Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment
Abstract
1. Introduction
2. Methodology
2.1. Study Area
2.1.1. Sample Collection
2.1.2. Enumeration of Presumptive Listeria in RTE Food Samples
2.2. Detection of L. monocytogenes
2.2.1. DNA Extraction
2.2.2. Molecular Characterization of L. monocytogenes Isolates
2.3. Antimicrobial Susceptibility Testing (AST)
2.3.1. Computation of Resistance Quotient (RQs) of L. monocytogenes Isolates
2.3.2. Antimicrobial Resistance Phenotyping, Multiple Antimicrobial Resistance Indexing of Isolates and Risk Evaluation
2.4. Data Analysis
3. Results
3.1. Occurrence of L. monocytogenes in RTE Foods
3.2. Antibiotic Susceptibility and Cluster Analysis of L. monocytogenes Isolates
3.2.1. Prevalence of Antimicrobial-Resistant L. monocytogenes and Computation of Resistance Quotient (RQs) of Isolates
3.2.2. Multiple Antimicrobial Resistance Phenotypes and Index (MARPs and MARI) of L. monocytogenes
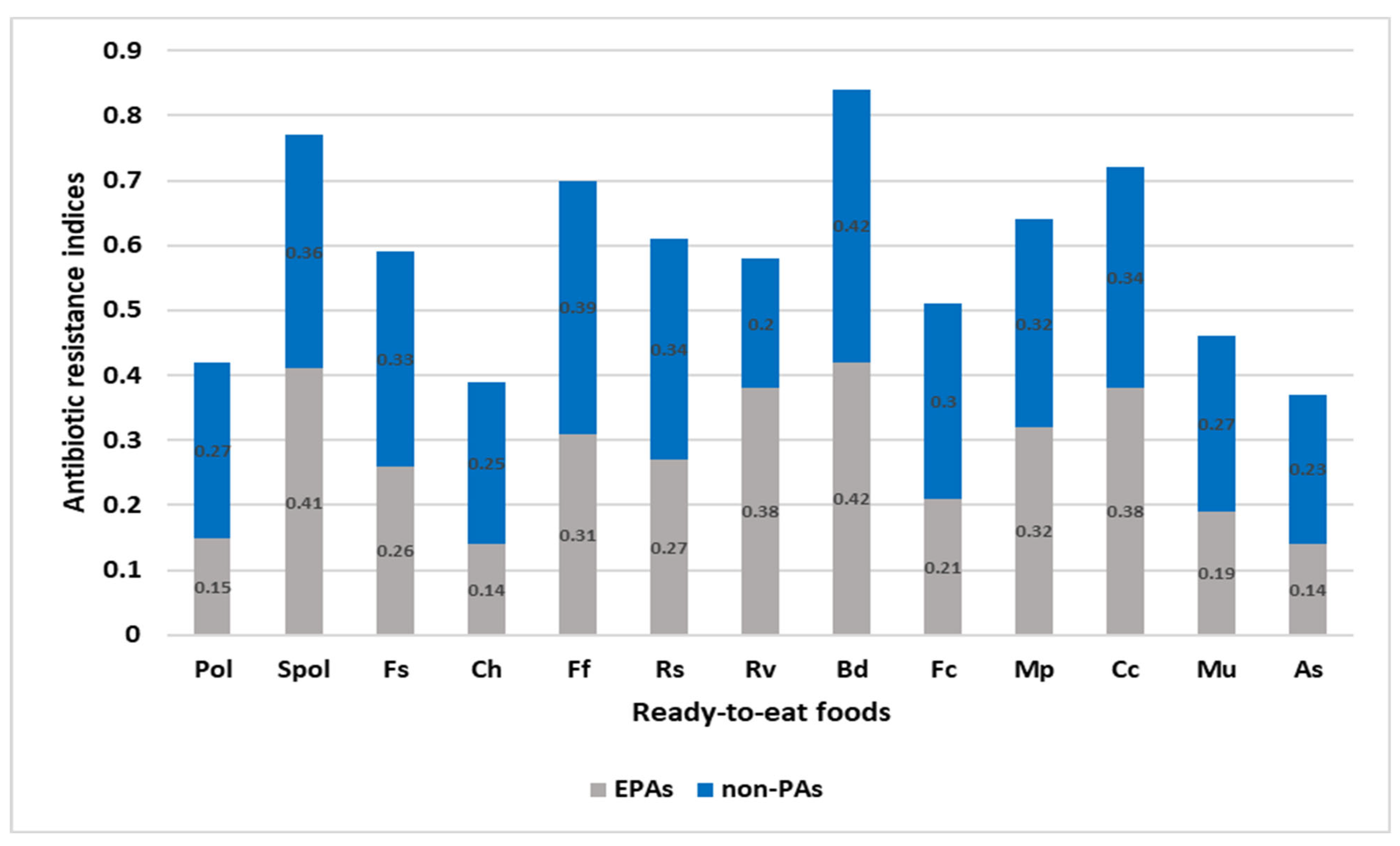
3.3. Evaluation of the RTEF and the EMPT Entrenched on the MAR and ARI of L. monocytogenes Isolates
EMPT from the Comparison of MARI of L. monocytogenes
3.4. RTEF from the Comparison of MARI of L. monocytogenes Isolates
RTEF from the Comparison of ARI across the Ready-to-Eat Foods
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef] [PubMed]
- Hashempour-Baltork, F.; Hosseini, H.; Shojaee-Aliabadi, S.; Torbati, M.; Alizadeh, A.M.; Alizadeh, M. Drug resistance and the prevention strategies in food borne bacteria: An update review. Adv. Pharm. Bull. 2019, 9, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Ilievska, N.; Pavlova, V.; Ilievska, J.; Kirovska, V.; Pavlovska, M. Review paper on the effects of antibiotic use in agricultural animals on the human health and formation of food born antibiotic resistant microorganisms. J. Hyg. Eng. Des. 2019, 27, 22–26. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Frieden, T. Antibiotic Resistance Threats in the United States, 2013|Antibiotic/Antimicrobial Resistance Report; U.S. Department of Health and Human Services, Centers for Disease Control and Prevention: Washington, DC, USA, 2013. [Google Scholar]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- World Bank. By 2050, Drug-Resistant Infections Could Cause Global Economic Damage on par with 2008 Financial Crisis. Press Release 2016. Available online: https://www.worldbank.org/en/news/press-release/2016/09/18/by-2050-drug-resistant-infections-could-cause-global-economic-damage-on-par-with-2008-financial-crisis (accessed on 7 February 2023).
- Bloom, G.; Merrett, G.B.; Wilkinson, A.; Lin, V.; Paulin, S. Antimicrobial resistance and universal health coverage. BMJ Glob. Health 2017, 2, e000518. [Google Scholar] [CrossRef]
- Davey, P.; Marwick, C.A.; Scott, C.L.; Charani, E.; Mcneil, K.; Brown, E.; Gould, I.M.; Ramsay, C.R.; Michie, S. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst. Rev. 2017. [CrossRef]
- Davey, P.; Brown, E.; Charani, E.; Fenelon, L.; Gould, I.M.; Holmes, A.; Ramsay, C.R.; Wiffen, P.J.; Wilcox, M. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst. Rev. 2013. [CrossRef]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Coetzee, J.; Corcoran, C.; Prentice, E.; Moodley, M.; Mendelson, M.; Poirel, L.; Nordmann, P.; Brink, A.J. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. South African Med. J. 2016, 106, 449–450. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Antibiotic resistance profile of Listeria monocytogenes recovered from ready-to-eat foods surveyed in South Africa. J. Food Prot. 2022, 85, 1807. [Google Scholar] [CrossRef]
- Jansen, W.; Müller, A.; Grabowski, N.T.; Kehrenberg, C.; Muylkens, B.; Al Dahouk, S. Foodborne diseases do not respect borders: Zoonotic pathogens and antimicrobial resistant bacteria in food products of animal origin illegally imported into the European Union. Vet. J. 2019, 244, 75–82. [Google Scholar] [CrossRef]
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods 2022, 11, 2966. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of the molecular epidemiology and genetic multiplicity of Listeria monocytogenes recovered from ready-to-eat foods following the South African listeriosis outbreak. Sci. Rep. 2022, 12, 20129. [Google Scholar] [CrossRef]
- Kayode, A.J.; Semerjian, L.; Osaili, T.; Olapade, O.; Okoh, A.I. Occurrence of Multidrug-Resistant Listeria monocytogenes in Environmental Waters: A Menace of Environmental and Public Health Concern. Front. Environ. Sci. 2021, 9, 737435. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Incidence and genetic diversity of multi-drug resistant Listeria monocytogenes isolates recovered from fruits and vegetables in the Eastern Cape Province, South Africa. Int. J. Food Microbiol. 2022, 363, 109515. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 28 October 2021).
- Ekundayo, T.C.; Okoh, A.I. Antimicrobial resistance in freshwater Plesiomonas shigelloides isolates: Implications for environmental pollution and risk assessment. Environ. Pollut. 2020, 257, 113493. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Nørrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Szymczak, B.; Szymczak, M.; Trafiałek, J. Prevalence of Listeria species and L. monocytogenes in ready-to-eat foods in the West Pomeranian region of Poland: Correlations between the contamination level, serogroups, ingredients, and producers. Food Microbiol. 2020, 91, 103532. [Google Scholar] [CrossRef]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 384. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Nandi, A.; Mitra, P.; Saha, K.; Patel, P.; Jha, E.; Panda, P.K.; Singh, S.K.; Dutt, A.; Mishra, Y.K.; et al. Theragnostic application of nanoparticle and CRISPR against food-borne multi-drug resistant pathogens. Mater. Today Bio 2022, 15, 100291. [Google Scholar] [CrossRef]
- Kunjachan, S.; Rychlik, B.; Storm, G.; Kiessling, F.; Lammers, T. Multidrug resistance: Physiological principles and nanomedical solutions. Adv. Drug Deliv. Rev. 2013, 65, 1852–1865. [Google Scholar] [CrossRef]
- Mpundu, P.; Mbewe, A.R.; Muma, J.B.; Mwasinga, W.; Mukumbuta, N.; Munyeme, M. A global perspective of antibiotic-resistant Listeria monocytogenes prevalence in assorted ready to eat foods: A systematic review. Vet. World 2021, 14, 2219. [Google Scholar] [CrossRef]
- Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, antibiogram and genetic characterization of Lsteria monocytogenes from food products in Egypt. Foods 2021, 10, 1381. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from a Pork Processing Plant and Its Respective Meat Markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef]
- Conter, M.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar] [CrossRef]
- Khatibi, S.A.; Hamidi, S.; Siahi-Shadbad, M.R. Current trends in sample preparation by solid-phase extraction techniques for the determination of antibiotic residues in foodstuffs: A review. Crit. Rev. Food Sci. Nutr. 2021, 6, 3361–3382. [Google Scholar] [CrossRef]
- González-Gutiérrez, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Microbial load and antibiotic resistance in raw beef preparations from northwest Spain. Food Sci. Nutr. 2020, 8, 777–785. [Google Scholar] [CrossRef]
- Rajaei, M.; Moosavy, M.H.; Gharajalar, S.N.; Khatibi, S.A. Antibiotic resistance in the pathogenic foodborne bacteria isolated from raw kebab and hamburger: Phenotypic and genotypic study. BMC Microbiol. 2021, 21, 272. [Google Scholar] [CrossRef]
- Du, X.J.; Zhang, X.; Wang, X.Y.; Su, Y.L.; Li, P.; Wang, S. Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control 2017, 74, 9–16. [Google Scholar] [CrossRef]
- Doménech, E.; Jimenez -Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Control 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Knöchel, S.; Hasman, H. Antimicrobial susceptibility of Listeria monocytogenes from food products. Foodborne Pathog. Dis. 2007, 4, 216–221. [Google Scholar] [CrossRef]
- Pagliano, P.; Arslan, F.; Ascione, T. Epidemiology and treatment of the commonest form of listeriosis: Meningitis and bacteraemia. Infez. Med. 2017, 25, 210–216. [Google Scholar] [PubMed]
- Charlier, C.; Perrodeau, É.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Kayode, A.J.; Igbinosa, E.O.; Okoh, A.I. Overview of listeriosis in the Southern African Hemisphere—Review. J. Food Saf. 2020, 40, e12732. [Google Scholar] [CrossRef]
- Opperman, C.J.; Bamford, C. Co-infection with Streptococcus pneumoniae and Listeria monocytogenes in an immunocompromised patient. S. Afr. Med. J. 2018, 108, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Lalloo, U.G.; Coovadia, Y.M.; Adhikari, M.; Poyiadji, O. Listeria monocytogenes meningitis at King Edward VIII Hospital, Durban. A 10-year experience, 1981–1990. S. Afr. Med. J. 1992, 81, 187–189. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; ISBN 1-56238-1-56238-805-3. [Google Scholar]
- De Gans, J.; Van De Beek, D. Dexamethason gunstig bij volwassenen met acute bacteriële meningitis; een gerandomiseerd placebogecontroleerd onderzoek. Ned. Tijdschr. Geneeskd. 2002, 146, 2235–2240. [Google Scholar]
- Temple, M.E.; Nahata, M.C. Treatment of listeriosis. Ann. Pharmacother. 2000, 34, 656–661. [Google Scholar] [CrossRef]
- Krawczyk-Balska, A.; Popowska, M.; Markiewicz, Z. Re-evaluation of the significance of penicillin binding protein 3 in the susceptibility of Listeria monocytogenes to β-lactam antibiotics. BMC Microbiol. 2012, 12, 57. [Google Scholar] [CrossRef]
- Van de Beek, D.; Cabellos, C.; Dzupova, O.; Esposito, S.; Klein, M.; Kloek, A.T.; Leib, S.L.; Mourvillier, B.; Ostergaard, C.; Pagliano, P.; et al. ESCMID guideline: Diagnosis and treatment of acute bacterial meningitis. Clin. Microbiol. Infect. 2016, 22, S37–S62. [Google Scholar] [CrossRef]
- Hof, H. An update on the medical management of listeriosis. Expert Opin. Pharmacother. 2004, 5, 1727–1735. [Google Scholar] [CrossRef]
- Minkowski, P.; Staege, H.; Groscurth, P.; Schaffner, A. Effects of trimethoprim and co-trimoxazole on the morphology of Listeria monocytogenes in culture medium and after phagocytosis. J. Antimicrob. Chemother. 2001, 48, 185–193. [Google Scholar] [CrossRef][Green Version]
- Michelet, C.; Leib, S.L.; Bentue-Ferrer, D.; Täuber, M.G. Comparative efficacies of antibiotics in a rat model of meningoencephalitis due to Listeria monocytogenes. Antimicrob. Agents Chemother. 1999, 43, 1651–1656. [Google Scholar] [CrossRef]
- Van de Velde, S.; Nguyen, H.A.; Van Bambeke, F.; Tulkens, P.M.; Grellet, J.; Dubois, V.; Quentin, C.; Saux, M.-C. Contrasting effects of human THP-1 cell differentiation on levofloxacin and moxifloxacin intracellular accumulation and activity against Staphylococcus aureus and Listeria monocytogenes. J. Antimicrob. Chemother. 2008, 62, 518–521. [Google Scholar] [CrossRef]
- Pagliano, P.; Brouwer, M.C. Comment: “Implementation of a Meningitis Care Bundle in the Emergency Room Reduces Mortality Associated With Acute Bacterial Meningitis”. Ann. Pharmacother. 2016, 50, 152. [Google Scholar] [CrossRef]
- Viale, P.; Scudeller, L.; Pea, F.; Tedeschi, S.; Lewis, R.; Bartoletti, M.; Sbrojavacca, R.; Cristini, F.; Tumietto, F.; Di Lauria, N.; et al. Implementation of a Meningitis Care Bundle in the Emergency Room Reduces Mortality Associated with Acute Bacterial Meningitis. Ann. Pharmacother. 2015, 49, 978–985. [Google Scholar] [CrossRef]
- Callapina, M.; Kretschmar, M.; Dietz, A.; Mosbach, C.; Hof, H.; Nichterlein, T. Systemic and intracerebral infections of mice with Listeria monocytogenes successfully treated with linezolid. J. Chemother. 2001, 13, 265–269. [Google Scholar] [CrossRef]
- Leiti, O.; Gross, J.W.; Tuazon, C.U. Treatment of brain abscess caused by Listeria monocytogenes in a patient with allergy to penicillin and trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 2005, 40, 907–908. [Google Scholar] [CrossRef]
- Carryn, S.; Van Bambeke, F.; Mingeot-Leclercq, M.-P.; Tulkens, P.M. Activity of β-lactams (ampicillin, meropenem), gentamicin, azithromycin and moxifloxacin against intracellular Listeria monocytogenes in a 24 h THP-1 human macrophage model. J. Antimicrob. Chemother. 2003, 51, 1051–1052. [Google Scholar] [CrossRef]
- Stepanović, S.; Lazarević, G.; Ješić, M.; Koš, R. Meropenem therapy failure in Listeria monocytogenes infection. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 484–486. [Google Scholar] [CrossRef]
- Thønnings, S.; Knudsen, J.D.; Schønheyder, H.C.; Søgaard, M.; Arpi, M.; Gradel, K.O.; Østergaard, C.; Jensen, U.S.; Koch, K.; Smit, J.; et al. Antibiotic treatment and mortality in patients with Listeria monocytogenes meningitis or bacteraemia. Clin. Microbiol. Infect. 2016, 22, 725–730. [Google Scholar] [CrossRef]
- Chenal-Francisque, V.; Charlier, C.; Mehvish, S.; Dieye, H.; Leclercq, A.; Courvalin, P.; Lecuit, M. Highly rifampin-resistant Listeria monocytogenes isolated from a patient with prosthetic bone infection. Antimicrob. Agents Chemother. 2014, 58, 1829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arsene, O.; Linassier, C.; Quentin, R.; Legras, A.; Colombat, P. Development of listeriosis during vancomycin therapy in a neutropenic patient. Scand. J. Infect. Dis. 1996, 28, 415–416. [Google Scholar] [CrossRef] [PubMed]
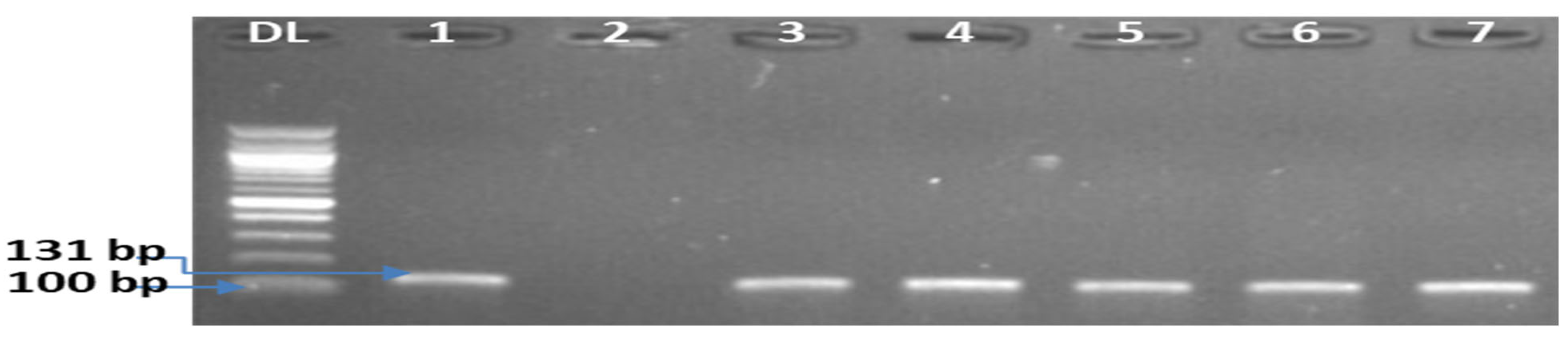
| Type of Samples | Samples Tested | Presumptive Counts of Listeria in RTE Food Samples (cfu/g) | RTE Foods Positive for L. monocytogenes (%) | L. monocytogenes in RTE Foods (%) | |||
|---|---|---|---|---|---|---|---|
| >100 | 10–100 | <10 | 0 | ||||
| Fruit salad | 20 | 0 | 3 | 7 | 10 | 10/20 (50) | 30 (15.46) |
| Fried fish (snoek) | 21 | 2 | 3 | 3 | 13 | 8 (38.10) | 21 (10.82) |
| Sliced polony | 21 | 3 | 5 | 5 | 8 | 13 (61.90) | 23 (11.85) |
| Polony | 19 | 0 | 1 | 9 | 9 | 10 (52.63) | 20 (10.30) |
| Russian sausage | 14 | 2 | 3 | 6 | 3 | 11 (78.57) | 14 (7.21) |
| Bread | 21 | 0 | 3 | 3 | 15 | 6 (28.57) | 11 (5.67) |
| Chips | 21 | 0 | 1 | 9 | 11 | 10 (47.62) | 16 (8.24 |
| Vetkoek | 21 | 0 | 0 | 0 | 21 | 0 | 0 |
| Cupcakes | 21 | 1 | 1 | 7 | 12 | 9 (42.86) | 10 (5.15) |
| Vienna sausages | 8 | 0 | 1 | 3 | 4 | 4 (50) | 4 (2.06) |
| Meat pie | 21 | 0 | 2 | 9 | 10 | 11 (52.38) | 22 (11.34) |
| Fried chicken | 5 | 0 | 1 | 1 | 3 | 2 (40) | 2 (1.03) |
| Assorted sandwiches | 14 | 0 | 1 | 5 | 8 | 6 (42.86) | 9 (4.63) |
| Muffins | 12 | 1 | 0 | 6 | 5 | 7 (58.33) | 12 (6.18) |
| Total (%) | 239 | 9 (3.77) | 25 (10.46) | 73 (30.54) | 132 (55.23) | 107 (44.77) | 194 (100) |
| RTE Food | N | P | AMP | SAM | AML | CN | AK | S | DOR | ETP | IPM | CRO | CTT | VA | E | CLA | CIP | W | RL | TS | OT | C | FOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pol | 20 | 30.00 | 0.00 | 0.00 | 30.00 | 5.00 | 10.00 | 30.00 | 15.00 | 5.00 | 35.00 | 45.00 | 65.00 | 30.00 | 30.00 | 5.00 | 5.00 | 50.00 | 50.00 | 5.00 | 60.00 | 5.00 | 15.00 |
| SPol | 23 | 60.87 | 39.13 | 30.44 | 47.82 | 17.39 | 8.70 | 52.17 | 4.35 | 60.87 | 21.74 | 47.83 | 60.87 | 60.87 | 47.83 | 26.09 | 13.04 | 65.22 | 69.57 | 34.78 | 65.22 | 26.09 | 8.70 |
| FS | 30 | 30.00 | 0.00 | 0.00 | 50.00 | 13.33 | 23.33 | 33.33 | 30.00 | 36.67 | 6.67 | 70.00 | 3.33 | 36.67 | 36.67 | 6.67 | 3.33 | 46.67 | 73.33 | 0.00 | 66.67 | 23.33 | 3.33 |
| Ch | 16 | 12.50 | 6.25 | 6.25 | 18.75 | 6.25 | 6.25 | 18.75 | 6.25 | 31.25 | 18.75 | 31.25 | 93.75 | 12.50 | 18.75 | 12.50 | 0.00 | 25.00 | 18.75 | 6.25 | 62.50 | 12.50 | 6.25 |
| FF | 21 | 57.14 | 0.00 | 0.00 | 33.33 | 9.52 | 38.1 | 47.62 | 23.81 | 33.33 | 19.05 | 76.19 | 95.23 | 47.62 | 47.62 | 19.05 | 0.00 | 66.67 | 52.38 | 19.05 | 61.91 | 9.52 | 0.00 |
| RS | 14 | 42.86 | 7.14 | 7.14 | 71.43 | 7.14 | 0.00 | 50.00 | 7.14 | 21.43 | 0.00 | 35.71 | 50.00 | 14.29 | 35.71 | 7.14 | 14.29 | 50.00 | 78.57 | 28.57 | 92.86 | 14.29 | 21.43 |
| RV | 4 | 75.00 | 25.00 | 25.00 | 100 | 0.00 | 0.00 | 25.00 | 0.00 | 75.00 | 0.00 | 0.00 | 0.00 | 100 | 0.00 | 0.00 | 0.00 | 75.00 | 75.00 | 25.00 | 75.00 | 0.00 | 0.00 |
| Bd | 11 | 54.55 | 0.00 | 0.00 | 54.55 | 27.27 | 36.36 | 72.72 | 36.36 | 27.27 | 0.00 | 72.72 | 72.27 | 81.82 | 90.91 | 27.27 | 0.00 | 100 | 90.91 | 18.18 | 45.46 | 9.09 | 0.00 |
| FC | 2 | 0.00 | 0.00 | 0.00 | 50.00 | 0.00 | 50.00 | 0.00 | 0.00 | 50.00 | 0.00 | 100 | 100 | 0.00 | 50.00 | 0.00 | 50.00 | 50.00 | 50.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ps | 22 | 40.91 | 4.55 | 0.00 | 50.00 | 4.55 | 36.36 | 45.45 | 9.09 | 50.00 | 18.18 | 63.64 | 81.82 | 40.91 | 50.00 | 22.73 | 9.09 | 59.09 | 59.09 | 27.27 | 72.73 | 13.64 | 0.00 |
| Cc | 10 | 70.00 | 0.00 | 0.00 | 40.00 | 0.00 | 10.00 | 60.00 | 40.00 | 50.00 | 10.00 | 50.00 | 70.00 | 50.00 | 50.00 | 10.00 | 10.00 | 90.00 | 80.00 | 10.00 | 70.00 | 10.00 | 10.00 |
| Mu | 12 | 33.33 | 0.00 | 0.00 | 25.00 | 0.00 | 8.33 | 25.00 | 16.67 | 8.33 | 0.00 | 25.00 | 83.33 | 25.00 | 16.67 | 16.67 | 0.00 | 41.67 | 66.67 | 8.33 | 66.67 | 0.00 | 25.00 |
| AS | 9 | 22.22 | 0.00 | 0.00 | 11.11 | 11.11 | 0.00 | 22.22 | 0.00 | 66.67 | 0.00 | 44.44 | 0.00 | 0.00 | 33.33 | 0.00 | 0.00 | 33.33 | 44.44 | 0.00 | 44.44 | 0.00 | 11.11 |
| MARPs (Prescribed Antibiotics) | No of Antibiotics | No Observed | MARI | MARPs (Non-Prescribed) | No of Antibiotics | No Observed | MARI | |
|---|---|---|---|---|---|---|---|---|
| 1 | AML/E/CLA | 3 | 1 | 0.25 | AK/S/OT’ | 3 | 1 | 0.30 |
| 2 | AML/ETP/RL | 3 | 1 | 0.25 | AK/OT/C | 3 | 1 | 0.30 |
| 3 | AMP/ETP/TS | 3 | 1 | 0.25 | AK/CRO/CTT | 3 | 2 | 0.30 |
| 4 | AML/IPM/RL | 3 | 3 | 0.25 | AK/CRO/CCT/OT | 4 | 7 | 0.40 |
| 5 | AML/W/RL | 3 | 2 | 0.25 | AK/S/CRO/CCT | 4 | 2 | 0.40 |
| 6 | AML/IPM/W | 3 | 1 | 0.25 | AK/S/CRO/CTT/VA | 5 | 5 | 0.50 |
| 7 | AML/DOR/RL | 3 | 1 | 0.25 | AK/S/CRO/CCT/OT | 5 | 4 | 0.50 |
| 8 | AML/RL/TS | 3 | 1 | 0.25 | AK/CRO/CCT/VA/OT | 5 | 3 | 0.50 |
| 9 | AML/DOR/IPM | 3 | 1 | 0.25 | AK/S/CRO/CCT/OT | 5 | 1 | 0.50 |
| 10 | AML/E/W/RL | 4 | 2 | 0.33 | AK/CRO/CCT/OT/C | 5 | 1 | 0.50 |
| 11 | AML/ETP/W/RL | 4 | 2 | 0.33 | AK/CRO/CCT/VA/OT/C | 6 | 1 | 0.60 |
| 12 | AML/IPM/E/W/RL | 5 | 2 | 0.41 | AK/S/CRO/CCT/VA/OT | 6 | 2 | 0.60 |
| 13 | AML/ETP/E/W/RL | 5 | 2 | 0.41 | CIP/OT/C | 3 | 1 | 0.30 |
| 14 | AML/E/W/RL/TS | 5 | 1 | 0.41 | CN/S/VA | 3 | 1 | 0.30 |
| 15 | AML/DOR/CLA/W/RL/TS | 6 | 2 | 0.50 | CCT/OT/C | 3 | 2 | 0.30 |
| 16 | AML/DOR/E/CLA/W/RL/TS | 7 | 1 | 0.58 | CN/S/CRO | 3 | 1 | 0.30 |
| 17 | AMP/SAM/AML/IPM/W/RL/TS | 7 | 1 | 0.58 | CRO/CTT/CIP | 3 | 1 | 0.30 |
| 18 | AMP/SAM/AML/DOR/CLA/W/RL/TS | 8 | 1 | 0.67 | CRO/CIP/OT | 3 | 1 | 0.30 |
| 19 | DOR/E/W/RL | 4 | 1 | 0.33 | CRO/CCT/C | 3 | 1 | 0.30 |
| 20 | DOR/IPM/ETP | 3 | 1 | 0.25 | CCT/VA/OT | 3 | 3 | 0.30 |
| 21 | DOR/ETP/E/CLA/W/RL/TS | 7 | 1 | 0.58 | CRO/CCT/VA | 3 | 1 | 0.30 |
| 22 | E/W/RL | 3 | 4 | 0.25 | CRO/CCT/OT | 3 | 21 | 0.30 |
| 23 | E/CLA/W | 3 | 1 | 0.25 | CCT/VA/FOS | 3 | 2 | 0.30 |
| 24 | E/W/RL/TS | 4 | 1 | 0.33 | CRO/VA/OT/C | 4 | 5 | 0.40 |
| 25 | E/CLA/W/RL/TS | 5 | 1 | 0.42 | CRO/CCT/VA/OT | 4 | 3 | 0.40 |
| 26 | IPM/E/W | 3 | 2 | 0.25 | CN/S/CCT/OT | 4 | 1 | 0.40 |
| 27 | IPM/ETP/E | 3 | 1 | 0.25 | CN/S/CRO/VA | 4 | 1 | 0.40 |
| 28 | IPM/E/W/RL | 4 | 1 | 0.33 | CRO/CCT/OT/C | 4 | 2 | 0.40 |
| 29 | IPM/ETP/E/RL | 4 | 1 | 0.33 | CN/CCT/VA/OT | 4 | 1 | 0.40 |
| 30 | IPM/W/RL/IPM | 4 | 1 | 0.33 | CCT/VA/CIP/OT | 4 | 1 | 0.40 |
| 31 | IPM/ETP/W/RL | 4 | 1 | 0.33 | CRO/CCT/CIP/OT | 4 | 1 | 0.40 |
| 32 | P/W/RL | 3 | 1 | 0.25 | CN/VA/OT/FOS | 4 | 1 | 0.40 |
| 33 | P/E/W/RL | 4 | 3 | 0.33 | CN/CCT/OT/FOS | 4 | 1 | 0.40 |
| 34 | P/AML/W/RL | 4 | 12 | 0.33 | CN/S/CCT/VA/OT | 5 | 1 | 0.50 |
| 35 | P/AML/E/RL | 4 | 1 | 0.33 | CN/S/CCT/OT/FOS | 5 | 1 | 0.50 |
| 36 | P/IPM/W/RL | 4 | 1 | 0.33 | CRO/CCT/OT/FOS | 4 | 1 | 0.40 |
| 37 | P/AML/IPM/W | 4 | 3 | 0.33 | CRO/VA/CIP/OT/C | 5 | 2 | 0.50 |
| 38 | P/AML/E/W/RL | 5 | 4 | 0.42 | CN/CRO/CTT/VA/OT | 5 | 1 | 0.50 |
| 39 | P/DOR/E/W/RL | 5 | 1 | 0.42 | CN/S/CRO/CTT/VA | 5 | 2 | 0.50 |
| 40 | P/IPM/E/W/RL | 5 | 1 | 0.42 | CN/AKS/VA/OT/FOS | 5 | 1 | 0.50 |
| 41 | P/AML/IMP/W/RL | 5 | 3 | 0.42 | CN/AK/S/CRO/CCT/OT | 6 | 1 | 0.60 |
| 42 | P/AML/DOR/W/RL | 5 | 1 | 0.42 | CN/AK/S/CRO/CCT/VA | 6 | 1 | 0.60 |
| 43 | P/AML/DOR/E/W/RL | 6 | 2 | 0.50 | CN/AK/S/CRO/CCT/OT/C | 7 | 1 | 0.70 |
| 44 | P/AML/E/CLA/W/RL | 6 | 2 | 0.50 | CN/CRO/CCT/VA/CIP/OT/C | 7 | 1 | 0.70 |
| 45 | P/AML/IPM/E/W/RL | 6 | 4 | 0.50 | S/VA/OT | 3 | 4 | 0.30 |
| 46 | P/IPM/ETP/E/W/RL | 6 | 1 | 0.50 | S/OT/FOS | 3 | 2 | 0.30 |
| 47 | P/DOR/IPM/E/W/RL | 6 | 1 | 0.50 | S/CCT/VA | 3 | 3 | 0.30 |
| 48 | P/AML/CLA/W/RL/TS | 6 | 2 | 0.50 | S/CCT/OT | 3 | 2 | 0.30 |
| 49 | P/AML/DOR/IPM/W/RL | 6 | 1 | 0.50 | S/CCT/FOS | 3 | 1 | 0.30 |
| 50 | P/AML/IPM/ETP/E/W | 6 | 1 | 0.50 | S/CRO/CCT | 3 | 1 | 0.30 |
| 51 | P/AML/E/CLA/W/RL/TS | 7 | 1 | 0.58 | S/VA/OT/C | 4 | 1 | 0.40 |
| 52 | P/AML/DOR/IPM/E/W/RL | 7 | 2 | 0.58 | S/CRO/VA/OT | 4 | 2 | 0.40 |
| 53 | P/AML/IPM/ETP/E/W/RL | 7 | 1 | 0.58 | S/CRO/CCT/OT | 4 | 2 | 0.40 |
| 54 | P/AML/DOR/CLA/W/RL/TS | 7 | 5 | 0.58 | S/CRO/CCT/VA | 4 | 3 | 0.40 |
| 55 | P/AMP/SAM/DOR/E/RL/TS | 7 | 1 | 0.58 | S/CRO/VA/OT/C | 5 | 1 | 0.50 |
| 56 | P/AML/DOR/E/CLA/W/RL/TS | 8 | 1 | 0.67 | S/CCT/VA/OT/C | 5 | 1 | 0.50 |
| 57 | P/AML/DOR/IPM/E/CLA/W/RL | 8 | 1 | 0.67 | S/VA/CIP/OT/C | 5 | 1 | 0.50 |
| 58 | P/AMP/SAM/IPM/E/CLA/W/RL/TS | 9 | 2 | 0.75 | S/CRO/CCT/OT/C | 5 | 1 | 0.50 |
| 59 | P/AMP/SAM/AML/E/CLA/W/RL/TS | 9 | 1 | 0.75 | S/CRO/CCT/VA/OT | 5 | 3 | 0.50 |
| 60 | P/AML/DOR/IPM/ETP/E/CLA/W/RL | 9 | 1 | 0.75 | S/CRO/CCT/VA/OT/C | 6 | 1 | 0.60 |
| 61 | P/AMP/SAM/AML/DOR/CLA/W/RL/TS | 9 | 1 | 0.75 | S/CCT/VA/CIP/OT/C | 6 | 1 | 0.60 |
| 62 | P/AMP/SAM/AML/IPM/E/CLA/W/RL/TS | 10 | 1 | 0.83 | S-CRO-VA-OT-FOS | 5 | 1 | 0.50 |
| 63 | P/AMP/SAM/AML/IPM/E/CLA/W/RL/TS | 10 | 1 | 0.83 | VA/OT/C | 3 | 1 | 0.30 |
| 64 | P/AMP/SAM/AML/IPM/ETP/E/CLA/W/RL/TS | 11 | 1 | 0.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kayode, A.J.; Okoh, A.I. Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment. Foods 2023, 12, 1346. https://doi.org/10.3390/foods12061346
Kayode AJ, Okoh AI. Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment. Foods. 2023; 12(6):1346. https://doi.org/10.3390/foods12061346
Chicago/Turabian StyleKayode, Adeoye John, and Anthony Ifeanyi Okoh. 2023. "Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment" Foods 12, no. 6: 1346. https://doi.org/10.3390/foods12061346
APA StyleKayode, A. J., & Okoh, A. I. (2023). Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment. Foods, 12(6), 1346. https://doi.org/10.3390/foods12061346






