Iturin A Strongly Inhibits the Growth and T-2 Toxin Synthesis of Fusarium oxysporum: A Morphological, Cellular, and Transcriptomics Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Experimental Strain
2.2. Colony Growth Analysis
2.3. Sporulation Capacity Analysis
2.4. Analysis of T-2 Toxin Synthesis
2.5. Scanning Electron Microscope (SEM) Analysis
2.6. Membrane Permeability Analysis
2.7. Analysis of Reactive Oxygen Species
2.8. Transcriptome Analysis
2.8.1. Isolation of Total RNA, cDNA Library Construction, and Sequencing
2.8.2. Data Analysis, Differentially Expressed Genes Screening, and Bioinformatics Analysis
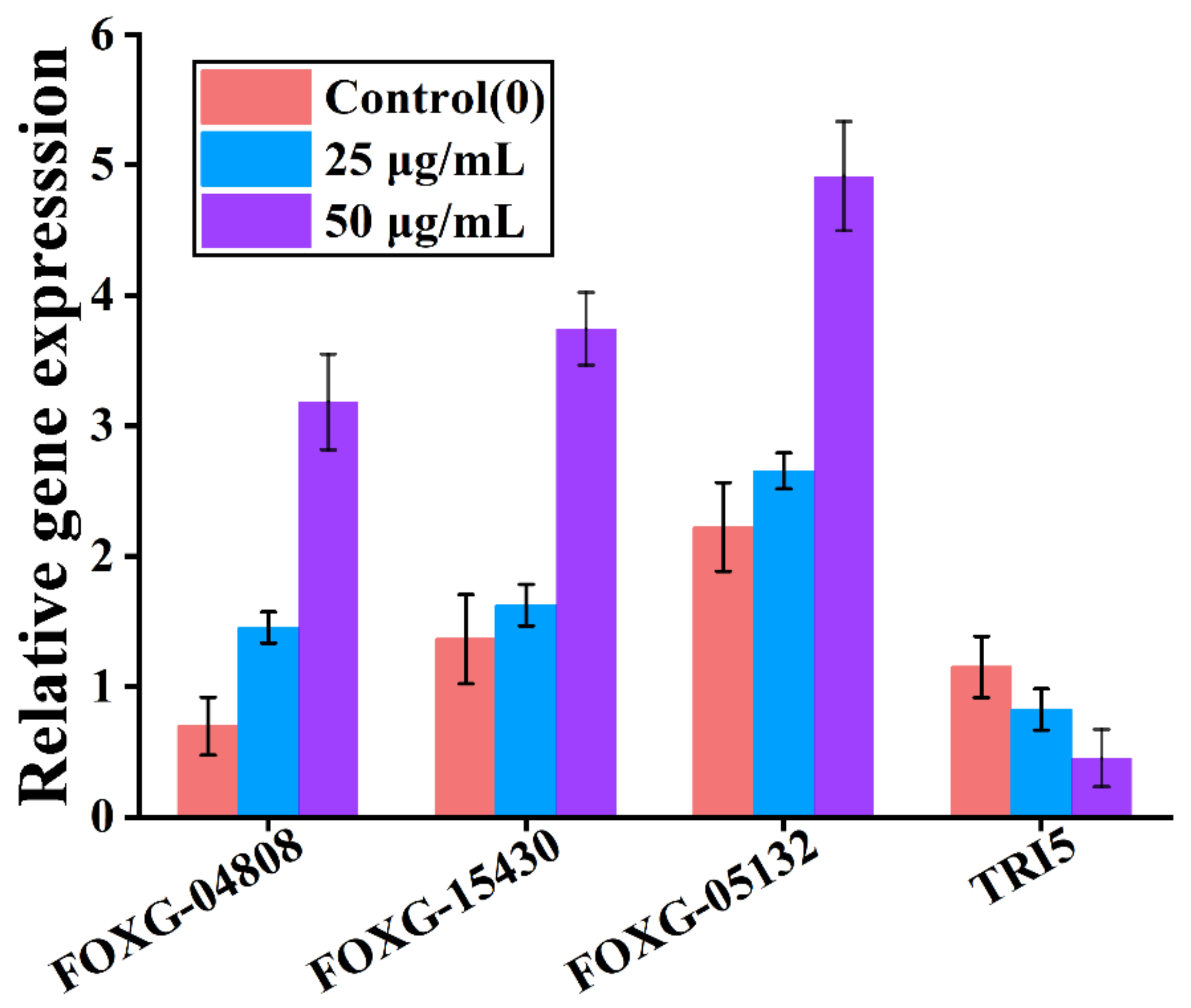
2.9. Quantitative Real-Time PCR Analysis
2.10. Statistical Analysis
3. Results
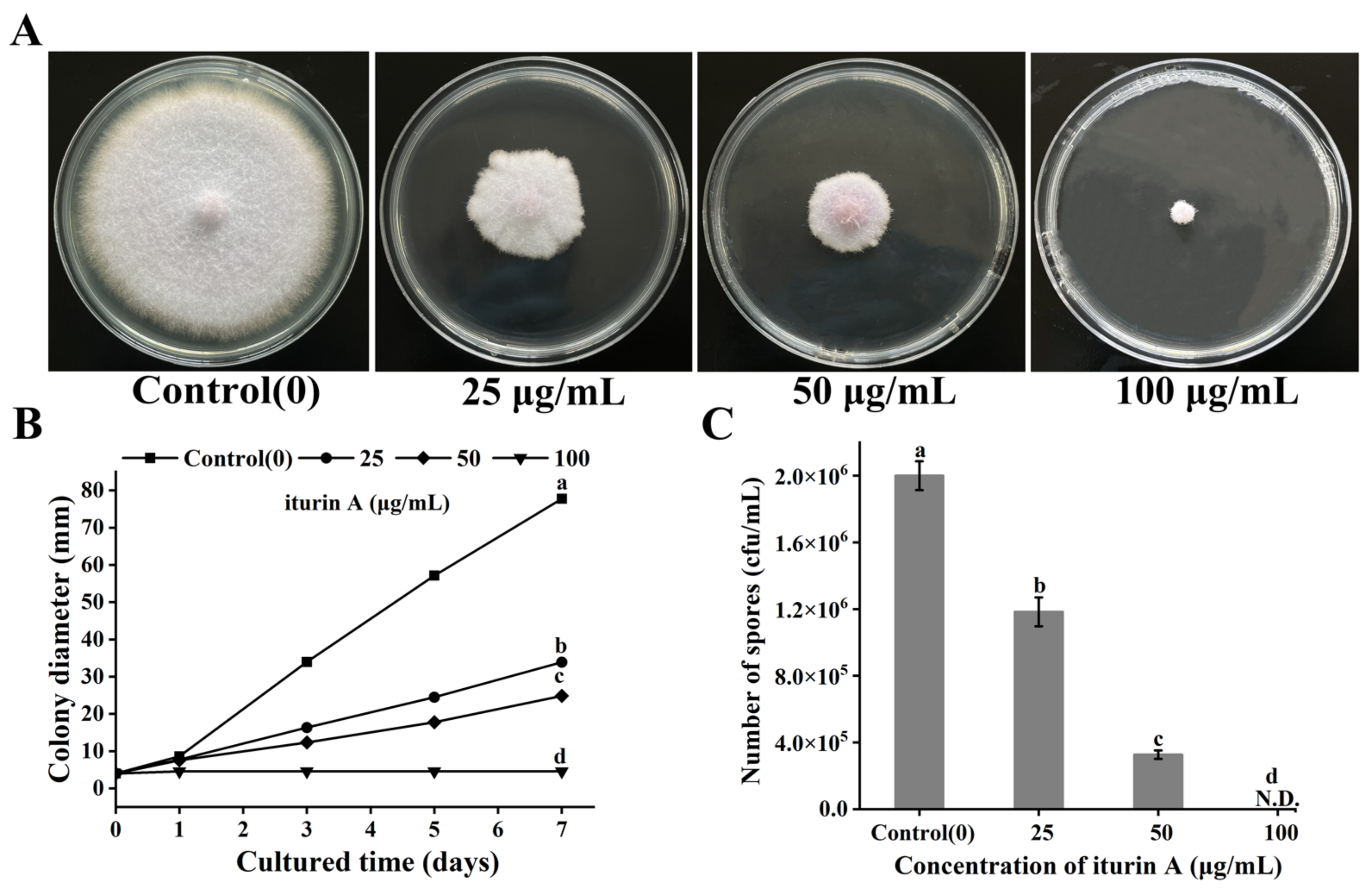
3.1. Effects of Iturin A on Colony Diameter
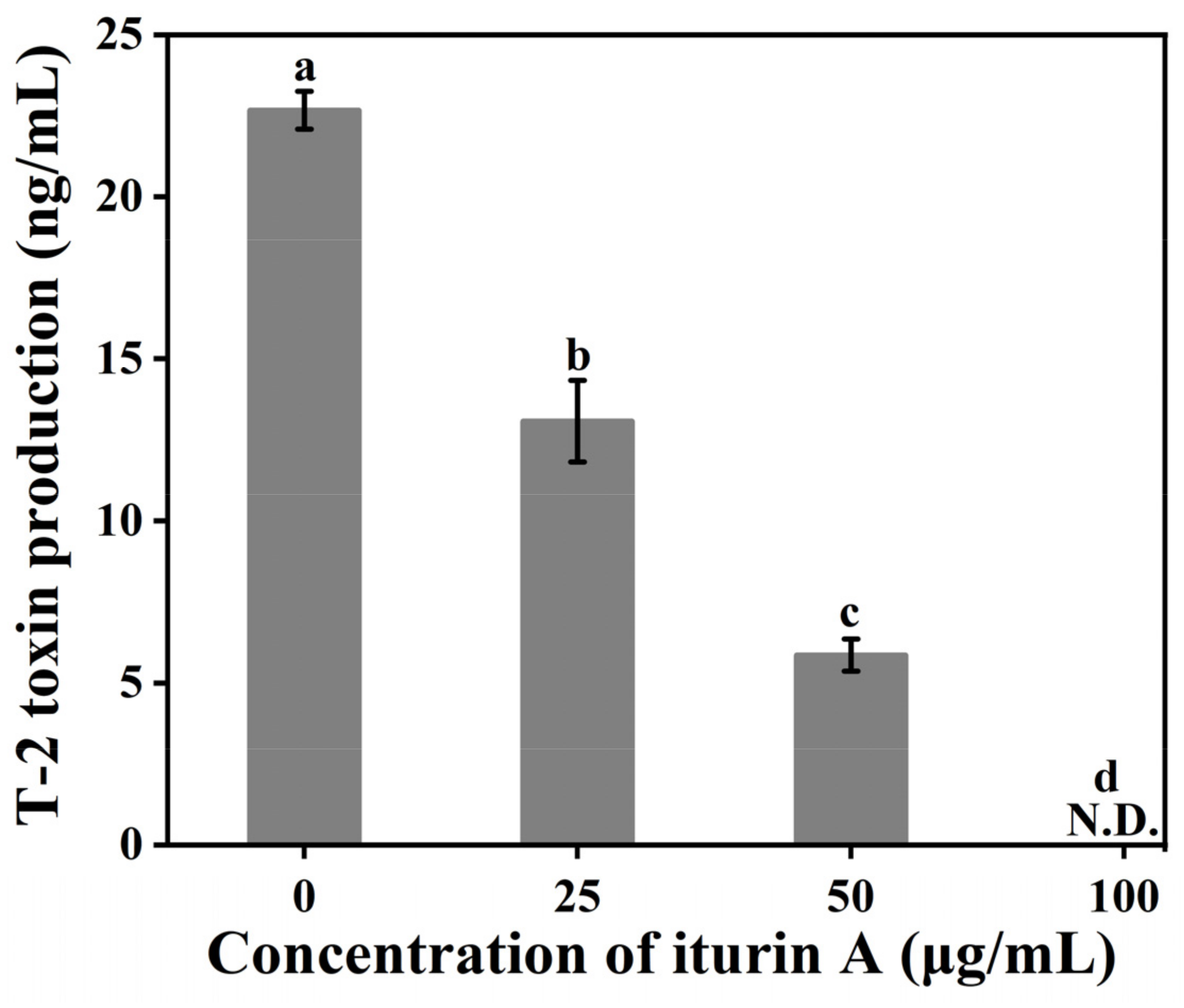
3.2. Effects of Iturin A on T-2 Toxin Synthesis
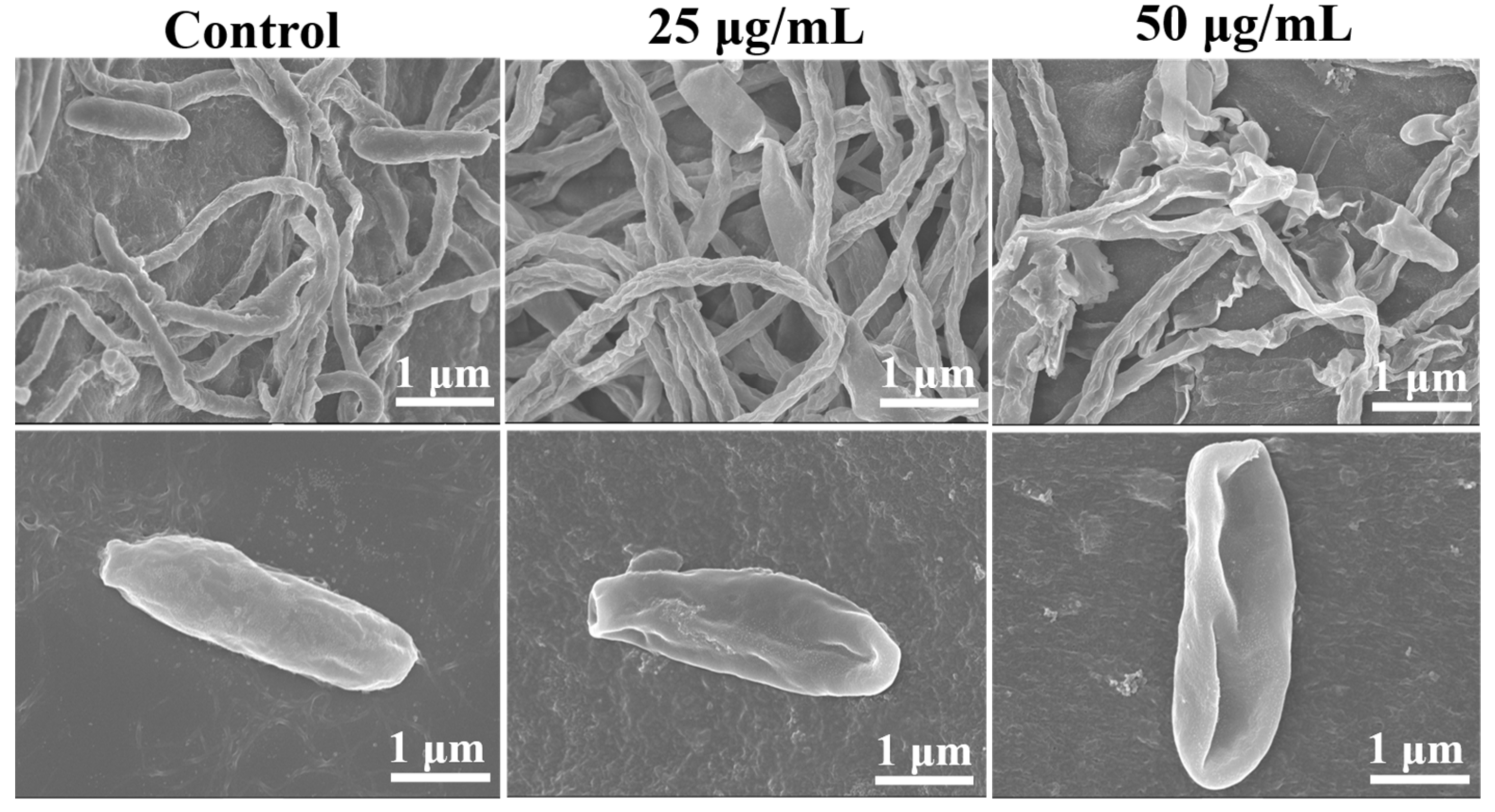
3.3. Effects of Iturin A on the Morphology of Hyphae and Spores
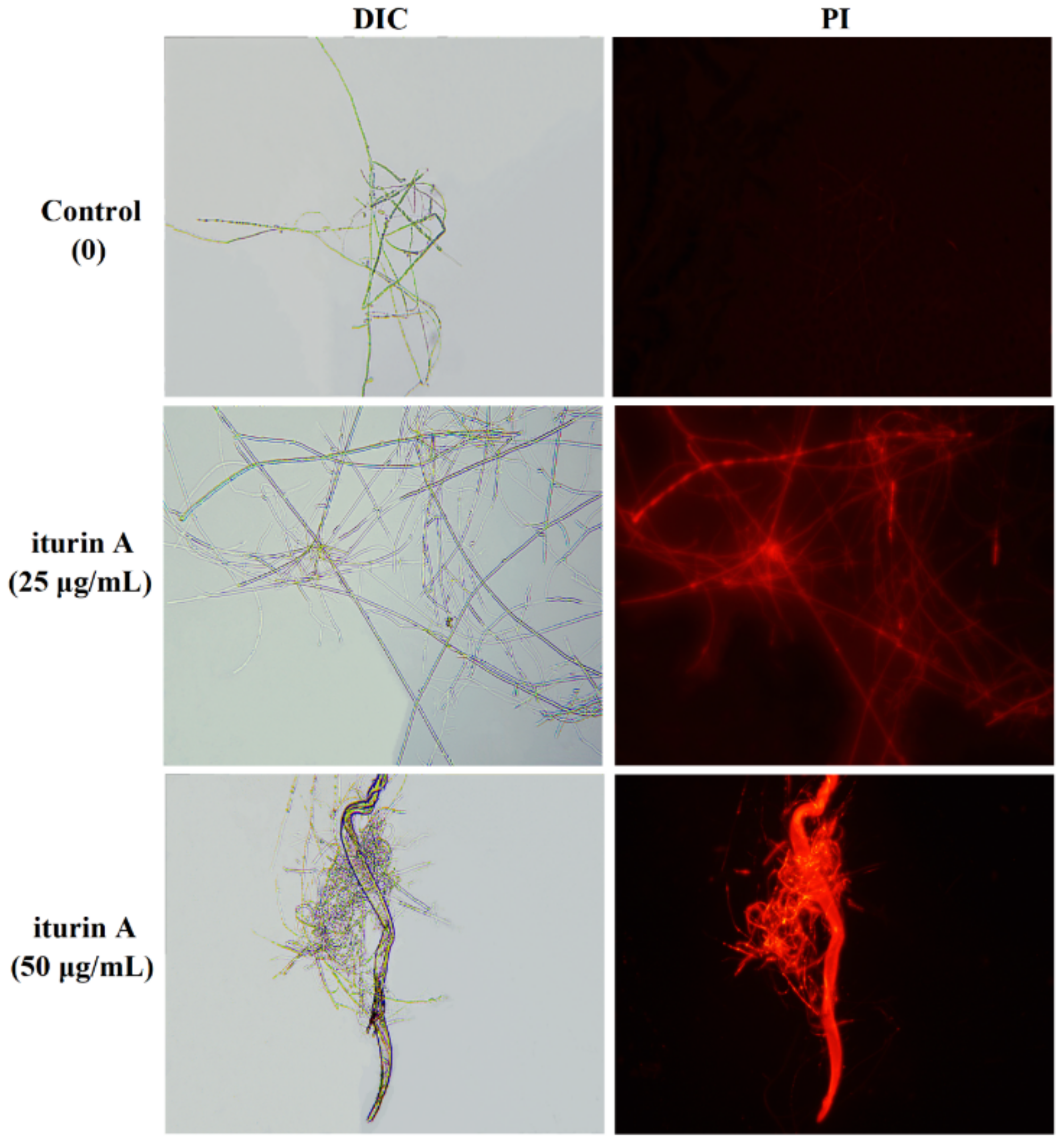
3.4. Effects of Iturin A on Cell Membrane Permeability
3.5. Effects of Iturin A on Reactive Oxygen Species
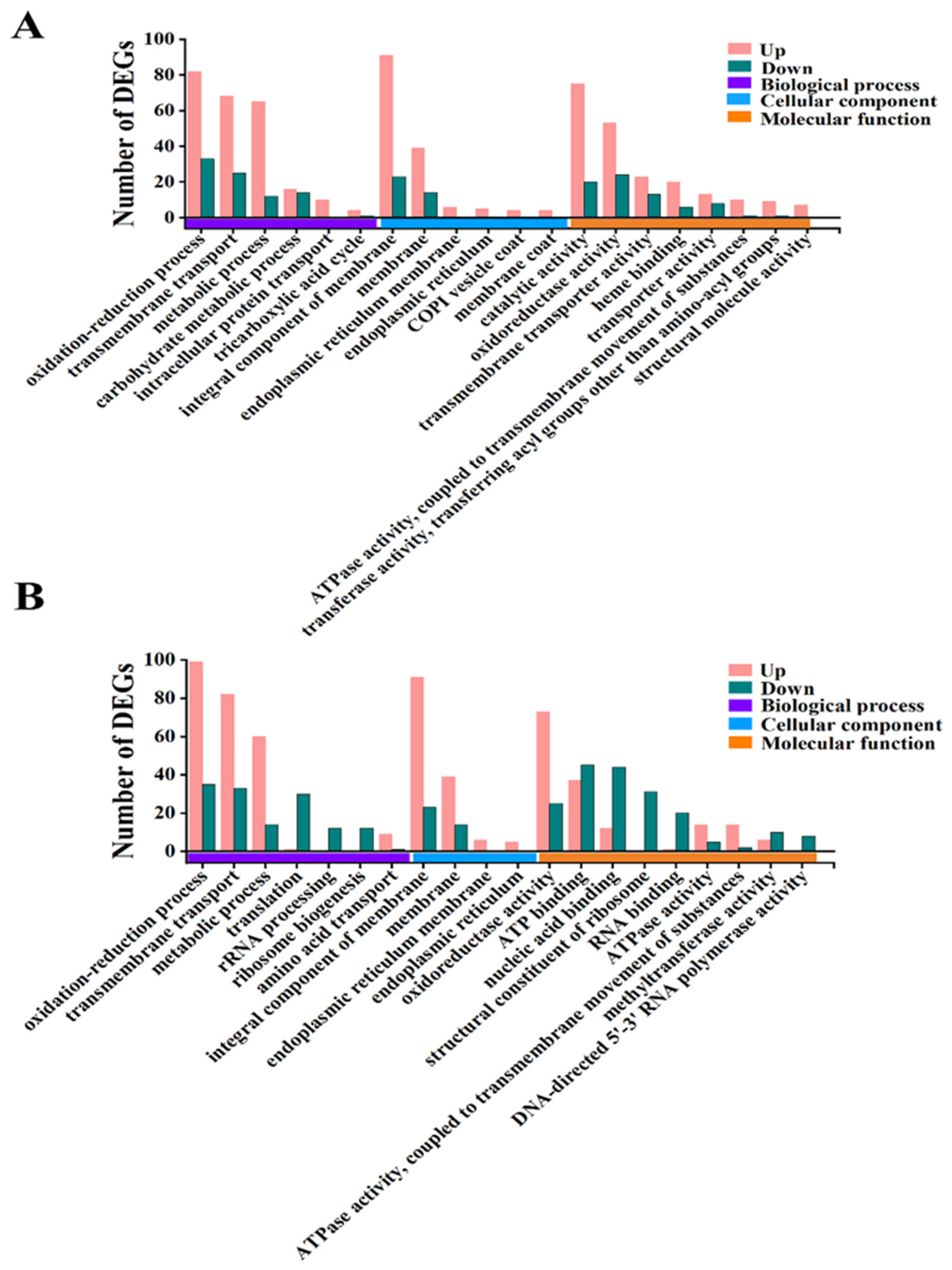
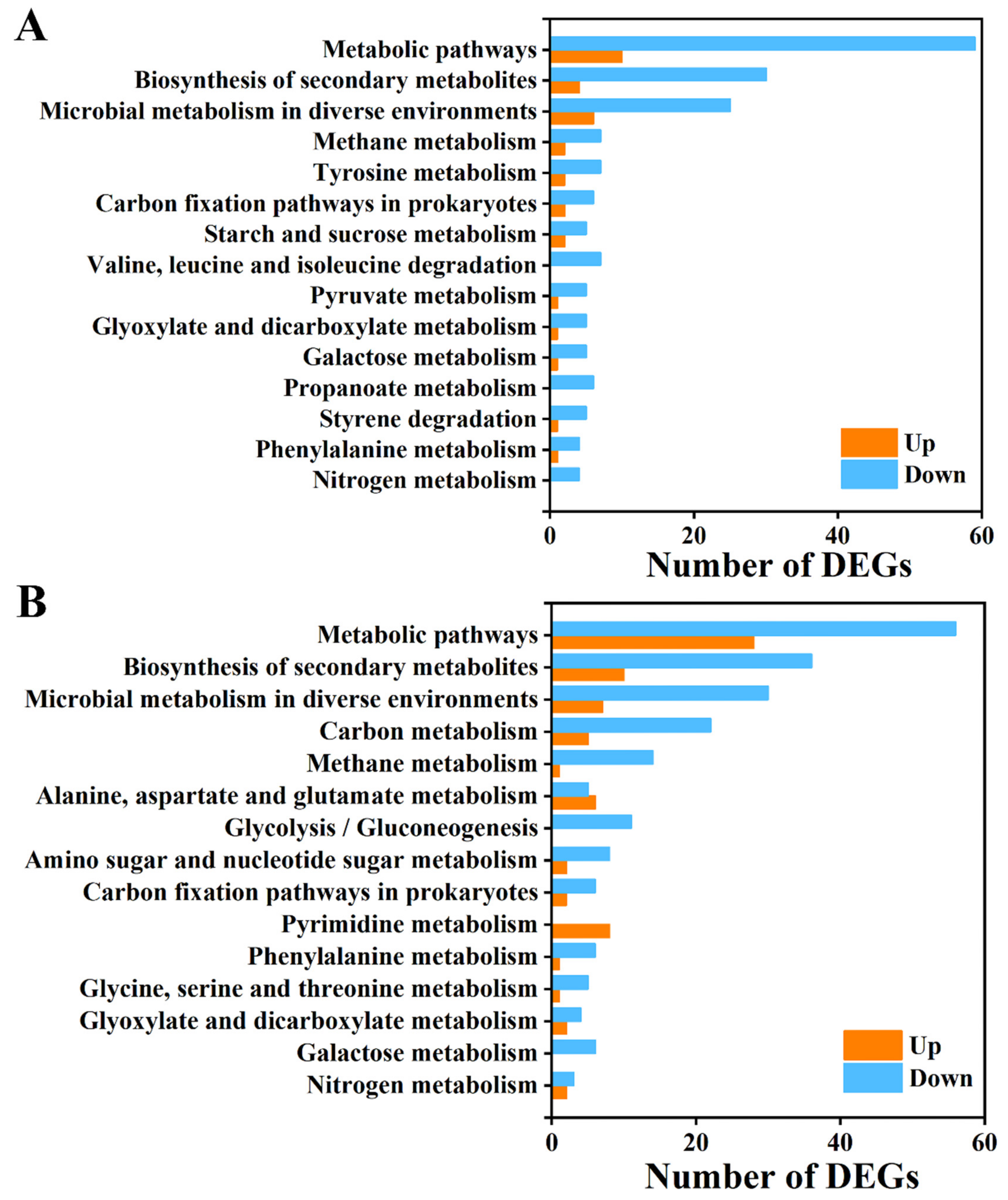
3.6. Analysis of GO Function and KEGG Pathway of DEGs
3.7. Functional Analysis of DEG
3.7.1. Mycelial Growth
3.7.2. T-2 Toxin Synthesis
3.7.3. Cell Integrity
3.7.4. Transmembrane Transport
3.7.5. Redox
3.7.6. Energy Metabolism
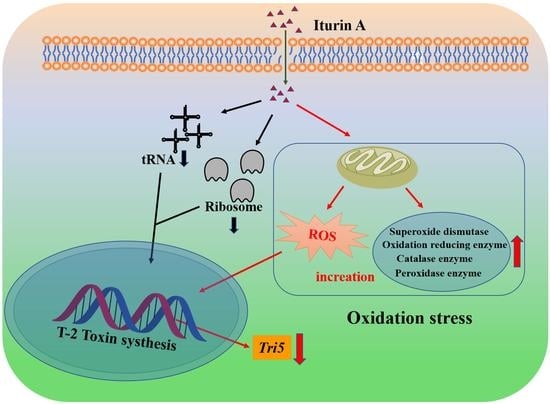
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piacenza, N.; Kaltner, F.; Maul, R.; Gareis, M.; Schwaiger, K.; Gottschalk, C. Distribution of T-2 toxin and HT-2 toxin during experimental feeding of yellow mealworm (Tenebrio molitor). Mycotoxin Res. 2021, 37, 11–21. [Google Scholar] [CrossRef]
- Gbashi, S.; Madala, N.E.; De Saeger, S.; De Boevre, M.; Njobeh, P.B. Numerical optimization of temperature-time degradation of multiple mycotoxins. Food Chem. Toxicol. 2019, 125, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Albarenque, S.M.; Doi, K. T-2 toxin-induced apoptosis in rat keratinocyte primary cultures. Exp. Mol. Pathol. 2005, 78, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Xu, X.; Hou, R.; Badawy, S.; Tao, Y.; Chen, D.; Yuan, Z. DNA methylation and RASSF4 expression are involved in T-2 toxin-induced hepatotoxicity. Toxicology 2019, 425, 152246. [Google Scholar] [CrossRef]
- Wang, J.; Fitzpatrick, D.W.; Wilson, J.R. Effects of the Trichothecene Mycotoxin T-2 Toxin on Neurotransmitters and Metabolites in Discrete Areas of the Rat Brain. Food Chem. Toxicol. 1998, 36, 947–953. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, Y.; Deng, Q.; Sun, L.; Wang, R.; Ye, L.; Gooneratne, R. Fungal diversity and mycotoxin contamination in dried fish products in Zhanjiang market, China. Food Control 2021, 121, 107614. [Google Scholar] [CrossRef]
- Medina, A.; Magan, N. Temperature and water activity effects on production of T-2 and HT-2 by Fusarium langsethiae strains from north European countries. Food Microbiol. 2011, 28, 392–398. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Podogrocki, M.; Ceremuga, M.; Stela, M.; Bijak, M. T-2 toxin—The most toxic trichothecene mycotoxin: Metabolism, toxicity, and decontamination strategies. Molecules 2021, 26, 6868. [Google Scholar] [CrossRef] [PubMed]
- Rovira, P.; Brugnini, G.; Rodriguez, J.; Cabrera, M.C.; Saadoun, A.; de Souza, G.; Rufo, C. Microbiological changes during long-storage of beef meat under different temperature and vacuum-packaging conditions. Foods 2023, 12, 694. [Google Scholar] [CrossRef]
- Duret, S.; Hoang, H.-M.; Guillier, L.; Derens-Bertheau, E.; Dargaignaratz, C.; Oriol, S.; Nguyen-the, C. Interactions between refrigeration temperatures, energy consumption in a food plant and microbiological quality of the food product: Application to refrigerated stuffed pasta. Food Control 2021, 126, 108076. [Google Scholar] [CrossRef]
- Skovgaard, N. Food irradiation research and technology. Int. J. Food Microbiol. 2017, 118, 228. [Google Scholar] [CrossRef]
- Del Olmo, A.; Calzada, J.; Nuñez, M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017, 57, 3084–3103. [Google Scholar] [CrossRef] [PubMed]
- Piper, J.D.; Piper, P.W. Benzoate and sorbate salts: A systematic review of the potential hazards of these invaluable preservatives and the expanding spectrum of clinical uses for sodium benzoate. Compr. Rev. Food Sci. F 2017, 16, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Wang, W.; Sun, L.; Wang, Y.; Liao, J.; Xu, D.; Gooneratne, R. A sensitive method for simultaneous quantitative determination of surfactin and iturin by LC-MS/MS. Anal. Bioanal. Chem. 2017, 409, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.W.; Arrebola, E.; Pérez-García, A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Jin, P.; Zheng, Y. Investigating the efficacy of Bacillus subtilis SM21 on controlling Rhizopus rot in peach fruit. Int. J. Food Microbiol. 2013, 164, 141–147. [Google Scholar] [CrossRef]
- Liu, C.; Sheng, J.; Chen, L.; Zheng, Y.; Lee, D.Y.; Yang, Y.; Shen, L. Biocontrol Activity of Bacillus subtilis Isolated from Agaricus bisporus Mushroom Compost against Pathogenic Fungi. J. Agric. Food Chem. 2015, 63, 6009–6018. [Google Scholar] [CrossRef]
- Nasir, M.N.; Besson, F. Specific interactions of mycosubtilin with cholesterol-containing artificial membranes. Langmuir 2011, 27, 10785–10792. [Google Scholar] [CrossRef]
- Jiang, C.; Shi, J.; Liu, Y.; Zhu, C. Inhibition of Aspergillus carbonarius and fungal contamination in table grapes using Bacillus subtilis. Food Control 2014, 35, 41–48. [Google Scholar] [CrossRef]
- Han, Q.; Wu, F.; Wang, X.; Qi, H.; Shi, L.; Ren, A.; Tang, C. The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ. Microbiol. 2015, 17, 1166–1188. [Google Scholar] [CrossRef]
- Cho, K.M.; Math, R.K.; Hong, S.Y.; Asraful Islam, S.M.; Mandanna, D.K.; Cho, J.J.; Yun, H.D. Iturin produced by Bacillus pumilus HY1 from Korean soybean sauce (kanjang) inhibits growth of aflatoxin producing fungi. Food Control 2009, 20, 402–406. [Google Scholar] [CrossRef]
- Aranda, F.J.; Teruel, J.A.; Ortiz, A. Further aspects on the hemolytic activity of the antibiotic lipopeptide iturin A. Biochim. Biophys. Acta (BBA)-Biomembr. 2005, 1713, 51–56. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Z.; Shi, Y.; Guo, D.; Pang, B.; Chen, X.; Shi, J. Bacillus subtilis inhibits Aspergillus carbonarius by producing iturin A, which disturbs the transport, energy metabolism, and osmotic pressure of fungal cells as revealed by transcriptomics analysis. Int. J. Food Microbiol. 2020, 330, 108783. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Tian, Z.H.; Luo, Y.; Cheng, Y.J.; Long, C.A. Antagonistic Activity and the Mechanism of Bacillus amyloliquefaciens DH-4 against Citrus Green Mold. Phytopathology 2018, 108, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Zhao, H.; Pang, B.; Qu, R.; Lian, Z.; Jiang, C.; Shi, J. Capability of iturin from Bacillus subtilis to inhibit Candida albicans in vitro and in vivo. Appl. Microbiol. Biot. 2019, 103, 4377–4392. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, L.; Guo, L.; Sun, H.; Huang, Q.; Shao, D.; Jiang, C.; Shi, J. Effects of Bacillus subtilis iturin A on HepG2 cells in vitro and vivo. AMB Express 2021, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Deng, Y.; Deng, Q.; Sun, L.; Fang, Z.; Wang, Y.; Zhao, J. Cysteine Inhibits the Growth of Fusarium oxysporum and Promotes T-2 Toxin Synthesis through the Gtr/Tap42 Pathway. Microbiol. Spectr. 2022, 10, e03682-22. [Google Scholar] [CrossRef]
- Yu, J.-J.; Ji, H.; Chen, Y.; Zhang, Y.-F.; Zheng, X.-C.; Li, S.-H.; Chen, Y. Analysis of the glycosylation products of peanut protein and lactose by cold plasma treatment: Solubility and structural characteristics. Int. J. Biol. Macromol. 2020, 158, 1194–1203. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, S.; Sun, K.; Hu, J. Identification and characterization of a cyclic lipopeptide iturin A from a marine-derived Bacillus velezensis 11-5 as a fungicidal agent to Magnaporthe oryzae in rice. J. Plant. Dis. Protect. 2019, 127, 15–24. [Google Scholar] [CrossRef]
- Yi, Y.; Luan, P.; Liu, S.; Shan, Y.; Hou, Z.; Zhao, S.; Li, R. Efficacy of Bacillus subtilis XZ18-3 as a Biocontrol Agent against Rhizoctonia cerealis on Wheat. Agriculture 2022, 12, 258. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Ambrico, A.; Trupo, M. Efficacy of cell free supernatant from Bacillus subtilis ET-1, an Iturin A producer strain, on biocontrol of green and gray mold. Postharvest Biol. Technol. 2017, 134, 5–10. [Google Scholar] [CrossRef]
- Veras, F.F.; Correa, A.P.; Welke, J.E.; Brandelli, A. Inhibition of mycotoxin-producing fungi by Bacillus strains isolated from fish intestines. Int. J. Food Microbiol. 2016, 238, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhao, J.; Zhang, B.; Shen, Q.; Shang, Q.; Li, P. Effect of a novel antifungal peptide P852 on cell morphology and membrane permeability of Fusarium oxysporum. Biochim. Biophys. Acta (BBA)-Biomembr. 2019, 1861, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wang, L.; Jiang, D.; Wang, M.; Liu, H.; Yu, H.; Yao, W. Transcriptomic analysis of inhibition by eugenol of ochratoxin A biosynthesis and growth of Aspergillus carbonarius. Food Control 2022, 135, 108788. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalić, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef]
- McCormick, S.P.; Hohn, T.M. Accumulation of Trichothecenes in Liquid Cultures of a Fusarium sporotrichioides Mutant Lacking a Functional Trichothecene C-15 Hydroxylase. Appl. Environ. Microb. 1997, 63, 1685–1688. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microb. 2002, 68, 2959–2964. [Google Scholar] [CrossRef]
- Tijerino, A.; Cardoza, R.; Moraga, J.; Malmierca, M.; Vicente, F.; Aleu, J.; Hermosa, R. Overexpression of the trichodiene synthase gene tri5 increases trichodermin production and antimicrobial activity in Trichoderma brevicompactum. Fungal. Genet. Biol. 2011, 48, 285–296. [Google Scholar] [CrossRef]
- Ma, Y.; Gao, K.; Yu, H.; Liu, W.; Qin, Y.; Xing, R.; Li, P. C-coordinated O-carboxymethyl chitosan Cu (II) complex exerts antifungal activity by disrupting the cell membrane integrity of Phytophthora capsici Leonian. Carbohyd. Polym. 2021, 261, 117821. [Google Scholar] [CrossRef]
- Ouyang, Q.; Tao, N.; Jing, G. Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genom. 2016, 17, 599. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yan, D.; Li, M.; Li, D.; Zhang, S.; Fan, G.; Peng, L.; Pan, S. Ergosterol depletion under bifonazole treatment induces cell membrane damage and triggers a ROS-mediated mitochondrial apoptosis in Penicillium expansum. Fungal Biol. 2022, 126, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yuan, K.; Zhou, Q.; Lu, C.; Du, L.; Liu, F. Mechanism of antifungal activity of Perilla frutescens essential oil against Aspergillus flavus by transcriptomic analysis. Food Control 2021, 123, 107703. [Google Scholar] [CrossRef]
- Crowley, J.H.; Smith, S.J.; Leak, F.W.; Parks, L.W. Aerobic isolation of an ERG24 null mutant of Saccharomyces cerevisiae. J. Bacteriol. 1996, 178, 2991–2993. [Google Scholar] [CrossRef]
- Nasir, M.N.; Besson, F. Interactions of the antifungal mycosubtilin with ergosterol-containing interfacial monolayers. Biochim. Biophys. Acta (BBA)-Biomembr. 2012, 1818, 1302–1308. [Google Scholar] [CrossRef]
- Xie, M.; Ma, N.; Bai, N.; Yang, L.; Yang, X.; Zhang, K.Q.; Yang, J. PKC-SWI6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 2455–2471. [Google Scholar] [CrossRef]
- Salinas, A.E.; Wong, M.G. Glutathione S-transferases-a review. Curr. Med. Chem. 1999, 6, 279–310. [Google Scholar] [CrossRef]
- Melov, S.; Coskun, P.; Patel, M.; Tuinstra, R.; Cottrell, B.; Jun, A.S.; Wallace, D.C. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc. Natl. Acad. Sci. USA 1999, 96, 846–851. [Google Scholar] [CrossRef]
- Qi, G.; Zhu, F.; Du, P.; Yang, X.; Qiu, D.; Yu, Z.; Zhao, X. Lipopeptide induces apoptosis in fungal cells by a mitochondria-dependent pathway. Peptides 2010, 31, 1978–1986. [Google Scholar] [CrossRef]

| Gene | Primer Extension Direction | Primer Sequence |
|---|---|---|
| FoEF1α | Forward | CGGTACTGGTGAGTTCGAGGCT |
| Reverse | TGTTGATGGCGACAATGAGGTT | |
| FOXG-05808 | Forward Reverse | AACAAGACTCCCGAGGATAAGG CATCATCAGGGCCAGCTACAT |
| FOXG-15530 | Forward Reverse | ACCGCATCACCACCCCTA TCGGAGGTAAGGCTTGACG |
| FOXG-05132 | Forward Reverse | ACTGCATCGATCTGCTCTACCT CGCCGTGAAGTTGCTGATA |
| Tri5 | Forward Reverse | CTTTCCCACCGAGTATTTTCT AATCTCATGGAGGCGGTATCG |
| Gene ID | log2 (IA/CK) | p-Value | Annotated Gene Function | Gene ID | log2 (IA/CK) | p-Value | Annotated Gene Function |
|---|---|---|---|---|---|---|---|
| DOWN-REGULATED | |||||||
| FOXG-00235 | −1.551 | 0.027 | DNA repair protein rhp51 | FOXG-05866 | −1.978 | 0.001 | MFS transporter, SP family, general alpha glucoside: H+ symporter |
| FOXG-05220 | −1.198 | 0.037 | myb-like DNA-binding protein FlbD | FOXG-05595 | −2.103 | 0.005 | cytochrome-b5 reductase |
| Tri5 | −1.521 | 0.256 | synthetic enzyme gene | FOXG-05595 | −2.052 | 0.006 | cytochrome P550 oxidoreductase |
| FOXG-05120 | −1.311 | 0.0322 | endoglucanase | FOXG-06551 | −1.957 | 0.002 | choline dehydrogenase |
| FOXG-15550 | −3.83 | 0 | beta-glucosidase | FOXG-16238 | −1.677 | 0.005 | alcohol dehydrogenase |
| FOXG-13566 | −2.392 | 0 | glucoamylase | FOXG-07519 | −1.581 | 0.002 | mannitol-1-phosphate dehydrogenase |
| FOXG-16920 | −1.205 | 0.037 | alpha-amylase | FOXG-12362 | −1.108 | 0.053 | acid phosphatase |
| FOXG-15621 | −5.085 | 0.015 | MFS transporter, AGZA family, xanthine/uracil permease | ||||
| UP-REGULATED | |||||||
| FOXG-08765 | 3.065 | 0.018 | sterol 25-C-methyltransferase | FOXG-01307 | 2.928 | 0.01 | glutathione S-transferase |
| FOXG-12787 | 2.756 | 0.0553 | sterol carrier protein 2 | FOXG-07591 | 2.658 | 0.01 | glutathione S-transferase |
| FOXG-11555 | 2.561 | 0.015 | cytochrome P450, family 51 (sterol 15-demethylase) | FOXG-03656 | 1.557 | 0.052 | glutathione S-transferase |
| FOXG-05355 | 1.352 | 0.375 | delta25(25(1))-sterol reductasedelta | FOXG-05798 | 2.755 | 0.168 | 3-isopropylmalate dehydrogenase |
| FOXG-10778 | 2.6 | 0.088 | MFS transporter, SP family, general alpha glucoside: H+ symporter | FOXG-05288 | 1.556 | 0.138 | ketol-acid reductoisomerase, mitochondrial |
| FOXG-13625 | 2.501 | 0.021 | MFS transporter, SP family, general alpha glucoside: H+ symporter | FOXG-07887 | 1.521 | 0.282 | acyl-CoA dehydrogenase |
| FOXG-09382 | 1.09 | 0.559 | vesicle transporter SEC22 | FOXG-00735 | 2.385 | 0.051 | acetyl-coenzyme A synthetase |
| FOXG-07958 | 3.395 | 0.015 | PiT family inorganic phosphate transporter | FOXG-02925 | 1.63 | 0.209 | acetyl-CoA acyltransferase |
| FOXG-05598 | 1.501 | 0.105 | Ca2+: H+ antiporter | FOXG-11385 | 1.51 | 0.138 | acetyl-CoA C-acetyltransferase |
| FOXG-07591 | 1.735 | 0.002 | glutathione S-transferase | FOXG-06200 | 1.312 | 0.221 | acetyl-CoA C-acetyltransferase |
| FOXG-05808 | 5.59 | 0.001 | catalase | FOXG-05118 | 1.595 | 0.151 | acyl-CoA dehydrogenase |
| FOXG-15530 | 3.23 | 0.001 | peroxidase | FOXG-02255 | 5.651 | 0.001 | endonuclease G, mitochondrial |
| FOXG-05132 | 2.871 | 0.023 | oxidoreductase | FOXG-05827 | −3.355 | 0.012 | acetyl-CoA acyltransferase |
| FOXG-12260 | 1.525 | 0.253 | catalase-peroxidase | FOXG-13050 | −2.138 | 0.005 | alcohol dehydrogenase |
| Gene ID | log2 (IA/CK) | p-Value | Annotated Gene Function | Gene ID | log2 (IA/CK) | p-Value | Annotated Gene Function |
|---|---|---|---|---|---|---|---|
| DOWN-REGULATED | |||||||
| FOXG-19811 | −5.152 | 0.035 | tRNA | FOXG-05075 | −1.38 | 0.005 | 60S ribosomal protein L27a |
| FOXG-00235 | −1.817 | 0 | DNA repair protein rhp51 | FOXG-10571 | −1.253 | 0.002 | 60S ribosomal protein L7 |
| FOXG-02005 | −1.615 | 0.002 | ribosome biogenesis protein ytm-1 | Tri5 | −2.362 | 0.202 | Synthetic enzyme gene |
| FOXG-10250 | −1.383 | 0.001 | ribosome biogenesis protein ERB1 | FOXG-01535 | −1.03 | 0.018 | ABC transporter |
| FOXG-02122 | −1.526 | 0.003 | 50S ribosomal protein S22 | FOXG-07519 | −1.252 | 0.005 | mannitol-1-phosphate dehydrogenase |
| FOXG-01619 | −1.327 | 0.006 | 50S ribosomal protein S29 | FOXG-07382 | −2.529 | 0.007 | acid phosphatase |
| FOXG-03375 | −1.253 | 0.007 | 50S ribosomal protein L15e | FOXG-10756 | −1.73 | 0.002 | acid phosphatase |
| FOXG-05325 | −1.253 | 0.008 | 50S ribosomal protein L22e | ||||
| UP-REGULATED | |||||||
| FOXG-11555 | 3.006 | 0.005 | cytochrome P550, family 51 (sterol 15-demethylase) | FOXG-03076 | 1.015 | 0.039 | superoxide dismutase |
| FOXG-08765 | 2.12 | 0.008 | sterol 25-C-methyltransferase | FOXG-17180 | 1.537 | 0.006 | catalase-peroxidase 2 |
| FOXG-10530 | 1.151 | 0.005 | C-5 sterol desaturase | FOXG-10652 | 1.651 | 0.005 | S-(hydroxymethyl)glutathione dehydrogenase |
| FOXG-12390 | 1.135 | 0.027 | MFS transporter, SP family, general alpha glucoside: H+ symporter | FOXG-08382 | 1.375 | 0.005 | glutathione S-transferase |
| FOXG-10778 | 2.321 | 0.001 | MFS transporter, SP family, general alpha glucoside: H+ symporter | FOXG-05237 | 1.373 | 0.011 | glutathione S-transferase |
| FOXG-13625 | 2.682 | 0 | MFS transporter, SP family, general alpha glucoside: H+ symporter | FOXG-13656 | 1.351 | 0.005 | glutathione S-transferase |
| FOXG-15360 | 1.291 | 0.013 | MFS transporter, SP family, sugar: H+ symporter | FOXG-07591 | 1.735 | 0.002 | glutathione S-transferase |
| FOXG-17535 | 1.073 | 0.016 | MFS transporter, SP family, sugar: H+ symporter | FOXG-01733 | 1.138 | 0.028 | pyruvate carboxylase |
| FOXG-12267 | 1.252 | 0.008 | MFS transporter, SP family, sugar: H+ symporter | FOXG-07887 | 1.521 | 0.282 | acyl-CoA dehydrogenase |
| FOXG-13707 | 1.599 | 0.003 | sodium/potassium-transporting ATPase subunit alpha | FOXG-00735 | 2.385 | 0.051 | acetyl-coenzyme A synthetase |
| FOXG-05132 | 3.859 | 0.002 | oxidoreductase | FOXG-11385 | 1.51 | 0.138 | acetyl-CoA acyltransferase |
| FOXG-15750 | 3.637 | 0.011 | oxidoreductase | FOXG-06200 | 1.312 | 0.221 | acetyl-CoA acyltransferase |
| FOXG-00572 | 1.672 | 0.001 | oxidoreductase | FOXG-08599 | 1.097 | 0.126 | acetyl-CoA C-acetyltransferase |
| FOXG-09098 | 1.526 | 0.009 | oxidoreductase | FOXG-05118 | 1.595 | 0.151 | acyl-CoA dehydrogenase |
| FOXG-07588 | 1.585 | 0.01 | oxidoreductase | FOXG-12010 | 2.312 | 0.035 | hexokinase |
| FOXG-05703 | 1.055 | 0.026 | oxidoreductase | FOXG-03031 | 1.238 | 0.02 | hexokinase |
| FOXG-05808 | 6.563 | 0.001 | catalase | FOXG-11536 | 1.577 | 0 | acetoacetate-CoA ligase |
| FOXG-02357 | 1.779 | 0.02 | catalase | FOXG-02255 | 5.758 | 0.02 | endonuclease G, mitochondrial |
| FOXG-16836 | 1.565 | 0.02 | catalase | FOXG-00035 | 1.521 | 0 | enolase |
| FOXG-02505 | 1.036 | 0.059 | catalase-1 | FOXG-06150 | 1.281 | 0.003 | 6-phosphofructokinase |
| FOXG-15530 | 5.757 | 0.001 | peroxidase | FOXG-02233 | 1.156 | 0.006 | triosephosphate isomerase |
| FOXG-17180 | 2.558 | 0 | peroxidase | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, M.; Deng, Q.; Qiu, M.; Deng, Y.; Sun, L.; Fang, Z.; Liao, J.; Zhao, J.; Gooneratne, R. Iturin A Strongly Inhibits the Growth and T-2 Toxin Synthesis of Fusarium oxysporum: A Morphological, Cellular, and Transcriptomics Study. Foods 2023, 12, 1278. https://doi.org/10.3390/foods12061278
Hua M, Deng Q, Qiu M, Deng Y, Sun L, Fang Z, Liao J, Zhao J, Gooneratne R. Iturin A Strongly Inhibits the Growth and T-2 Toxin Synthesis of Fusarium oxysporum: A Morphological, Cellular, and Transcriptomics Study. Foods. 2023; 12(6):1278. https://doi.org/10.3390/foods12061278
Chicago/Turabian StyleHua, Meifang, Qi Deng, Mei Qiu, Yijia Deng, Lijun Sun, Zhijia Fang, Jianmeng Liao, Jian Zhao, and Ravi Gooneratne. 2023. "Iturin A Strongly Inhibits the Growth and T-2 Toxin Synthesis of Fusarium oxysporum: A Morphological, Cellular, and Transcriptomics Study" Foods 12, no. 6: 1278. https://doi.org/10.3390/foods12061278
APA StyleHua, M., Deng, Q., Qiu, M., Deng, Y., Sun, L., Fang, Z., Liao, J., Zhao, J., & Gooneratne, R. (2023). Iturin A Strongly Inhibits the Growth and T-2 Toxin Synthesis of Fusarium oxysporum: A Morphological, Cellular, and Transcriptomics Study. Foods, 12(6), 1278. https://doi.org/10.3390/foods12061278