Microbiological Quality and Safety of Fresh Turkey Meat at Retail Level, Including the Presence of ESBL-Producing Enterobacteriaceae and Methicillin-Resistant S. aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Turkey Meat Samples and Microbiological Analysis
2.2. Isolation and Identification
2.3. Phenotypic Confirmation of ESBL Producers
2.4. Phenotypic Confirmation of Methicillin Resistance of S. aureus
2.5. Resistance of E. coli and Klebsiella spp. Isolates
2.6. Resistance of Macrococcus spp. and Staphylococci Isolates
2.7. Statistical Analysis
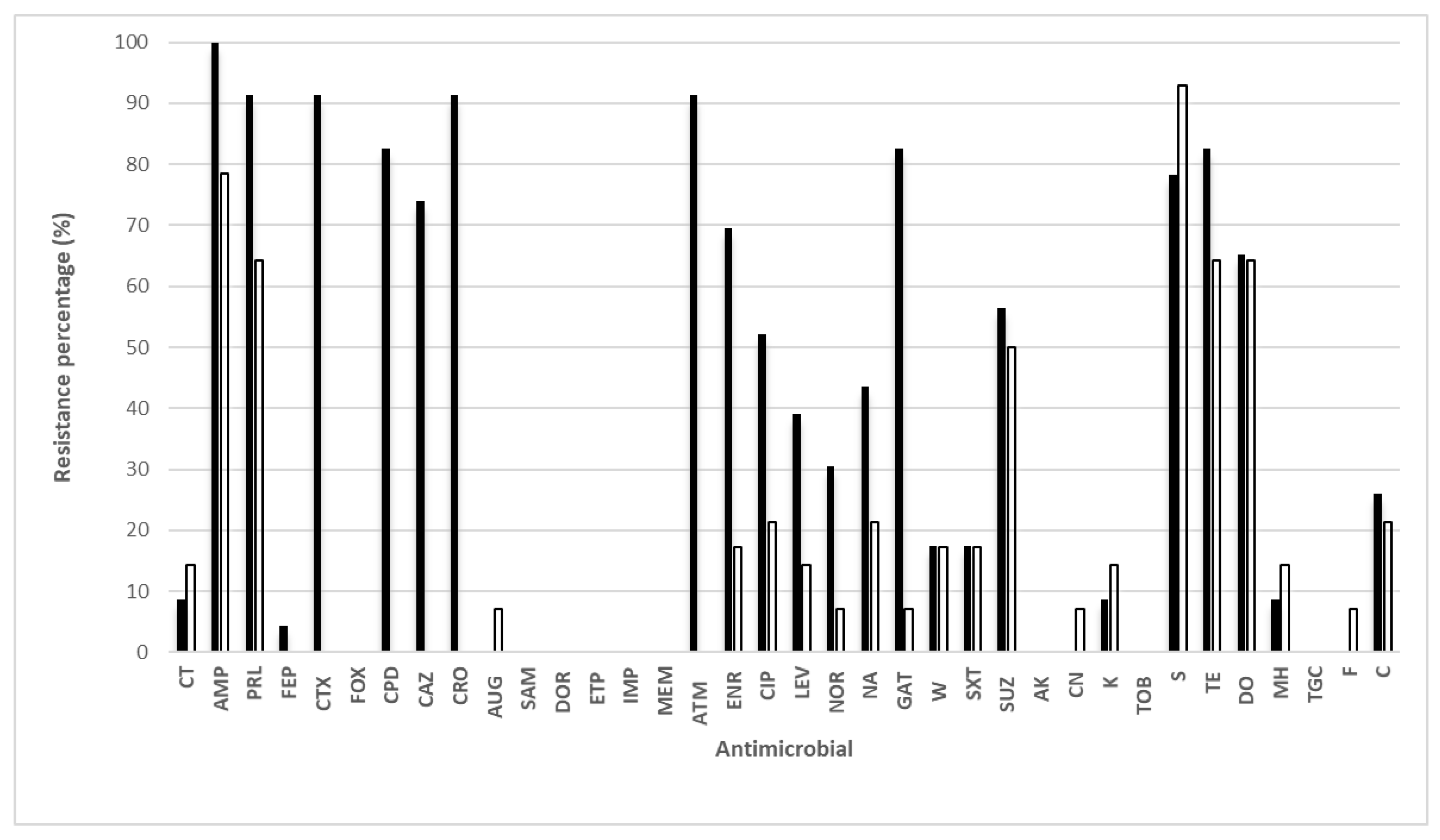
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jukna, V.; Klementavičiūtėm, J.; Meškinytė-Kaušilienėm, E.; Pečiulaitienė, N.; Samborskytėm, M.; Ambrasūnas, L. Comparative evaluation of quality and composition of ostrich, turkey and broiler meat. Biotechnol. Anim. Husb. 2012, 28, 385–392. [Google Scholar] [CrossRef]
- Chai, S.J.; Cole, D.; Nisler, A.; Mahon, B.E. Poultry: The most common food in outbreaks with known pathogens, United States, 1998–2012. Epidemiol. Infect. 2017, 145, 316–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, B.; Crilly, N.; Pendleton, S.; Andino, A.; Wallis, A.; Zhang, N.; Hanning, I. Impact of rearing conditions on the microbiological quality of raw retail poultry meat. J. Food Sci. 2013, 78, M1232–M1235. [Google Scholar] [CrossRef] [PubMed]
- Perez Arnedo, I.; Gonzalez-Fandos, E. Prevalence of Campylobacter spp. in poultry in three Spanish farms, a slaughterhouse and a further processing plant. Foods 2019, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Perez-Arnedo, I.; Cantalejo, M.J.; Martinez-Laorden, A.; Gonzalez-Fandos, E. Effect of processing on the microbiological quality and safety of chicken carcasses at slaughterhouse. Int. J. Food Sci. Technol. 2021, 56, 1855–1864. [Google Scholar] [CrossRef]
- Höll, L.; Behr, J.; Vogel, R.F. Identification and growth dynamics of meat spoilage microorganisms in modified atmosphere packaged poultry meat by MALDI-TOF MS. Food Microbiol. 2016, 60, 84–91. [Google Scholar] [CrossRef]
- Rouger, A.; Odile, T.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Desmonts, M.H.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 19, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Matthews, K.R.; Kniel, K.E.; Montville, T.J. Food Microbiology, 4th ed.; ASM Press: Washington, DC, USA, 2017. [Google Scholar]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef] [Green Version]
- Stellato, G.; Utter, D.R.; Voorhis, A.; De Angelis, M.; Murat Eren, A.; Ercolini, D.A. Few Pseudomonas oligotypes dominate in the meat and dairy processing environment. Front. Microbiol. 2017, 8, 264. [Google Scholar] [CrossRef] [Green Version]
- Kunert-Filho, H.C.; Furian, T.Q.; Sesterhenn, R.; Chitolina, G.Z.; Willsmann, D.E.; Borges, K.A.; Salle, C.T.P.; Moraes, H.L.D.S.; do Nascimento, V.P. Bacterial community identification in poultry carcasses using high-throughput next generation sequencing. Int. J. Food Microbiol. 2022, 364, 109533. [Google Scholar] [CrossRef]
- Oakley, B.B.; Morales, C.A.; Berrang, M.E.; Meinersmann, R.J.; Glenn, E.; Tillman; Wise, M.G.; Siragusa, G.R.; Hiett, K.L.; Bruce, S. The poultry-associated microbiome: Network analysis and farm-to-fork characterizations. PLoS ONE 2013, 8, e57190. [Google Scholar] [CrossRef] [Green Version]
- Saenz-García, C.E.; Castañeda-Serrano, P.; Silva, E.M.; Alvarado, C.Z.; Nava, G.M. Insights into the identification of the specific spoilage organisms in chicken meat. Foods 2020, 9, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.Y.; Wang, H.H.; Han, Y.W.; Xing, T.; Ye, K.P.; Xu, X.L.; Zhou, G.H. Evaluation of the spoilage potential of bacteria isolated from chilled chicken in vitro and in situ. Food Microbiol. 2017, 63, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Han, Y.Q.; Cao, J.X.; Xu, X.L.; Zhou, G.H.; Zhang, W.Y. The spoilage of air-packaged broiler meat during storage at normal and fluctuating storage temperatures. Poult. Sci. 2012, 91, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Jaber, H.; Ijoub, R.; Zaher, A.; Chakit, M.; Rhaiem, N.; Bourkhiss, B.; Ouhssine, M. Microbiological study of turkey meat marketed in Kenitra (North-oust of Morocco). J. Nutr. Food Sci. 2017, 7, 1000620. [Google Scholar]
- Saucier, L.; Gendron, C.; Gariépy, C. Shelf life of ground poultry meat stored under modified atmosphere. Poult. Sci. 2000, 79, 1851–1856. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Sofos, J. Microbial contamination of carcasses and cuts. In Encyclopedia of Meat Science, 2nd ed.; Jennen, W., Devine, C., Dikeman, M., Eds.; Elsevier Academic: Oxford, UK, 2004; pp. 727–737. [Google Scholar]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 2016, 22, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillaspy, A.G.; Iandolo, J.J. Staphylococcus. In Encyclopedia of Food Microbiology; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: London, UK, 2014; Volume 3, pp. 482–486. [Google Scholar]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London: Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tacklng%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 1 April 2022).
- Gonzalez-Fandos, E.; Martinez-Laorden, A.; Abad-Fau, A.; Sevilla, E.; Bolea, R.; Serrano, M.J.; Mitjana, O.; Bonastre, C.; Laborda, A.; Falceto, M.V.; et al. Effect of intramuscularly administered oxytetracycline or enrofloxacin on vancomycin-resistant enterococci, extended spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae in pigs. Animals 2022, 12, 622. [Google Scholar] [CrossRef]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Grande, H.; Weaver, B.; Papp, K.; Horwinski, J.; Koch, B.; Hungate, B.A.; Liu, C.M.; et al. Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 2018, 18, 174. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Jiménez, D.; García-Meniño, I.; Fernández, J.; García, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef] [PubMed]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- De Boer, E.; Zwartkruis-Nahuis, J.T.; Wit, B.; Huijsdens, X.W.; de Neeling, A.J.; Bosch, T.; van Oosterom, R.A.A.; Vila, A.; Heuvelink, A.E. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 2009, 134, 52–56. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [Green Version]
- Ministerio de Agricultura, Pesca y Alimentación. Informe del Consumo de Alimentación en España 2019; MAPA: Madrid, Spain, 2019. [Google Scholar]
- Serrano, M.J.; Elorduy, J.; Zabaleta, I.; Istamboulie, G.; González-Fandos, E.; Bousquet-Mélou, A.; Mata, L.; Aymard, C.; Martínez-Laorden, A.; Da Silva-Guedes, J.; et al. Antimicrobial residue assessment in 5357 commercialized meat samples from the Spain-France cross-border area: A new approach for effective monitoring. Food Control 2022, 138, 109033. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI document M 100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Cotting, K.; Strauss, C.; Rodriguez-Campos, S.; Rostaher, A.; Fischer, N.M.; Roosje, P.J.; Favrot, C.; Perreten, V. Macrococcus canis and M. caseolyticus in dogs: Occurrence, genetic diversity and antibiotic resistance. Vet. Dermatol. 2017, 28, 559–e133. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation EU 2010, Regulation No 37/2010 of 22 December 2009 on pharmacologically active substances and their classification regarding maximum residue limits in food stuffs of animal origin. Off. J. Eur. Union 2010, 15, 1–72.
- Augustyńska-Prejsnar, A.; Hanus, P.; Sokołowicz, Z.; Kačániová, M. Assessment of technological characteristics and microbiological quality of marinated turkey meat with the use of dairy products and lemon juice. Anim. Biosci. 2021, 34, 2003–2011. [Google Scholar] [CrossRef]
- Gonzalez-Fandos, E.; Herrera, B. Efficacy of acetic acid against Listeria monocytogenes attached in poultry skin during refrigerated storage. Foods 2014, 3, 527–540. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Thippareddi, H.; Wang, L. Balamurugan. Meat and poultry. In Food Microbiology Fundamentals and Frontiers, 5th ed.; Doyle, M.P., Diez-Gonzalez, F., Hill, C., Eds.; ASM Press: Washington, DC, USA, 2019; pp. 125–177. [Google Scholar]
- Labadie, J. Consequences of packaging on bacterial growth. Meat is an ecological niche. Meat Sci. 1999, 52, 299–305. [Google Scholar] [CrossRef]
- Pennacchia, C.; Ercolini, D.; Villani, F. Development of a real-time PCR assay for the specific detection of Brochothrix thermosphacta in fresh and spoiled raw meat. Int. J. Food Microbiol. 2009, 134, 230–236. [Google Scholar] [CrossRef]
- Vihavainen, E.; Lundstrom, H.S.; Susiluoto, T.; Koort, J.; Paulin, L.; Auvinen, P.; Bjorkroth, J. Role of broiler carcasses and processing plant air in contamination of modified-atmosphere-packaged broiler products with psychrotrophic lactic acid bacteria. Appl. Environ. Microbiol. 2007, 73, 1136–1145. [Google Scholar] [CrossRef] [Green Version]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Hinton, A.; Cason, J.A.; Ingram, K.D. Tracking spoilage bacteria in commercial poultry processing and refrigerated storage of poultry carcasses. Int. J. Food Microbiol. 2004, 91, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Naik, O.A.; Shashidhar, R.; Rath, D.; Bandekar, J.R.; Rath, A. Metagenomic analysis of total microbial diversity and antibiotic resistance of culturable microorganisms in raw chicken meat and mung sprouts (Phaseolus aureus) sold in retail markets of Mumbai, India. Curr. Sci. 2017, 113, 71–79. [Google Scholar] [CrossRef]
- Schofield, B.J.; Andreani, N.A.; Günther, C.S.; Law, G.R.; McMahon, G.; Swainson, M.; Goddard, M.R. Livestock microbial landscape patterns: Retail poultry microbiomes significantly vary by region and season. Food Microbiol. 2022, 101, 103878. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wu, W.; Yu, Q.; Hou, M.; Gao, F.; Li, X.; Dai, R. Bacterial diversity analysis of pork longissimus lumborum following long term ohmic cooking and water bath cooking by amplicon sequencing of 16S rRNA gene. Meat Sci. 2017, 123, 97–104. [Google Scholar] [CrossRef]
- Burch, J.; Tatineni, S.; Enofe, I.; Laird-Fick, H. Brevundimonas diminuta coinfection as source of pyogenic liver abscess. BMJ Case Rep. 2021, 14, e236235. [Google Scholar] [CrossRef]
- Jałowiecki, Ł.; Żur, J.; Chojniak, J.; Ejhed, H.; Płaza, G. Properties of antibiotic-resistant bacteria isolated from onsite wastewater treatment plant in relation to biofilm formation. Curr. Microbiol. 2018, 7, 639–649. [Google Scholar] [CrossRef] [Green Version]
- Rahdar, H.A.; Mahmoudi, S.; Bahador, A.; Ghiasvand, F.; Sadeghpour Heravi, F.; Feizabadi, M.M. Molecular identification and antibiotic resistance pattern of actinomycetes isolates among immunocompromised patients in Iran, emerging of new infections. Sci. Rep. 2021, 11, 10745. [Google Scholar] [CrossRef]
- Serrano, M.J.; Mitjana, O.; Bonastre, C.; Laborda, A.; Falceto, M.V.; García-Gonzalo, D.; Abilleira, E.; Elorduy, J.; Bousquet-Mélou, A.; Mata, L.; et al. Is Blood a good indicator for detecting antimicrobials in meat? Evidence for the development of in vivo surveillance methods. Antibiotics 2020, 9, 175. [Google Scholar] [CrossRef] [Green Version]
- Kačániová, M.; Mellen, M.; Vukovic, N.L.; Maciej Kluz, M.; Puchalski, C.; Peter Hašcík, P.; Kunová, S. Combined effect of vacuum packaging, fennel and savory essential oil treatment on the quality of chicken thighs. Microorganisms 2019, 7, 134. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, H.M.; Hassan, M.A.; Nahla, A.; Mohga, A.E.A. Prevalence and molecular characterization of Pseudomonas species in frozen imported meat. Benha Vet. Med. J. 2016, 31, 220–224. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef]
- Abu-Ghazaleh, N.; Chua, W.J.; Gopalan, V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J. Gastroenterol. Hepatol. 2021, 36, 75–88. [Google Scholar] [CrossRef] [Green Version]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of antimicrobial resistance and virulence genes in Enterococcus spp. isolated from retail meats in Alberta, Canada. Int. J. Food Microbiol. 2012, 156, 222–230. [Google Scholar] [CrossRef]
- Gobbetti, M.; Calosso, M. Streptococcus. In Encyclopedia of Food Microbiology; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: London, UK, 2014; Volume 3, pp. 553–555. [Google Scholar]
- Schulz, J.; Dumke, J.; Hinse, D.; Dreier, J.; Habig, C.; Kemper, N. Organic Turkey flocks: A reservoir of Streptococcus gallolyticus subspecies gallolyticus. PLoS ONE 2015, 10, A775. [Google Scholar] [CrossRef]
- Giannitsioti, E.; Chirouze, C.; Bouvet, A.; Beguinot, I.; Delahaye, F.; Mainardi, J.L.; Celard, M.; Mihaila-Amrouche, L.; Moing, V.L.; Hoen, B. Characteristics and regional variations of group D streptococcal endocarditis in France. Clin. Microbiol. Infect. 2007, 13, 770–776. [Google Scholar] [CrossRef]
- Hernandez-Macedo, M.A.; Barancelli, G.B.; Contreras-Castillo, C.J. Microbial deterioration of vacuum-packaged chilled beef cuts and techniques for microbiota detection and characterization: A review. Braz. J. Microbiol. 2011, 42, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Miranda, J.M.; Guarddon, M.; Vázquez, B.I.; Fente, C.A.; Barros-Velázquez, J.; Cepeda, A.; Franco, C.M. Antimicrobial resistance in Enterobacteriaceae strains isolated from organic chicken, conventional chicken and conventional turkey meat: A comparative survey. Food Control 2008, 19, 412–416. [Google Scholar] [CrossRef]
- Säde, E.; Murros, A.; Björkroth, J. Predominant enterobacteria on modified atmosphere packaged meat and poultry. Food Microbiol. 2013, 34, 252–258. [Google Scholar] [CrossRef]
- Godziszewska, J.; Guzek, D.; Pogorzelska, E.; Brodowska, M.; Górska-Horczyczak, E.; Sakowska, A.; Wojtasik-Kalinowska, I.; Gantner, M.; Wierzbicka, A. A simple method of the detection of pork spoilage caused by Rahnella aquatilis. LWT Food Sci. Technol. 2017, 84, 248–255. [Google Scholar] [CrossRef]
- Germanou, D.; Spernovasilis, N.; Papadopoulos, A.; Christodoulou, S.; Agouridis, A.P. Infections caused by Moellerella wisconsensis: A case report and a systematic review of the literature. Microorganisms 2022, 10, 892. [Google Scholar] [CrossRef]
- Athanasakopoulou, Z.; Sofia, M.; Giannakopoulos, A.; Papageorgiou, K.; Chatzopoulos, D.C.; Spyrou, V.; Petridou, E.; Petinaki, E.; Billinis, C. ESBL-Producing Moellerella wisconsensis. The contribution of wild birds in the dissemination of a zoonotic pathogen. Animals 2022, 12, 340. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- EMA (European Medicine Agency). Categorisation of Antibiotics for Use in Animals for Prudent and Responsible Use. 2020. Available online: https://www.ema.europa.eu/en/documents/report/infographic-categorisation-antibiotics-use-animals-pru-dent-responsible-use_en.pdf (accessed on 1 August 2022).
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [Green Version]
- Ludden, C.; Moradigaravand, D.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Naydenova, P.; Hernandez-Garcia, J.; Wood, P.; Hadjirin, N.; Radakovic, M.; et al. A one health study of the genetic relatedness of Klebsiella pneumoniae and their mobile elements in the east of England. Clin. Infect. Dis. 2020, 70, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Syed, M.A.; Ullah, H.; Tabassum, S.; Fatima, B.; Woodley, T.A.; Ramadan, H.; Jackson, C.R. Staphylococci in poultry intestines: A comparison between farmed and household chickens. Poult. Sci. 2020, 99, 4549–4557. [Google Scholar] [CrossRef]
- Casanova, C.; Iselin, L.; von Steiger, N.; Droz, S.; Sendi, P. Staphylococcus hyicus bacteremia in a farmer. J. Clin. Microbiol. 2011, 49, 4377–4378. [Google Scholar] [CrossRef] [Green Version]
- Kamath, U.; Singer, C.; Isenberg, H.D. Clinical significance of Staphylococcus warneri bacteremia. J. Clin. Microbiol. 1992, 30, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Meservey, A.; Sullivan, A.; Wu, C.; Lantos, P.M. Staphylococcus sciuri peritonitis in a patient on peritoneal dialysis. Zoonoses Public Health 2020, 67, 93–95. [Google Scholar] [CrossRef]
- Ramnarain, J.; Yoon, J.; Runnegar, N. Staphylococcus pasteuri infective endocarditis: A case report. IDCases 2019, 18, e00656. [Google Scholar] [CrossRef]
- Soldera, J.; Nedel, W.L.; Cardoso, P.R.; d’Azevedo, P.A. Bacteremia due to Staphylococcus cohnii -ssp. urealyticus caused by infected pressure ulcer: Case report and review of the literature. Sao Paulo Med. J. 2013, 131, 59–61. [Google Scholar] [CrossRef] [Green Version]
- Tous Romero, F.; Gutierrez Garcia-Rodrigo, C.; Tamariz, V.; Llamas Martin, R. Acute Infection by Staphylococcus simulans in the hand of a man. JAMA Dermatol. 2016, 152, 1060. [Google Scholar] [CrossRef]
- Aslantaş, Ö. High occurence of methicillin resistant Staphylococcus sciuri (MRSS) and first detection of mecC from broiler flocks in Turkey. Isr. J. Vet. Med. 2020, 75, 185–192. [Google Scholar]
- Keller, J.E.; Schwendener, S.; Neuenschwander, J.; Overesch, G.; Perreten, V. Prevalence and characterization of methicillin-resistant Macrococcus spp. in food producing animals and meat in Switzerland in 2019. Band 2022, 164, 153–164. [Google Scholar] [CrossRef]
- Mazhar, S.; Hill, C.; McAuliffe, O. The Genus Macrococcus: An insight into its biology, evolution, and relationship with Staphylococcus. Adv. Appl. Microbiol. 2018, 105, 1–50. [Google Scholar]
- Hanson, B.M.; Dressler, A.E.; Harper, A.L.; Scheibel, R.P.; Wardyn, S.E.; Roberts, L.K.; Kroegera, J.S.; Smitha, T.C. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 2011, 4, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Waters, A.E.; Contente-Cuomo, T.; Buchhagen, J.; Liu, C.M.; Watson, L.; Pearce, K.; Foster, J.T.; Bowers, J.; Driebe, E.M.; Engelthaler, D.M.; et al. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 2011, 52, 1227–1230. [Google Scholar] [CrossRef]
- Kraushaar, B.; Ballhausen, B.; Leeser, D.; Tenhagen, B.A.; Käsbohrer, A.; Fetsch, A. Antimicrobial resistances and virulence markers in Methicillin-resistant Staphylococcus aureus from broiler and turkey: A molecular view from farm to fork. Vet. Microbiol. 2017, 200, 25–32. [Google Scholar] [CrossRef]
- Tegegne, H.A.; Koláčková, I.; Florianová, M.; Gelbíčová, T.; Madec, J.Y.; Haenni, M.; Karpíšková, R. Detection and molecular characterisation of methicillin-resistant Staphylococcus aureus isolated from raw meat in the retail market. J. Glob. Antimicrob. Resist. 2021, 26, 233–238. [Google Scholar] [CrossRef]
- Pyzik, E.; Marek, A.; Stepien-Pysniak, D.; Urban-Chmiel, R.; Jarosz, L.S.; Jagiello-Podebska, I. Detection of antibiotic resistance and classical enterotoxin genes in coagulase-negative staphylococci isolated from poultry in Poland. J. Vet. Res. 2019, 63, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Regecová, I.; Pipová, M.; Jevinová, P.; Kmeť, V.; Výrostková, J.; Sopková, D. Antimicrobial resistance of Coagulase-Negative species of staphylococci isolated from the meat of wild pheasants (phasianus colchicus). Ital. J. Anim. Sci. 2014, 13, 627–630. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [Green Version]
- Mezher, Z.; Saccares, S.; Marcianò, R.; De Santis, P.; Flores Rodas, E.D.; De Angelis, V.; Condoleo, R. Occurrence of Campylobacter spp. in poultry meat at retail and processing plants’ levels in Central Italy. Ital. J. Food Saf. 2016, 5, 47–49. [Google Scholar] [CrossRef] [Green Version]
- Korsak, D.; Mackiw, E.; Rozynek, E.; Zyłowska, M. Prevalence of Campylobacter spp. in retail chicken, turkey, pork, and beef meat in Poland between 2009 and 2013. J. Food Protect. 2015, 78, 1024–1028. [Google Scholar] [CrossRef]
- Narvaez-Bravo, C.; Mutschall, S.K.; Aslam, M. Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int. J. Food Microbiol. 2017, 253, 43–47. [Google Scholar] [CrossRef]
- Cohen, N.; Ennaji, H.; Bouchrif, B.; Hassar, M.; Karib, H. Comparative study of microbiological quality of raw poultry meat at various seasons and for different slaughtering processes in Casablanca (Morocco). J. Appl. Poult. Res. 2007, 16, 502–508. [Google Scholar] [CrossRef]
| Microbial Group and Species | Number of Isolates | Percentage (%) |
|---|---|---|
| Lactic acid bacteria Lactobacillus spp. (30) * Carnobacterium divergens (31) Carnobacterium maltaromaticum (13) Lactococcus lactis (5) Lactococcus raffinolactis (1) Leuconostoc mesenteroides (4) Leuconostoc carnosum (2) Leuconostoc citreum (1) | 87 | 37.66% |
| Brocchotrix thermosphacta | 53 | 22.94% |
| Pseudomonas spp. P. fragi (10) P. lundensis (4) P. brenneri (2) P. libanensis (2) P. fluorescens (1) P. extremorientalis (1) P. taetrolens (1) | 21 | 9.09% |
| Enterobacteriaceae Serratia liquefaciens (5) Serratia proteamaculnas (5) Serratia marcescens (1) Escherichia coli (2) Hafnia alvei (2) Ewingella americana (1) Raoultella planticola (1 Rahnella inusitata (1) Erwinia rhapontici (1)) | 19 | 8.23% |
| Microccaceae Kocuria varians (5) Kocuria salsicia (4) Kocuria rizhophila (1) Staphylococcus saprophyticus (3) Staphylococcus warneri (1) Macrococcus caseolyticus (2) Micrococcus luteus (1) Rothia nasimurium (1) | 18 | 7.79% |
| Enterococci Enterococcus faecalis (3) | 3 | 1.30% |
| Other Gram-negative bacteria Chryseobacterium scophtalnum (4) Chryseobacterium aquaticum (1) Chryseobacterium rhizosphaerae (1) Chryseobacterium piscium (1) Chryseobacterium sigense (1) Acinetobacter gulliouiae (2) Acinetobacter lwoffii (2) Acinetobacter johnsonii (1) Acinetobacter harbonensis (1) Brevundimonas diminuta (1) Stenotrophomonas rhizophila (5) Wautersiella falsenii (2) Psychrobacter pulmonis (1) | 23 | 9.96% |
| Other Gram-positive bacteria Microbacterium liquefaciens (3) Microbacterium maritypicum (2) Rhodococcus erythropolis (1) Bacillus endophyticus (1) | 7 | 3.03% |
| Total | 231 | 100 |
| Species | Number of Isolates | Percentage (%) |
|---|---|---|
| Pseudomonas libanensis | 31 | 31 |
| Pseudomonas extremorientalis | 14 | 14 |
| Pseudomonas fluorescens | 12 | 12 |
| Pseudomonas antarctica | 7 | 7 |
| Pseudomonas rhodesiae | 6 | 6 |
| Pseudomonas azotoformans | 5 | 5 |
| Pseudomonas veronii | 5 | 5 |
| Pseudomonas brenneri | 5 | 5 |
| Pseudomonas orientalis | 3 | 3 |
| Pseudomonas synxantha | 3 | 3 |
| Pseudomonas marginalis | 2 | 2 |
| Pseudomonas cedrina | 2 | 2 |
| Pseudomonas trialis | 2 | 2 |
| Pseudomonas kilorensis | 1 | 1 |
| Pseudomonas proteolítica | 1 | 1 |
| Pseudomonas koreensis | 1 | 1 |
| Total Pseudomonas spp. | 100 | 100 |
| Specie | Number of Isolates | Percentage (%) |
|---|---|---|
| E faecium | 16 | 38.10 |
| E. faecalis | 10 | 23.81 |
| E. gallinarum | 7 | 16.67 |
| E gilvus | 5 | 11.90 |
| E. cassiliflavus | 3 | 7.14 |
| E. hirae | 1 | 2.38 |
| Total enterococci | 42 | 100 |
| Specie | Number of Isolates | Percentage (%) |
|---|---|---|
| Serratia liquefaciens | 22 | 16.42 |
| Hafnia alvei | 19 | 14.18 |
| Escherichia coli | 19 | 14.18 |
| Ewingella americana | 18 | 13.43 |
| Buttiauxella gaviniae | 16 | 11.94 |
| Rahnella aquatilis | 8 | 5.97 |
| Serratia proteamaculans | 7 | 5.22 |
| Buttiauxella warmboldiae | 5 | 3.73 |
| Buttiauxella agrestis | 4 | 2.99 |
| Serratia fonticola | 4 | 2.99 |
| Klebsiella pneumoniae | 3 | 2.24 |
| Klebsiella oxytoca | 2 | 1.49 |
| Moellerella wisconsensis | 2 | 1.49 |
| Kluywera intermedia | 2 | 1.49 |
| Enterobacter cloacae | 1 | 0.75 |
| Pantoea aglomerans | 1 | 0.75 |
| Yersinia enterocolitica | 1 | 0.75 |
| Total Enterobacteriacceae | 134 | 100 |
| Medium of Isolation (Number of Isolates) | Antibiotic Resistance Phenotype 1 (Number of Isolates) | Retailer 3 |
|---|---|---|
| ChromID ESBL (23) | TE-S-ENR-CIP-NA-PRL-AMP (1) | SA |
| TE-S-PRL-CT-K-SUZ-SXT-W (1) | SC | |
| ENR-PRL-AMP-ATM-CAZ-CPD-CTX-CRO (1) | SD | |
| TE-PRL-AMP-ATM-CPD-CTX-CRO-DO (1) | HB | |
| AMP-ATM-CAZ-CPD-CTX-CRO-DO S (1) | SA | |
| TE-S-SUZ-PRL-AMP-ATM-CTX- CRO-C (1) | HA | |
| TE-S-PRL-AMP-ATM-CAZ-CPD-CTX-CAZ-CRO-DO K (1) | SE | |
| TE-S-PRL-AMP-ATM-CAZ-CPD-CTX-CRO-DO (1) | SE | |
| TE-S-PRL-AMP-ATM-CAZ-CPD-CTX-CRO-DO-SUZ (1) | HA | |
| TE-S-ENR-AMP-SUZ-ATM-CAZ-CPD-CTX-CRO-SXT-W (1) | SG | |
| TE-S-ENR-AMP- PRL-ATM-CAZ-CPD-CTX-CRO-DO (1) 2- | SF | |
| TE-S-PRL-ENR-CIP-AMP-ATM-CAZ-CPD-CTX-CRO-DO (1) | SE | |
| ENR-CIP-NA-PRL-AMP-ATM-CTX-CPD-CAZ-CRO-GAT-LEV-NOR (1) | SF | |
| AMP-ATM-CTX-CAZ-CPD-CTX-CRO-C-CIP-ENRO-NA-PRL-S-SUZ (1) | SG | |
| TE-S-ENR-CIP-PRL-AMP-SUZ-ATM-CAZ-CPD-CTX-CRO-DO-C (1) | HB | |
| TE-S-ENR-CIP-NA-PRL-AMP-SUZ-ATM-CAZ-CPD-CTX-CRO-DO-C-LEV (1) | HB | |
| TE-S-ENR-CIP-NA-PRL-AMP-SUZ-ATM-CAZ-CPD-CTX-CRO-DO-LEV-NOR (1) | HA | |
| TE-S-ENR-CIP-NA-PRL-AMP-ATM-CAZ-CPD-CTX-CRO-DO-GAT-LEV-NOR-(1) | SA | |
| TE-S-ENR-CIP-NA-PRL-K-SUZ-AMP-ATM-CAZ-CPD-CTX-CRO-DO-MH (1) | SB | |
| TE-S-ENR-CIP-NA-PRL-AMP-CT-SUZ-SXT-W-ATM-CTX-CRO-DO-LEV-NOR (1) | SC | |
| TE-S-ENR-CIP-NA-PRL-AMP--SUZ-SXT-W-ATM-CPD-CTX-CRO-LEV-NOR-FEP (1) | HB | |
| TE-S-ENR-CIP-NA-PRL-AMP-SUZ-ATM-XAZ-CPD-CTX-CRO-DO-LEV-NOR-GAT (1) | HB | |
| TE-S-ENR-CIP-NA-PRL-AMP-SUZ-CAZ-CPD-CRO-DO-C-LEV-NOR-FEP-GAT (1) | SA | |
| MacConkey agar (10) | S-ENR-PRL-DO (1) | SG |
| S-ENR-NA-PRL-AMP (1) | SB | |
| TE-AMP-CT-DO-F-AUG (1) | HA | |
| TE-S-PRL-AMP-SUZ-SXT-DO (1) 2 | SF | |
| TE-S-PRL-AMP- SUZ-SXT-W-DO (1) | SA | |
| TE-S-PRL-AMP-SUZ-SXT-W-DO (1) | SE | |
| TE-S-AMP-K-SUZ-SXT-W-DO-C-MH-CN (1) | SC | |
| TE-S-ENR-CIP-NA-CT-K-SUZ-DO-C-LEV-NOR (1) | HB | |
| TE-ENR-CIP-NA-PRL-AMP-SUZ-DO-C-LEV-GAT (1) | SG | |
| TE-S-ENR-CIP-NA-PRL-AMP-SUZ-SXT-W-DO-MH-(1) | SE |
| Species (Number of Isolates) | Antibiotic Resistance Phenotype 1 (Number of Isolates) | Retailer 6 |
|---|---|---|
| Klebsiella oxytoca (2) | CT-AMP 2 | HA |
| AMP 2 | TA | |
| Klebsiella pneumoniae (8) | CT-AMP-PRL-FEP-CAZ-CPD-CTX-CRO-ATM-ENRO-CIP-NA-SUZ-SXT-W-S (1) 2,3,4 | SF |
| CT-AMP-PRL-FEP-CAZ-CPD-CTX-CRO-ATM-ENRO-CIP-SUZ-SXT-W-S-K-TOB-TE-DO (1) 2,3,4 | SF | |
| CT-AMP-PRL-FEP-CAZ-CPD-CTX-CRO-ATM-ENRO-CIP-NA-SUZ-SXT-W-S- K-TOB-TE-DO-F (1) 2,3,4 | SF | |
| AMP-PRL-FEP-CPD-CTX-CRO-ATM-ENRO-CIP-SUZ-SXT-W-S-K-TOB-TE (2) 3,4,5 | SF | |
| AMP-PRL-FEP-CAZ-CPD-CTX-CRO-ATM-ENRO-CIP-SUZ-SXT-W-S- K-TOB-TE-DO-F-TGC-LEV-NOR (1) 3,4,5 | SF | |
| CT-AMP-PRL-CAZ-CPD-CRO-ENRO-CIP-SUZ-S-TE-DO-F-TGC-SAM-FOX-ETP-AK-MH (1) 4,5 | SA | |
| AMP-PRL-FEP-CAZ-CPD-CTX-CRO-ATM-ENRO-CIP-SUZ-SXT-W-S- K-TOB-TE-DO (1) 4,5 | HA |
| Species | Number of Isolates | Percentage (%) |
|---|---|---|
| Staphylococcus saprophyticus | 39 | 31.45 |
| Staphylococcus equorum | 17 | 13.7 |
| Macrococcus caseolyticus | 15 | 12.1 |
| Staphylococcus aureus | 10 | 8.1 |
| Staphylococcus epidermidis | 8 | 6.45 |
| Staphylococcus vitulinus | 8 | 6.45 |
| Staphylococcus lentus | 5 | 4.03 |
| Staphylococcus cohnii | 4 | 3.23 |
| Staphylococcus warneri | 4 | 3.23 |
| Staphylococcus xylosus | 4 | 3.23 |
| Staphylococcus fleurettii | 3 | 2.41 |
| Staphylococcus pasteuri | 2 | 1.61 |
| Staphylococcus sciuri | 2 | 1.61 |
| Staphylococcus capitis | 1 | 0.8 |
| Staphylococcus hyicus | 1 | 0.8 |
| Staphylococcus simlulans | 1 | 0.8 |
| Total | 124 | 100 |
| Species (Number of Isolates) | Antibiotic Resistance Phenotype 1 (Number of Isolates) | Retailer 3 (Number of Isolates) |
|---|---|---|
| Macrococcus caseolyticus (8) | susceptible to all antibiotics tested (4) 2 | SB (3) SC (1) |
| FOX (1) 2 | SC (1) | |
| MY (1) 2 | SA (1) | |
| PUM (1) 2 | SB (1) | |
| MY-PUM-FAD-LZD-P-RD-TZD-TY-VA-ERY-CMN (1) 2 | SE (1) |
| Methicillin-Resistant Isolates (Number of Isolates) | Antibiotic Resistance Phenotype 1 (Number of Isolates) | Retailer 4 |
|---|---|---|
| No (5) | P-PNG-TE-DO (1) 2 | SF |
| P-PNG-TE-MH-ENR-NOR (1) 2 | SG | |
| P-PNG-TE-ENRO-CIP-GAT-NOR-LEV (1) 2 | SG | |
| P-PNG-TE-DO-ENRO-CIP-GAT-NOR-LEV- MY- TZD-TY-ERY-CMN-S (1) 2 | HA | |
| P-PNG-TE-DO-ENRO-CIP-NOR-SUZ (1) 3 | HB | |
| Yes (4) | P-PNG-TE-DO-FAD-FOX (1) 2 | SE |
| P-PNG- TE- ENRO-CIP-GAT-NOR-LEV-MY-TY-ERY-CMN-FOX (1) 3 | SD | |
| P-PNG-TE-ENRO-CIP-GAT-NOR-LEV-MY-TY-ERY-CMN-FOX-S-SUZ-AK-C-K (1) 3 | HB | |
| P-PNG-TE-MH-MY-TY-ERY-CMN-FOX-S-SUZ-AK-K-PUM-TOB-CPT-CN-QD-RD-FAD (1) 3 | HA |
| Specie (Number of Isolates) | Antibiotic Resistance Phenotype 1 (Number of Isolates) | Retailer 4 (Number of Isolates) |
|---|---|---|
| Staphylococcus capitis (1) | PUM-P-ENR (1) 2 | SD (1) |
| Staphylococcus cohnii (3) | MY-ERY-TE (1) 2 | SF (1) |
| P-MY-TE-DO-FAD (1) 2 | HB (1) | |
| MY-ERY-TE-TY-CMN-C-W (1) 2 | HB (1) | |
| Staphylococcus epidermidis (4) | PUM-P (1) 2 | HA (1) |
| PUM-ERY (1) 2 | SB (1) | |
| P-ERY (1) 2 | SA (1) | |
| PUM-P-ERY (1) 2 | SG (1) | |
| Staphylococcus equorum (7) | susceptible to all antibiotics tested (1) 2 | SB (1) |
| ERY (4) 2 | SC (4) | |
| S (1) 2 | TA (1) | |
| MY-TE (1) 2 | SC (1) | |
| Staphylococcus fleurettii (1) | P-MY (1) 2 | HB (1) |
| Staphylococcus hyicus (3) | susceptible to all antibiotics tested (1) 2 | SG (1) |
| Staphylococcus lentus (3) | MY-TE- DO-CMN-ENRO (1) 2 | SA (1) |
| MY-TE-DO-S-CMN (1) 2 | SC (1) | |
| MY-TE-DO-CMN-W-ENRO-SUZ (1) 2 | SC (1) | |
| Staphylococcus pasteuri (2) | PUM-P-MY-ERY-TE-CMN-S-SUZ FOX-AK-TOB-CPT (1) 2 | SA (1) |
| Staphylococcus saprophyticus (21) | susceptible to all antibiotics tested (2) 2 | HB (1) TA (1) |
| ERY (1) 2 | SF (M1) | |
| P (2) 2 | SA (1) TA (1) | |
| FAD (1) 2 | HA (1) | |
| FAD-P (2) 2 | SC (1) TA (1) | |
| TE (1) 2 | SA (1) | |
| TE-DO (4) 2 | SA (2) SC (2) | |
| MY-TE (1) 2 | SC (1) | |
| MY-TE-DO (1) 2 | SA (1) | |
| P-TE-DO-FAD (1) 2 | SA (1) | |
| MY-PUM-FAD-P (1) 2 | HA (1) | |
| P-ERY-CMN (1) 2 | TA (1) | |
| MY-PUM-TE-AK-CPT (1) 2 | SA (1) | |
| MY-PUM-FAD-P-S-SUZ-AK-K-CPT (1) 2 | SA (1) | |
| MY-PUM-P-TE-FOX-AK-K-CPT-CN (1) 2 | SC (1) | |
| Staphylococcus sciuri (2) | FAD (1) 2 | SB (1) |
| MY-TE-FAD (1) 2 | SB (1) | |
| Staphylococcus simulans (1) | susceptible to all antibiotics tested (1) 2 | SG (1) |
| Staphylococcus vitulinus (4) | susceptible to all antibiotics tested (3) 2 | HB (1) |
| TE (1) 2 | HB (1) | |
| Staphylococcus warneri (4) | P-ERY (1) 2 | SB (1) |
| PUM-P (1) 2 | TA (1) | |
| TOB-CN-K (2) 2,3 | SA (1) | |
| Staphylococcus xylosus (1) | susceptible to all antibiotics tested (1) 2 | SA (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Laorden, A.; Arraiz-Fernández, C.; González-Fandos, E. Microbiological Quality and Safety of Fresh Turkey Meat at Retail Level, Including the Presence of ESBL-Producing Enterobacteriaceae and Methicillin-Resistant S. aureus. Foods 2023, 12, 1274. https://doi.org/10.3390/foods12061274
Martínez-Laorden A, Arraiz-Fernández C, González-Fandos E. Microbiological Quality and Safety of Fresh Turkey Meat at Retail Level, Including the Presence of ESBL-Producing Enterobacteriaceae and Methicillin-Resistant S. aureus. Foods. 2023; 12(6):1274. https://doi.org/10.3390/foods12061274
Chicago/Turabian StyleMartínez-Laorden, Alba, Celia Arraiz-Fernández, and Elena González-Fandos. 2023. "Microbiological Quality and Safety of Fresh Turkey Meat at Retail Level, Including the Presence of ESBL-Producing Enterobacteriaceae and Methicillin-Resistant S. aureus" Foods 12, no. 6: 1274. https://doi.org/10.3390/foods12061274
APA StyleMartínez-Laorden, A., Arraiz-Fernández, C., & González-Fandos, E. (2023). Microbiological Quality and Safety of Fresh Turkey Meat at Retail Level, Including the Presence of ESBL-Producing Enterobacteriaceae and Methicillin-Resistant S. aureus. Foods, 12(6), 1274. https://doi.org/10.3390/foods12061274








