing. Trends Food Sci. Technol. 2021, 118, 106–124. [Google Scholar] [CrossRef]
- Magnus, I.; Virte, M.; Thienpont, H.; Smeesters, L. Combining optical spectroscopy and machine learning to improve food classification. Food Control 2021, 130, 108342. [Google Scholar] [CrossRef]
- Kollia, I.; Stevenson, J.; Kollias, S. AI-Enabled Efficient and Safe Food Supply Chain. Electronics 2021, 10, 1223. [Google Scholar] [CrossRef]
- Lee, A.; Park, S.; Yoo, J.; Kang, J.; Lim, J.; Seo, Y.; Kim, B.; Kim, G. Detecting Bacterial Biofilms Using Fluorescence Hyperspectral Imaging and Various Discriminant Analyses. Sensors 2021, 21, 2213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, G.; Peng, J. Risk Assessment and Monitoring of Green Logistics for Fresh Produce Based on a Support Vector Machine. Sustainability 2020, 12, 7569. [Google Scholar] [CrossRef]
- O’Hagan, S.; Knowles, J.; Kell, D.B. Exploiting genomic knowledge in optimising molecular breeding programmes: Algorithms from evolutionary computing. PLoS ONE 2012, 7, e48862. [Google Scholar] [CrossRef]
- Reščič, N.; Eftimov, T.; Koroušić Seljak, B.; Luštrek, M. Optimising an FFQ using a machine learning pipeline to teach an efficient nutrient intake predictive model. Nutrients. 2020, 12, 3789. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.L.; Menichetti, G.; Loscalzo, J. The unmapped chemical complexity of our diet. Nat. Food 2020, 1, 33–37. [Google Scholar] [CrossRef]
- Chungcharoen, T.; Donis-Gonzalez, I.; Phetpan, K.; Udompetaikul, V.; Sirisomboon, P.; Suwalak, R. Machine learning-based prediction of nutritional status in oil palm leaves using proximal multispectral images. Comput. Electron. Agric. 2022, 198, 107019. [Google Scholar] [CrossRef]
- Habib, M.T.; Mia, M.J.; Uddin, M.S.; Ahmed, F. An in-depth exploration of automated jackfruit disease recognition. J. King Saud Univ. -Comput. Inf. Sci. 2020, 34, 1200–1209. [Google Scholar] [CrossRef]
- Qiu, Z.; Ma, F.; Li, Z.; Xu, X.; Ge, H.; Du, C. Estimation of nitrogen nutrition index in rice from UAV RGB images coupled with machine learning algorithms. Comput. Electron. Agric. 2021, 189, 106421. [Google Scholar] [CrossRef]
- Kim, Y.K.; Baek, I.; Lee, K.M.; Qin, J.; Kim, G.; Shin, B.K.; Chan, D.E.; Herrman, T.J.; Cho, S.-K.; Kim, M.S. Investigation of reflectance, fluorescence, and Raman hyperspectral imaging techniques for rapid detection of aflatoxins in ground maize. Food Control 2022, 132, 108479. [Google Scholar] [CrossRef]
- Ndraha, N.; Hsiao, H.I.; Hsieh, Y.Z.; Pradhan, A.K. Predictive models for the effect of environmental factors on the abundance of Vibrio parahaemolyticus in oyster farms in Taiwan using extreme gradient boosting. Food Control 2021, 130, 108353. [Google Scholar] [CrossRef]
- Parent, L.E.; Jamaly, R.; Atucha, A.; Jeanne Parent, E.; Workmaster, B.A.; Ziadi, N.; Parent, S.É. Current and next-year cranberry yields predicted from local features and carryover effects. PLoS ONE 2021, 16, e0250575. [Google Scholar] [CrossRef]
- Hengl, T.; Miller, M.A.; Križan, J.; Shepherd, K.D.; Sila, A.; Kilibarda, M.; Antonijević, O.; Glušica, L.; Dobermann, A.; Crouch, J. African soil properties and nutrients mapped at 30 m spatial resolution using two-scale ensemble machine learning. Sci. Rep. 2021, 11, 6130. [Google Scholar] [CrossRef]
- Mangmee, S.; Reamtong, O.; Kalambaheti, T.; Roytrakul, S.; Sonthayanon, P. MALDI-TOF mass spectrometry typing for predominant serovars of non-typhoidal Salmonella in a Thai broiler industry. Food Control 2020, 113, 107188. [Google Scholar] [CrossRef]
- Bouzembrak, Y.; Marvin, H.J. Impact of drivers of change; including climatic factors, on the occurrence of chemical food safety hazards in fruits and vegetables: A Bayesian Network approach. Food Control 2019, 97, 67–76. [Google Scholar] [CrossRef]
- Ataş, M.; Yardimci, Y.; Temizel, A. A new approach to aflatoxin detection in chili pepper by machine vision. Comput. Electron. Agric. 2012, 87, 129–141. [Google Scholar] [CrossRef]
- Saetta, D.; Buddenhagen, K.; Noha, W.; Willman, E.; Boyer, T. HUltraviolet/visible absorbance trends for beverages under simulated rinse conditions and development of data-driven prediction model. Food Control 2023, 146, 109530. [Google Scholar] [CrossRef]
- Liu, N.; Bouzembrak, Y.; Van den Bulk, L.M.; Gavai, A.; van den Heuvel, L.J.; Marvin, H.J. Automated food safety early warning system in the dairy supply chain using machine learning. Food Control 2022, 136, 108872. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, C.; Chi, K.; Gao, Q.; Bai, X.; Xu, Y.; Guo, N. Development of a machine learning-based predictor for identifying and discovering antioxidant peptides based on a new strategy. Food Control 2022, 131, 108439. [Google Scholar] [CrossRef]
- Cardoso, V.G.K.; Poppi, R.J. Cleaner and faster method to detect adulteration in cassava starch using Raman spectroscopy and one-class support vector machine. Food Control 2021, 125, 107917. [Google Scholar] [CrossRef]
- Alfian, G.; Syafrudin, M.; Farooq, U.; Ma’arif, M.R.; Syaekhoni, M.A.; Fitriyani, N.L.; Lee, J.; Rhee, J. Improving efficiency of RFID-based traceability system for perishable food by utilizing IoT sensors and machine learning model. Food Control 2020, 110, 107016. [Google Scholar] [CrossRef]
- Davies, T.; Louie, J.C.Y.; Scapin, T.; Pettigrew, S.; Wu, J.H.; Marklund, M.; Coyle, D.H. An Innovative Machine Learning Approach to Predict the Dietary Fiber Content of Packaged Foods. Nutrients 2021, 13, 3195. [Google Scholar] [CrossRef]
- Westhues, C.C.; Mahone, G.S.; da Silva, S.; Thorwarth, P.; Schmidt, M.; Richter, J.C.; Simianer, H.; Beissinger, T.M. Prediction of maize phenotypic traits with genomic and environmental predictors using gradient boosting frameworks. Front. Plant Sci. 2021, 12, 699589. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, Y.; Cheng, Q.; Jiang, S.; Wang, Q.; Xiao, Y.; Ma, C.; Yan, J.; Wang, X. LightGBM: Accelerated genomically designed crop breeding through ensemble learning. Genome Biol. 2021, 22, 271. [Google Scholar] [CrossRef] [PubMed]
- Shete, S.; Srinivasan, S.; Gonsalves, T.A. TasselGAN: An Application of the generative adversarial model for creating field-based maize tassel data. Plant Phenomics 2020, 2020, 8309605. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Aghamirzaie, D.; Elmarakeby, H.; Collakova, E.; Li, S.; Grene, R.; Heath, L.S. A machine learning approach to predict gene regulatory networks in seed development in Arabidopsis. Front. Plant Sci. 2016, 7, 1936. [Google Scholar] [CrossRef]
- Ma, C.; Xin, M.; Feldmann, K.A.; Wang, X. Machine learning–based differential network analysis: A study of stress-responsive transcriptomes in Arabidopsis. Plant Cell 2014, 26, 520–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Hu, F.B. Precision nutrition for prevention and management of type 2 diabetes. Lancet Diabetes Endocrinol. 2018, 6, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Triantafyllidis, A.K.; Tsanas, A. Applications of machine learning in real-life digital health interventions: Review of the literature. J. Med. Internet Res. 2019, 21, e12286. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Elinav, E. Harnessing smartphones to personalize nutrition in a time of global pandemic. Nutrients 2021, 13, 422. [Google Scholar] [CrossRef]
- Alfian, G.; Rhee, J.; Ahn, H.; Lee, J.; Farooq, U.; Ijaz, M.F.; Syaekhoni, M.A. Integration of RFID, wireless sensor networks, and data mining in an e-pedigree food traceability system. J. Food Eng. 2017, 212, 65–75. [Google Scholar] [CrossRef]
- Sundaravadivel, P.; Kesavan, K.; Kesavan, L.; Mohanty, S.P.; Kougianos, E. Smart-Log: A deep-learning based automated nutrition monitoring system in the IoT. IEEE Trans. Consum. Electron. 2018, 64, 390–398. [Google Scholar] [CrossRef]
- Lei, Z.; Yang, S.; Liu, H.; Aslam, S.; Liu, J.; Tekle, H.; Bugingo, E.; Zhang, D. Mining of nutritional ingredients in food for disease analysis. IEEE Access 2018, 6, 52766–52778. [Google Scholar] [CrossRef]
- Chen, Y.; Hsu, C.Y.; Liu, L.; Yang, S. Constructing a nutrition diagnosis expert system. Expert Syst. Appl. 2012, 39, 2132–2156. [Google Scholar] [CrossRef]
- Guo, T.; Yu, X.; Li, X.; Zhang, H.; Zhu, C.; Flint-Garcia, S.; McMullen, M.D.; Holland, J.B.; Szalma, S.J.; Yu, J. Optimal designs for genomic selection in hybrid crops. Mol. Plant. 2019, 12, 390–401. [Google Scholar] [CrossRef]
- Liu, J.; Kang, Y.; Liu, K.; Yang, X.; Sun, M.; Hu, J. Maize Carotenoid Gene Locus Mining Based on Conditional Gaussian Bayesian Network. IEEE Access 2020, 8, 15223–15231. [Google Scholar] [CrossRef]
- Wang, J.; Yue, H. Food safety pre-warning system based on data mining for a sustainable food supply chain. Food Control 2017, 73, 223–229. [Google Scholar] [CrossRef]
- Kirk, D.; Kok, E.; Tufano, M.; Tekinerdogan, B.; Feskens, E.J.; Camps, G. Machine Learning in Nutrition Research. Adv. Nutr. 2022, 13, 2573–2589. [Google Scholar] [CrossRef] [PubMed]
- Gunasekara, C.; Zhang, K.; Deng, W.; Brown, L.; Wei, H. TGMI: An efficient algorithm for identifying pathway regulators through evaluation of triple-gene mutual interaction. Nucleic Acids Res. 2018, 46, e67. [Google Scholar] [CrossRef] [PubMed]
- Frelat, R.; Lopez-Ridaura, S.; Giller, K.E.; Herrero, M.; Douxchamps, S.; Djurfeldt, A.A.; McMullen, M.D.; Holland, J.B.; Szalma, S.J.; Van Wijk, M.T. Drivers of household food availability in sub-Saharan Africa based on big data from small farms. Proc. Natl. Acad. Sci. USA 2016, 113, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, T.; Zhu, Y.; Qiu, M. A case study of sensor data collection and analysis in smart city: Provenance in smart food supply chain. Int. J. Distrib. Sens. Netw. 2013, 9, 382132. [Google Scholar] [CrossRef]
- Misra, N.N.; Dixit, Y.; Al-Mallahi, A.; Bhullar, M.S.; Upadhyay, R.; Martynenko, A. IoT, big data and artificial intelligence in agriculture and food industry. IEEE Internet Things J. 2020, 9, 6305–6324. [Google Scholar] [CrossRef]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar-Bowles, J. The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef]
- Rai, K.K. Integrating speed breeding with artificial intelligence for developing climate-smart crops. Mol. Biol. Rep. 2022, 49, 11385–11402. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, M.; Wang, X.; Tong, C.; Huang, S.; Tehrim, S.; Liu, Y.; Hua, W.; Liu, S. Bolbase: A comprehensive genomics database for Brassica oleracea. BMC Genom. 2013, 14, 664. [Google Scholar] [CrossRef]
- Al-Adhaileh, M.H.; Aldhyani, T.H. Artificial intelligence framework for modeling and predicting crop yield to enhance food security in Saudi Arabia. Peer J. Comput. Sci. 2022, 8, e1104. [Google Scholar]
- McLennon, E.; Dari, B.; Jha, G.; Sihi, D.; Kankarla, V. Regenerative agriculture and integrative permaculture for sustainable and technology driven global food production and security. Agron. J. 2021, 113, 4541–4559. [Google Scholar]
- Qian, J.; Dai, B.; Wang, B.; Zha, Y.; Song, Q. Traceability in food processing: Problems, methods, and performance evaluations—A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 679–692. [Google Scholar]
- Katiyar, S.; Khan, R.; Kumar, S. Artificial bee colony algorithm for fresh food distribution without quality loss by delivery route optimization. J. Food Qual. 2021, 2021, 4881289. [Google Scholar] [CrossRef]
- Chai, J.J.; O’Sullivan, C.; Gowen, A.A.; Rooney, B.; Xu, J.L. Augmented/mixed reality technologies for food: A review. Trends Food Sci. Technol. 2022, 124, 182–194. [Google Scholar]
- Zhao, W.; Lai, X.S.; Liu, D.Y.; Zhang, Z.Y.; Ma, P.P.; Wang, Q.S.; Zhang, Z.; Pan, Y.C. Applications of Support Vector Machine in Genomic Prediction in Pig and Maize Populations. Front. Genet. 2020, 11, 598318. [Google Scholar] [CrossRef]
- Khan, P.W.; Byun, Y.C.; Park, N. IoT-blockchain enabled optimized provenance system for food industry 4.0 using advanced deep learning. Sensors 2020, 20, 2990. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, F.; Xu, Y.; Wang, Q.; Yan, J.; Wang, J.; Wang, X.; Wang, X. MODAS: Exploring maize germplasm with multi-omics data association studies. Sci. Bulletin. 2022, 67, 903–906. [Google Scholar]
- Morgenstern, J.D.; Rosella, L.C.; Costa, A.P.; de Souza, R.J.; Anderson, L.N. Perspective: Big data and machine learning could help advance nutritional epidemiology. Adv. Nutr. 2021, 12, 621–631. [Google Scholar]
- Kittichotsatsawat, Y.; Jangkrajarng, V.; Tippayawong, K.Y. Enhancing coffee supply chain towards sustainable growth with big data and modern agricultural technologies. Sustainability 2021, 13, 4593. [Google Scholar] [CrossRef]
- Oscar, T.P. Neural network model for thermal inactivation of Salmonella Typhimurium to elimination in ground chicken: Acquisition of data by whole sample enrichment, miniature most-probable-number method. J. Food Prot. 2017, 80, 104–112. [Google Scholar]
- Kyaw, K.S.; Adegoke, S.C.; Ajani, C.K.; Nwabor, O.F.; Onyeaka, H. Toward in-process technology-aided automation for enhanced microbial food safety and quality assurance in milk and beverages processing. Crit. Rev. Food Sci. Nutr. 2022, 9, 1–21. [Google Scholar]
- Erdogdu, F.; Sarghini, F.; Marra, F. Mathematical modeling for virtualization in food processing. Food Eng. Rev. 2017, 9, 295–313. [Google Scholar]
- Nogales, A.; Díaz-Morón, R.; García-Tejedor, Á.J. A comparison of neural and non-neural machine learning models for food safety risk prediction with European Union RASFF data. Food Control 2022, 134, 108697. [Google Scholar]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A deep learning-based approach for automated yellow rust disease detection from high-resolution hyperspectral UAV images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef]
- Liu, Y.; Pu, H.; Sun, D.W. Efficient extraction of deep image features using convolutional neural network (CNN) for applications in detecting and analysing complex food matrices. Trends Food Sci. Technol. 2021, 113, 193–204. [Google Scholar]
- Kaur, P.; Harnal, S.; Tiwari, R.; Upadhyay, S.; Bhatia, S.; Mashat, A.; Alabdali, A.M. Recognition of leaf disease using hybrid convolutional neural network by applying feature reduction. Sensors 2022, 22, 575. [Google Scholar]
- Wolanin, A.; Mateo-García, G.; Camps-Valls, G.; Gómez-Chova, L.; Meroni, M.; Duveiller, G.; Liangzhi, Y.; Guanter, L. Estimating and understanding crop yields with explainable deep learning in the Indian Wheat Belt. Environ. Res. Lett. 2020, 15, 024019. [Google Scholar]
- Wongchai, A.; rao Jenjeti, D.; Priyadarsini, A.I.; Deb, N.; Bhardwaj, A.; Tomar, P. Farm monitoring and disease prediction by classification based on deep learning architectures in sustainable agriculture. Ecological Modelling. 2022, 474, 110167. [Google Scholar]
- Hu, W.J.; Fan, J.; Du, Y.X.; Li, B.S.; Xiong, N.; Bekkering, E. MDFC–ResNet: An agricultural IoT system to accurately recognize crop diseases. IEEE Access 2020, 8, 115287–115298. [Google Scholar] [CrossRef]
- Zambrano, F.; Vrieling, A.; Nelson, A.; Meroni, M.; Tadesse, T. Prediction of drought-induced reduction of agricultural productivity in Chile from MODIS, rainfall estimates, and climate oscillation indices. Remote Sens. Environ. 2018, 219, 15–30. [Google Scholar]
- Xiao, Z.; Wang, J.; Han, L.; Guo, S.; Cui, Q. Application of Machine Vision System in Food Detection. Front. Nutr. 2022, 9, 888245. [Google Scholar]
- Zhu, L.; Spachos, P.; Pensini, E.; Plataniotis, K.N. Deep learning and machine vision for food processing: A survey. Curr. Res. Food Sci. 2021, 4, 233–249. [Google Scholar]
- Shao, W.; Hou, S.; Jia, W.; Zheng, Y. Rapid Non-Destructive Analysis of Food Nutrient Content Using Swin-Nutrition. Foods 2022, 11, 3429. [Google Scholar] [PubMed]
- Chen, X.; Johnson, E.; Kulkarni, A.; Ding, C.; Ranelli, N.; Chen, Y.; Xu, R. An Exploratory Approach to Deriving Nutrition Information of Restaurant Food from Crowdsourced Food Images: Case of Hartford. Nutrients 2021, 13, 4132. [Google Scholar]
- Veeramani, B.; Raymond, J.W.; Chanda, P. DeepSort: Deep convolutional networks for sorting haploid maize seeds. BMC Bioinform. 2018, 19, 289. [Google Scholar]
- Zhai, Q.; Ye, C.; Li, S.; Liu, J.; Guo, Z.; Chang, R.; Hua, J. Rice nitrogen nutrition monitoring classification method based on the convolution neural network model: Direct detection of rice nitrogen nutritional status. PLoS ONE 2022, 17, e0273360. [Google Scholar]
- Dey, B.; Haque, M.M.U.; Khatun, R.; Ahmed, R. Comparative performance of four CNN-based deep learning variants in detecting Hispa pest, two fungal diseases, and NPK deficiency symptoms of rice (Oryza sativa). Comput. Electron. Agric. 2022, 202, 107340. [Google Scholar]
- Rong, D.; Xie, L.; Ying, Y. Computer vision detection of foreign objects in walnuts using deep learning. Comput. Electron. Agric. 2019, 162, 1001–1010. [Google Scholar]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar]
- Chakravartula, S.S.N.; Moscetti, R.; Bedini, G.; Nardella, M.; Massantini, R. Use of convolutional neural network (CNN) combined with FT-NIR spectroscopy to predict food adulteration: A case study on coffee. Food Control 2022, 135, 108816. [Google Scholar]
- Estrada-Pérez, L.V.; Pradana-López, S.; Pérez-Calabuig, A.M.; Mena, M.L.; Cancilla, J.C.; Torrecilla, J.S. Thermal imaging of rice grains and flours to design convolutional systems to ensure quality and safety. Food Control 2021, 121, 107572. [Google Scholar] [CrossRef]
- Vo, S.A.; Scanlan, J.; Turner, P. An application of Convolutional Neural Network to lobster grading in the Southern Rock Lobster supply chain. Food Control 2020, 113, 107184. [Google Scholar]
- Izquierdo, M.; Lastra-Mejías, M.; González-Flores, E.; Pradana-López, S.; Cancilla, J.C.; Torrecilla, J.S. Visible imaging to convolutionally discern and authenticate varieties of rice and their derived flours. Food Control 2020, 110, 106971. [Google Scholar]
- Hafiz, R.; Haque, M.R.; Rakshit, A.; Uddin, M.S. Image-based soft drink type classification and dietary assessment system using deep convolutional neural network with transfer learning. J. King Saud Univ. -Comput. Inf. Sci. 2022, 34, 1775–1784. [Google Scholar]
- Ma, P.; Lau, C.P.; Yu, N.; Li, A.; Liu, P.; Wang, Q.; Sheng, J. Image-based nutrient estimation for Chinese dishes using deep learning. Food Res. Int. 2021, 147, 110437. [Google Scholar] [PubMed]
- Ahn, D.; Choi, J.Y.; Kim, H.C.; Cho, J.S.; Moon, K.D.; Park, T. Estimating the composition of food nutrients from hyperspectral signals based on deep neural networks. Sensors 2019, 19, 1560. [Google Scholar] [PubMed]
- Yang, S.; Zheng, L.; He, P.; Wu, T.; Sun, S.; Wang, M. High-throughput soybean seeds phenotyping with convolutional neural networks and transfer learning. Plant Methods 2021, 17, 50. [Google Scholar] [PubMed]
- Zingaretti, L.M.; Gezan, S.A.; Ferrão, L.F.V.; Osorio, L.F.; Monfort, A.; Muñoz, P.R.; Whitaker, V.M.; Pérez-Enciso, M. Exploring deep learning for complex trait genomic prediction in polyploid outcrossing species. Front. Plant Sci. 2020, 11, 25. [Google Scholar]
- Ma, W.; Qiu, Z.; Song, J.; Li, J.; Cheng, Q.; Zhai, J.; Ma, C. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 2018, 248, 1307–1318. [Google Scholar]
- Tay, W.; Kaur, B.; Quek, R.; Lim, J.; Henry, C.J. Current developments in digital quantitative volume estimation for the optimisation of dietary assessment. Nutrients 2020, 12, 1167. [Google Scholar] [PubMed]
- Huang, L.; Zhao, J.; Chen, Q.; Zhang, Y. Nondestructive measurement of total volatile basic nitrogen (TVB-N) in pork meat by integrating near infrared spectroscopy, computer vision and electronic nose techniques. Food Chem. 2014, 145, 228–236. [Google Scholar] [PubMed]
- Delloye, C.; Weiss, M.; Defourny, P. Retrieval of the canopy chlorophyll content from Sentinel-2 spectral bands to estimate nitrogen uptake in intensive winter wheat cropping systems. Remote Sens. Environ. 2018, 216, 245–261. [Google Scholar]
- Das, B.; Nair, B.; Reddy, V.K.; Venkatesh, P. Evaluation of multiple linear, neural network and penalised regression models for prediction of rice yield based on weather parameters for west coast of India. Int. J. Biometeorol. 2018, 62, 1809–1822. [Google Scholar] [PubMed]
- Geng, Z.; Zhao, S.; Tao, G.; Han, Y. Early warning modeling and analysis based on analytic hierarchy process integrated extreme learning machine (AHP-ELM): Application to food safety. Food Control 2017, 78, 33–42. [Google Scholar]
- Al-Mahasneh, M.; Aljarrah, M.; Rababah, T.; Alu’datt, M. Application of hybrid neural fuzzy system (ANFIS) in food processing and technology. Food Eng. Rev. 2016, 8, 351–366. [Google Scholar]
- Anandhakrishnan, T.; Jaisakthi, S.M. Deep Convolutional Neural Networks for image based tomato leaf disease detection. Sustain. Chem. Pharm. 2022, 30, 100793. [Google Scholar]
- Chamundeeswari, G.; Srinivasan, S.; Bharathi, S.P.; Priya, P.; Kannammal, G.R.; Rajendran, S. Optimal deep convolutional neural network based crop classification model on multispectral remote sensing images. Microprocess. Microsyst. 2022, 94, 104626. [Google Scholar]
- Zhao, X.; Que, H.; Sun, X.; Zhu, Q.; Huang, M. Hybrid convolutional network based on hyperspectral imaging for wheat seed varieties classification. Infrared Phys. Technol. 2022, 125, 104270. [Google Scholar]
- Putra, A.N.; Setyawan, R.A.; Teixeira, H.M.; Khumairoh, U. Forecasting Rice Status for a Food Crisis Early Warning System Based on Satellite Imagery and Cellular Automata in Malang, Indonesia. Sustainability 2022, 14, 8972. [Google Scholar]
- Pham, T.N.; Tran, L.V.; Dao, S.V.T. Early disease classification of mango leaves using feed-forward neural network and hybrid metaheuristic feature selection. IEEE Access 2020, 8, 189960–189973. [Google Scholar] [CrossRef]
- Tao, J.B.; Liu, W.B.; Tan, W.X.; Kong, X.B.; Meng, X.U. Fusing multi-source data to map spatio-temporal dynamics of winter rape on the Jianghan Plain and Dongting Lake Plain, China. J. Integr. Agric. 2019, 18, 2393–2407. [Google Scholar]
- Raj Bhagya, G.V.S.; Dash, K.K. Comprehensive study on applications of artificial neural network in food process modeling. Crit. Rev. Food Sci. Nutr. 2022, 62, 2756–2783. [Google Scholar] [CrossRef]
- Kondakci, T.; Zhou, W. Recent applications of advanced control techniques in food industry. Food Bioprocess Technol. 2017, 10, 522–542. [Google Scholar]
- Bortolini, M.; Faccio, M.; Ferrari, E.; Gamberi, M.; Pilati, F. Fresh food sustainable distribution: Cost, delivery time and carbon footprint three-objective optimization. J. Food Eng. 2016, 174, 56–67. [Google Scholar]
- Okut, H.; Wu, X.L.; Rosa, G.J.; Bauck, S.; Woodward, B.W.; Schnabel, R.D.; Taylor, J.F.; Gianola, D. Predicting expected progeny difference for marbling score in Angus cattle using artificial neural networks and Bayesian regression models. Genet. Sel. Evol. 2013, 45, 34. [Google Scholar]
- González-Camacho, J.M.; de Los Campos, G.; Pérez, P.; Gianola, D.; Cairns, J.E.; Mahuku, G.; Babu, R.; Crossa, J. Genome-enabled prediction of genetic values using radial basis function neural networks. Theor. Appl. Genet. 2012, 125, 759–771. [Google Scholar]
- Lv, J.; Wang, Y.; Ni, P.; Lin, P.; Hou, H.; Ding, J.; Chang, Y.; Hu, J.; Wang, S.; Bao, Z. Development of a high-throughput SNP array for sea cucumber (Apostichopus japonicus) and its application in genomic selection with MCP regularized deep neural networks. Genomics 2022, 114, 110426. [Google Scholar]
- Li, C.; Nie, J.; Zhang, Y.; Shao, S.; Liu, Z.; Rogers, K.M.; Zhang, W.; Yuan, Y. Geographical origin modeling of Chinese rice using stable isotopes and trace elements. Food Control 2022, 138, 108997. [Google Scholar]
- Shi, P.; Wang, Y.; Xu, J.; Zhao, Y.; Yang, B.; Yuan, Z.; Sun, Q. Rice nitrogen nutrition estimation with RGB images and machine learning methods. Comput. Electron. Agric. 2021, 180, 105860. [Google Scholar]
- Kuzuoka, K.; Kawai, K.; Yamauchi, S.; Okada, A.; Inoshima, Y. Chilling control of beef and pork carcasses in a slaughterhouse based on causality analysis by graphical modelling. Food Control 2020, 118, 107353. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Yang, G.; Yang, X.; Fan, L. Estimation of the yield and plant height of winter wheat using UAV-based hyperspectral images. Sensors 2020, 20, 1231. [Google Scholar] [CrossRef]
- Tian, H.; Wang, P.; Tansey, K.; Zhang, S.; Zhang, J.; Li, H. An IPSO-BP neural network for estimating wheat yield using two remotely sensed variables in the Guanzhong Plain, PR China. Comput. Electron. Agric. 2020, 169, 105180. [Google Scholar] [CrossRef]
- Geng, Z.; Shang, D.; Han, Y.; Zhong, Y. Early warning modeling and analysis based on a deep radial basis function neural network integrating an analytic hierarchy process: A case study for food safety. Food Control 2019, 96, 329–342. [Google Scholar] [CrossRef]
- Wang, J.; Yue, H.; Zhou, Z. An improved traceability system for food quality assurance and evaluation based on fuzzy classification and neural network. Food Control 2017, 79, 363–370. [Google Scholar] [CrossRef]
- Silva, C.E.T.; Filardi, V.L.; Pepe, I.M.; Chaves, M.A.; Santos, C.M.S. Classification of food vegetable oils by fluorimetry and artificial neural networks. Food Control 2015, 47, 86–91. [Google Scholar] [CrossRef]
- Sadhu, T.; Banerjee, I.; Lahiri, S.K.; Chakrabarty, J. Modeling and optimization of cooking process parameters to improve the nutritional profile of fried fish by robust hybrid artificial intelligence approach. J. Food Process Eng. 2020, 43, e13478. [Google Scholar] [CrossRef]
- Kang, J.; Wang, D.H. Food safety risk prediction method based on brain neural network. Fresenius Environ. Bulletin. 2020, 29, 2459–2468. [Google Scholar]
- Fu, Y.H.; Liu, J.; Mao, Y.C.; Cao, W.; Huang, J.Q.; Zhao, Z.G. Experimental study on quantitative inversion model of heavy metals in soda saline-alkali soil based on RBF neural network. Spectrosc. Spectr. Anal. 2022, 42, 1595–1600. [Google Scholar]
- Tagkopoulos, I.; Brown, S.F.; Liu, X.; Zhao, Q.; Zohdi, T.I.; Earles, J.M.; Nitin, N.; Runcie, D.E.; Lemay, D.G.; Smith, A.D.; et al. Special report: AI Institute for next generation food systems (AIFS). Comput. Electron. Agric. 2022, 196, 106819. [Google Scholar] [CrossRef]
- Zhou, P.; Li, Z.; Magnusson, E.; Gomez Cano, F.; Crisp, P.A.; Noshay, J.M.; Grotewold, E.; Hirsch, C.N.; Briggs, S.P.; Springer, N.M. Meta gene regulatory networks in maize highlight functionally relevant regulatory interactions. Plant Cell 2020, 32, 1377–1396. [Google Scholar] [CrossRef]
- Yan, J.; Wang, X. Machine learning bridges omics sciences and plant breeding. Trends Plant Sci. 2022, 28, 199–210. [Google Scholar] [CrossRef]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; De Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Huber, F.; Yushchenko, A.; Stratmann, B.; Steinhage, V. Extreme Gradient Boosting for yield estimation compared with Deep Learning approaches. Comput. Electron. Agric. 2022, 202, 107346. [Google Scholar] [CrossRef]
- Nosratabadi, S.; Ardabili, S.; Lakner, Z.; Mako, C.; Mosavi, A. Prediction of food production using machine learning algorithms of multilayer perceptron and ANFIS. Agriculture 2021, 11, 408. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, L.; Li, Y.; Li, D. Model predictive control and its application in agriculture: A review. Comput. Electron. Agric. 2018, 151, 104–117. [Google Scholar] [CrossRef]
- Nayak, J.; Vakula, K.; Dinesh, P.; Naik, B.; Pelusi, D. Intelligent food processing: Journey from artificial neural network to deep learning. Comput. Sci. Rev. 2020, 38, 100297. [Google Scholar] [CrossRef]




| Topic | Keyword | Author(s) | Journal |
|---|---|---|---|
| Remote sensing | Food security | Maimaitijiang et al., 2019 [58] |
|
| Random forest | Hao et al., 2015 [59] |
| |
| Vreugdenhil et al., 2018 [60] |
| ||
| Impact | Han et al., 2020 [61] |
| |
| Climate change | Teluguntla et al., 2018 [62] |
| |
| Cao et al., 2020 [63] |
| ||
| Time series Vegetation Index | Duke et al., 2022 [64] |
| |
| Ma et al., 2021 [65] |
| ||
| Remote sensing | Hu et al., 2021 [66] |
| |
| Tao et al., 2019 [67] |
| ||
| Food quality | Machine learning | Saha and Manickavasagan, 2021 [68] |
|
| Jiménez-Carvelo et al., 2019 [69] |
| ||
| Classification | Ropodi et al., 2016 [43] |
| |
| Pu et al., 2014 [70] |
| ||
| Prediction | Bhargava and Bansal, 2021 [71] |
| |
| Lopes et al., 2019 [72] |
| ||
| Barbon et al., 2018 [73] |
| ||
| Identification | Kim et al., 2013 [74] |
| |
| Lin et al., 2022 [75] |
| ||
| Crusiol et al., 2022 [76] |
| ||
| Food quality | Talukdar et al., 2022 [77] |
| |
| Liao et al., 2021 [78] |
| ||
| Quality | Rezapour et al., 2021 [79] |
| |
| Guo et al., 2021 [80] |
| ||
| Löw et al., 2018 [81] |
| ||
| Support Vector Machine | Çetin, 2022 [82] |
| |
| Meenu et al., 2021 [83] |
| ||
| Magnus et al., 2021 [84] |
| ||
| Kollia et al., 2021 [85] |
| ||
| Lee et al., 2021 [86] |
| ||
| Zhang et al., 2020 [87] |
| ||
| O’Hagan et al., 2012 [88] |
| ||
| Reščič et al., 2021 [89] |
| ||
| Barabási et al., 2020 [90] |
| ||
| Chungcharoen et al., 2022 [91] |
| ||
| Habib et al. 2022 [92] |
| ||
| Qiu et al., 2021 [93] |
| ||
| Kim et al., 2022 [94] |
| ||
| Ndraha et al., 2021 [95] |
| ||
| Parent et al., 2021 [96] |
| ||
| Hengl et al., 2021 [97] |
| ||
| Mangmee et al., 2020 [98] |
| ||
| Bouzembrak et al., 2019 [99] |
| ||
| Atas et al., 2012 [100] |
| ||
| Saetta et al., 2023 [101] |
| ||
| Liu et al., 2022 [102] |
| ||
| Shen et al., 2022 [103] |
| ||
| Cardoso and Poppi, 2021 [104] |
| ||
| Alfian et al., 2020 [105] |
| ||
| Davies et al., 2021 [106] |
| ||
| Westhues et al., 2021 [107] |
| ||
| Yan et al., 2021 [108] |
| ||
| Shete et al., 2020 [109] |
| ||
| Ni et al., 2016 [110] |
| ||
| Ma et al., 2014 [111] |
| ||
| Personalized nutrition | Nutrition | Zeevi et al., 2015 [44] |
|
| Wang and Hu, 2018 [112] |
| ||
| Risk | Triantafyllidis, and Tsanas, 2019 [113] |
| |
| Zmora and Elinav, 2021 [114] |
| ||
| Health | Alfian et al., 2017 [115] |
| |
| Sundaravadivel et al., 2018 [116] |
| ||
| Lei et al., 2018 [117] |
| ||
| Data mining | Chen et al., 2012 [118] |
| |
| Guo et al., 2019 [119] |
| ||
| Liu et al., 2020 [120] |
| ||
| Disease | Wang and Yue, 2017 [121] |
| |
| Kirk et al., 2022 [122] |
| ||
| Gunasekara et al., 2018 [123] |
| ||
| Bigdata | System | Frelat et al., 2016 [124] |
|
| Zhang et al., 2013 [125] |
| ||
| Model | Misra et al., 2020 [126] |
| |
| Jung et al.,2021 [127] |
| ||
| Rai, 2022 [128] |
| ||
| Yu et al., 2013 [129] |
| ||
| Big data | Al-Adhaileh and Aldhyani, 2022 [130] |
| |
| McLennon et al., 2021 [131] |
| ||
| Kumar et al., 2021 [8] |
| ||
| Artificial intelligence | Qian et al., 2020 [132] |
| |
| Katiyar et al., 2022 [133] |
| ||
| Chai et al.,2022 [134] |
| ||
| Management | Zhao et al., 2020 [135] |
| |
| Khan et al., 2020 [136] |
| ||
| Liu et al., 2022 [137] |
| ||
| Morgenstern et al., 2021 [138] |
| ||
| Food Safety | Food safety | Kittichotsatsawat et al., 2021 [139] |
|
| Growth | Oscar, 2017 [140] |
| |
| Network | Kyaw et al., 2022 [141] |
| |
| Temperature | Erdogdu et al., 2017 [142] |
| |
| Nogales et al., 2022 [143] |
| ||
| Deep learning | Deep learning | Zhou et al., 2019 [13] |
|
| Zhang et al., 2019 [144] |
| ||
| Liu et al., 2021 [145] |
| ||
| Kaur et al., 2022 [146] |
| ||
| Wolanin et al., 2020 [147] |
| ||
| Wongchai et al., 2022 [148] |
| ||
| Hu et al., 2020 [149] |
| ||
| Feature extraction | Zambrano et al., 2018 [150] |
| |
| Xiao et al., 2022 [151] |
| ||
| Zhu et al., 2021 [152] |
| ||
| Shao et al., 2022 [153] |
| ||
| Chen et al., 2021 [154] |
| ||
| Veeramani et al., 2018 [155] |
| ||
| Zhai et al., 2022 [156] |
| ||
| Image | Dey et al., 2022 [157] |
| |
| Rong et al., 2019 [158] |
| ||
| Too et al., 2019 [159] |
| ||
| Chakravartula et al., 2022 [160] |
| ||
| Estrada-Pérez et al., 2021 [161] |
| ||
| Vo et al., 2020 [162] |
| ||
| Izquierdo et al., 2020 [163] |
| ||
| Convolutional neural network | Hafiz et al., 2022 [164] |
| |
| Ma et al., 2021 [165] |
| ||
| Ahn et al., 2019 [166] |
| ||
| Yang et al., 2021 [167] |
| ||
| Zingaretti et al., 2020 [168] |
| ||
| Ma et al., 2018 [169] |
| ||
| Tay et al., 2020 [170] |
| ||
| Artificial neural network | Neural network | Huang et al., 2014 [171] |
|
| Delloye et al., 2018 [172] |
| ||
| Das et al., 2018 [173] |
| ||
| Geng et al., 2017 [174] |
| ||
| Al-Mahasneh et al., 2016 [175] |
| ||
| Anandhakrishnan and Jaisakthi 2022 [176] |
| ||
| Chamundeeswari et al., 2022 [177] |
| ||
| Artificial neural network | Zhao et al., 2022 [178] |
| |
| Sujarwo et al., 2022 [179] |
| ||
| Pham et al., 2020 [180] |
| ||
| Tao et al., 2019 [181] |
| ||
| Raj and Dash, 2022 [182] |
| ||
| Kondakci and Zhou, 2017 [183] |
| ||
| Bortolini et al., 2016 [184] |
| ||
| Okut et al., 2013 [185] |
| ||
| González-Camacho et al., 2012 [186] |
| ||
| Performance | Lv et al., 2022 [187] |
| |
| Li et al., 2022 [188] |
| ||
| Shi et al., 2021 [189] |
| ||
| Kuzuoka et al., 2020 [190] |
| ||
| Tao et al., 2020 [191] |
| ||
| Tian et al., 2020 [192] |
| ||
| Geng, 2019 [193] |
| ||
| Wang et al., 2017 [194] |
| ||
| Silva et al., 2015 [195] |
| ||
| Sadhu et al., 2020 [196] |
|
| Field | Sample | Functionality | Method(s) | Result(s) |
|---|---|---|---|---|
| Molecular Breeding | Crops (Yan et al., 2021) [108] | Genomic prediction | LightGBM | LightGBM exhibited superior performance of genomic selection prediction. |
| Maize (Liu et al., 2022). [137] | Germplasm exploitation | MODAS | MODAS can accelerate association analysis of genotypic data. | |
| Pig and maize (Zhao, et al., 2020) [135] | Genomic prediction | SVM | The prediction model based on SVM outperformed BayesR and GBLUP in two data sets. | |
| Sea cucumber (Lv et al., 2022) [187] | Genomic prediction | DNN-MCP RR-GBLUP Bayes B DNN | DNN-MCP can greatly improve genomic prediction ability. | |
| Agricul-tural Production | Tomato (Anandhakrishnan and Jaisakthi, 2022) [174] | Leaf disease recognition | DCNN | DCNN model gained an accuracy of 98.40% for the testing set. |
| Farm crop (Wongchai et al., 2022) [148] | Crop disease prediction | DAL_CL RNN | Experimental results showed an accuracy of 96%. | |
| Rice (Qiu et al., 2021) [93] | Nitrogen Nutrition Index | AB, ANN, KNN, PLSR, RF SVM | The RF algorithms performed the best, with the R2 ranging from 0.88 to 0.96 and RMSE ranging from 0.03 to 0.07. | |
| Soybean (Crusiol et al., 2022) [76] | Monitoring of yield | PLSR, SVR | Field-based SVR models presented the highest accuracies for yield mapping. | |
| Grape leaves (Kaur et al., 2022) [146] | Identification of leaf diseases | Hy-CNN TL, LR | For leaf disease recognition, Hy-CNN has the highest accuracy of 98.7%. | |
| Corn (Ma et al., 2021) [65] | Prediction of corn yield | BNN | The BNN model can predict corn yield in normal and abnormal years with extreme weather. | |
| Crop (Hu et al., 2021) [66] | Crop type mapping | RF-r | The spatial consistency between the sub-pixel crop distribution map generated by temporal MODIS and the medium-high resolution reference map reached 0.75. | |
| Rice (Guo et al., 2021) [80] | Yields prediction | MLR, BPNN, SVM, RF | In yield predictions, SVM obtained the highest precisions. | |
| Crop (Tao et al., 2019) [67] | Cropping intensity mapping | BNPK | For cropping intensity index mapping, BNPK model can settle intra-class variations. | |
| Agricultural productivity (Zambrano et al., 2018) [150] | Prediction of agricultural productivity | OLR, DL | OLR, compared to DL, only showed a slightly smaller accuracy. | |
| Food Processing and Distribu-tion | Antioxidant peptide (Shen et al., 2022) [103] | Feature extraction | LR, LDA, SVM, KNN | The ML-based predictor was effective in mining the multifunctional peptides. |
| Walnut (Rong et al., 2019) [158] | Objectives detection | CNN | The proposed method obtained an accuracy of 95% for foreign object detection. | |
| Walnut (Magnus et al., 2021) [84] | Non-destructive food classification | ELM, SVM LDA, QDA PLS-DA | An ML-based algorithm, compared to classical techniques, improved the performance metric by up to 80%. | |
| Bacterial biofilms (Lee et al., 2021) [86] | Detection of bacterial biofilms | DT, KNN LDA, PLS-DA | KNN algorithm proved a high performance in predicting the biofilm region. | |
| Barley flour (Lopes et al., 2019) [72] | Barley flour classification | CVS, SVM, KNN, J48, RF | The accuracy of this method ranged from 75.00% to 100.00%. | |
| Milk (Liu et al., 2022) [102] | Anomaly detection | BN | Food safety problems in the supply chain could be predicted by detecting severe changes in related fields. | |
| Coffee (Chakravartula et al., 2022) [160] | Coffee adulterant quantification | CNN | The results confirmed the feasibility of the CNN algorithm with excellent performances (R2 > 0.98). | |
| Kimchi supply chain (Alfian et al., 2017) [115] | Food traceability system | MLP | In the case of missing sensor data, MLP proved to be the best model with high prediction accuracy. | |
| Fresh food (Bortolini et al., 2016) [184] | Fresh food distribution | LP | The expert system outperformed the traditional cost minimization model. | |
| Food Nutrition | Daily diet (Shao et al., 2022) [153] | Nutritional evaluation | ST, FFM | Swin-Nutrition provided a novel non-destructive detection technology. |
| Restaurant food (Chen et al., 2021) [154] | Nutrition assessment. | Calorie Mama (DL model) | The DL model obtained an accuracy of 75.1%. | |
| Soft drinks (Hafiz et al., 2022) [164] | Classification and dietary assessment | DCNN with transfer learning | The DCNN-based transfer learning model showed an accuracy of 98.51%. | |
| Infant diet (Sundaravadivel et al., 2018) [116] | Automated nutrition monitoring | Bayesian network | Smart-Log predicted 8172 foods for 1000 meals with 98.6 percent accuracy. | |
| Chinese dishes (Ma et al., 2021) [165] | Nutrient estimation | DCNN | The DCNN model showed the highest performance for protein estimation. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, S.; Zhang, Y.; Feng, Y.; Liu, J.; Zhu, H. Artificial Intelligence in Food Safety: A Decade Review and Bibliometric Analysis. Foods 2023, 12, 1242. https://doi.org/10.3390/foods12061242
Liu Z, Wang S, Zhang Y, Feng Y, Liu J, Zhu H. Artificial Intelligence in Food Safety: A Decade Review and Bibliometric Analysis. Foods. 2023; 12(6):1242. https://doi.org/10.3390/foods12061242
Chicago/Turabian StyleLiu, Zhe, Shuzhe Wang, Yudong Zhang, Yichen Feng, Jiajia Liu, and Hengde Zhu. 2023. "Artificial Intelligence in Food Safety: A Decade Review and Bibliometric Analysis" Foods 12, no. 6: 1242. https://doi.org/10.3390/foods12061242
APA StyleLiu, Z., Wang, S., Zhang, Y., Feng, Y., Liu, J., & Zhu, H. (2023). Artificial Intelligence in Food Safety: A Decade Review and Bibliometric Analysis. Foods, 12(6), 1242. https://doi.org/10.3390/foods12061242







