Revealing the Mechanism of Aroma Production Driven by High Salt Stress in Trichomonascus ciferrii WLW
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Source
2.2. Salt Tolerance and Aroma Production Capacity in Salt Conditions Test
2.3. Chemical Analysis
2.4. Library Construction and Sequencing
2.5. Transcriptome Data Analysis
2.6. Data Analysis
3. Results and Discussion
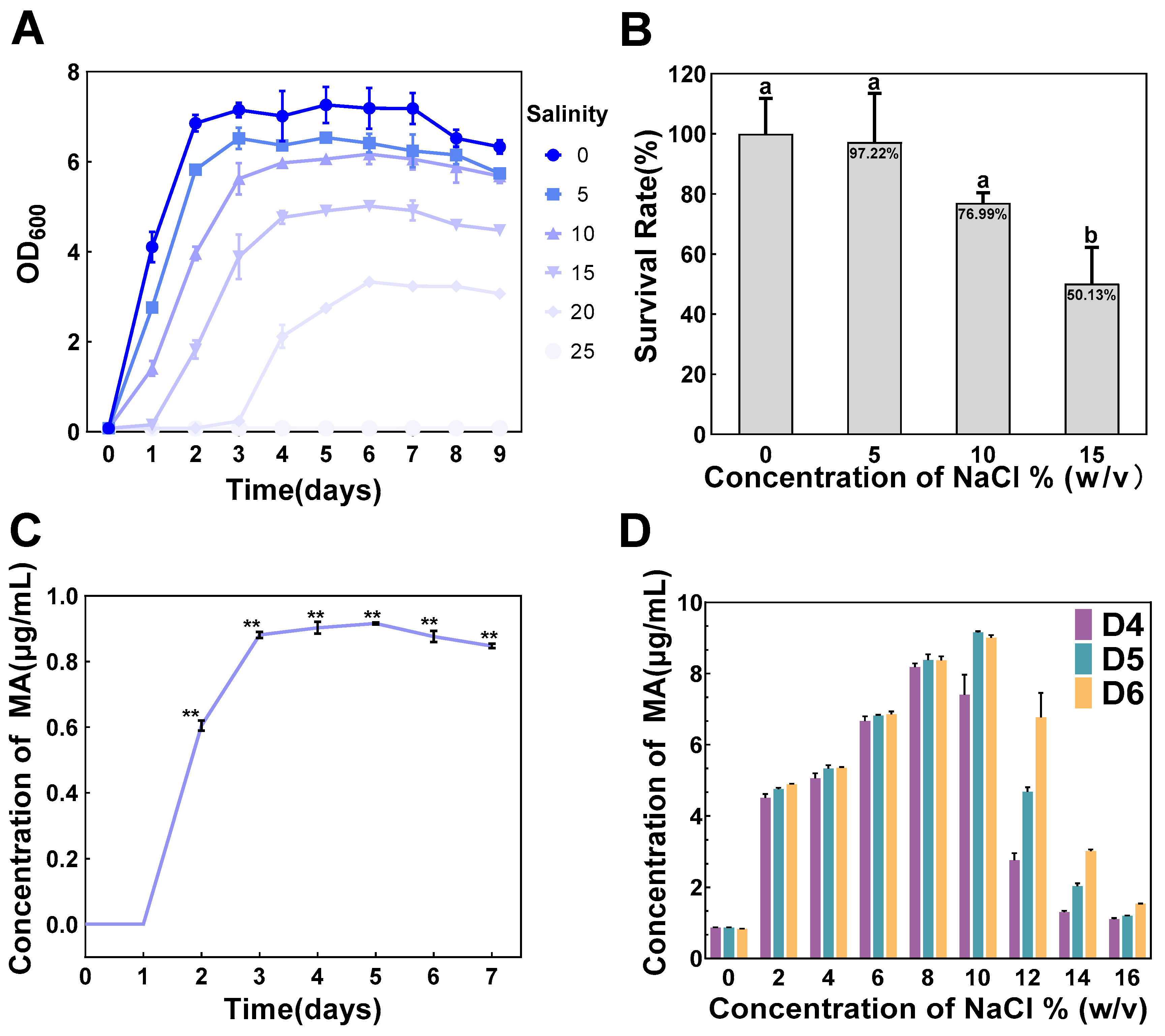
3.1. Characterization of Salt Tolerance and MA Production in T. ciferrii WLW
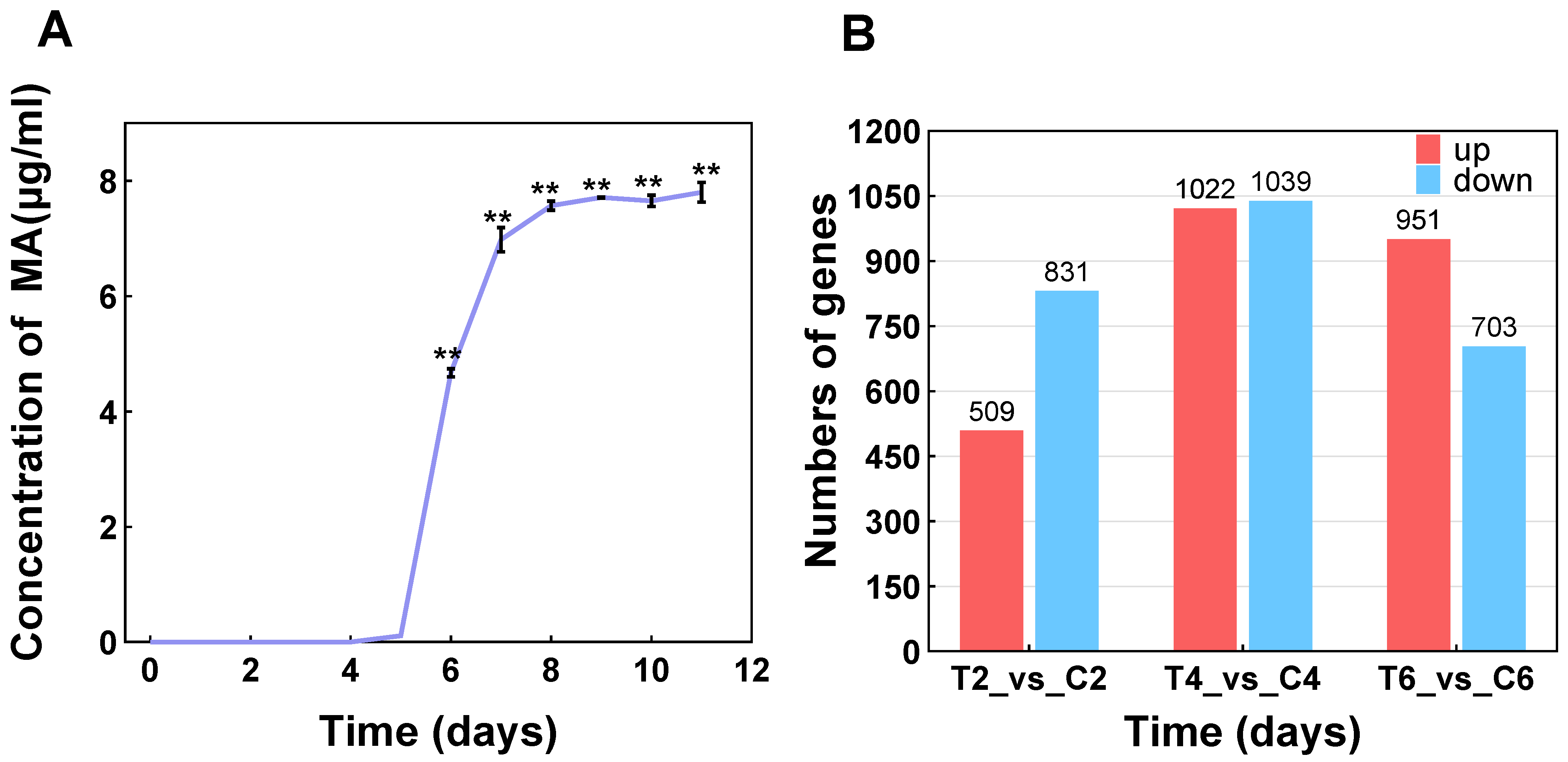
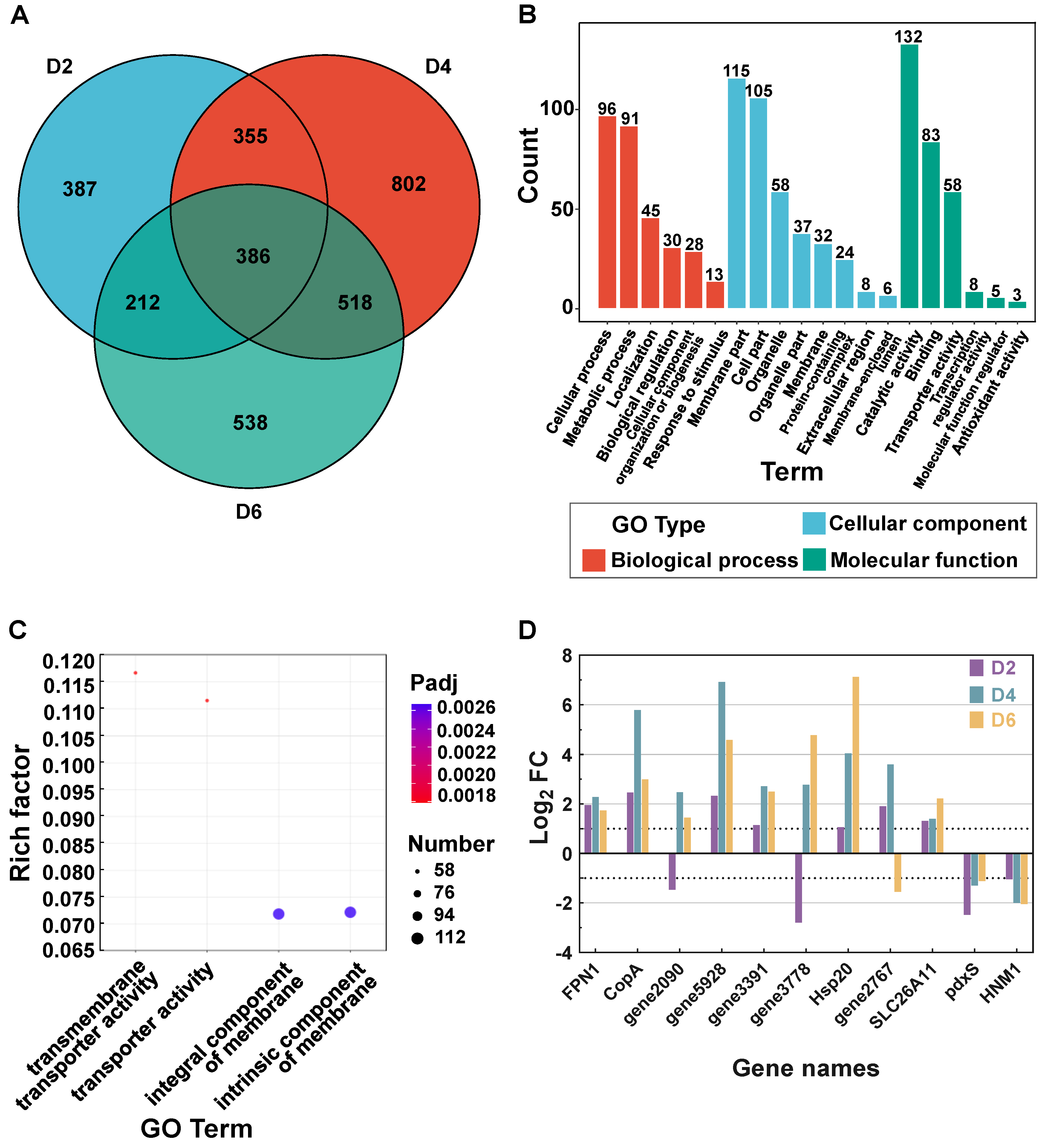
3.2. Transcriptome Assembly and Difference Analysis
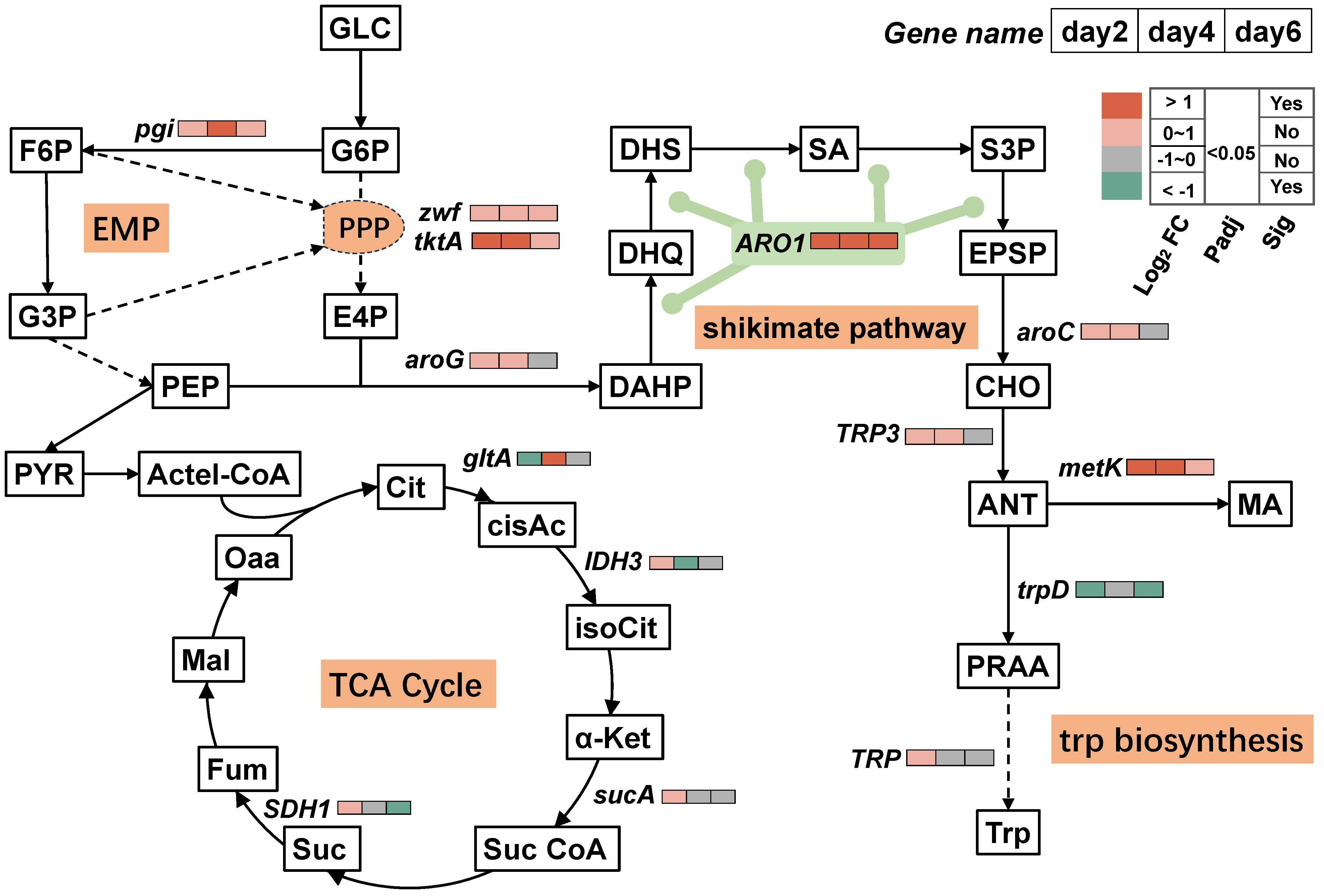
3.3. Mechanisms of T. ciferrii WLW Response to Salt Stress
3.4. Timing Analysis of the MA-Synthesis-Pathway-Associated DEGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, B.; Li, H.R.; Hu, Z.H.; Zhang, Y.H.; Sun, M.; Qiu, S.K.; Zeng, B. Difference in microbial community and taste compounds between Mucor-type and Aspergillus-type Douchi during koji-making. Food Res. Int. 2019, 121, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.Y.; Guo, Y.; Hu, X.Y.; Liu, W.H.; Liu, B.Q.; Yang, J.; Tu, Z.C.; Huang, Y.H. Flavor improvement of fermented soybean foods by co-fermentation with Bacillus velezensis and Lactiplantibacillus plantarum. LWT-Food Sci. Technol. 2023, 186, 9. [Google Scholar] [CrossRef]
- Wang, J.; De Luca, V. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 2005, 44, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Cho, J.S.; Lee, S.Y. Microbial production of methyl anthranilate, a grape flavor compound. PNAS 2019, 116, 10749–10756. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.R.; Liang, J.; Liu, X.H.; Gao, K.X.; Zhang, Y.; Li, A.; Chen, C.; Hou, L.A.; Yang, Y. Graphene oxide/methyl anthranilate modified anti-biofouling membrane possesses dual functions of anti-adhesion and quorum quenching. J. Membr. Sci. 2023, 668, 10. [Google Scholar] [CrossRef]
- Sulzbach, M.; da Silva, M.A.S.; Gonzatto, M.P.; Marques, M.M.O.; Böettcher, G.N.; Silvestre, W.P.; Silvad, J.; Pauletti, G.F.; Schwarz, S.F. Effect of distillation methods on the leaf essential oil of some Citrus cultivars. J. Essent. Oil Res. 2021, 33, 452–463. [Google Scholar] [CrossRef]
- Wang, B.; Diao, Y.W.; Yuan, J.Q.; Zhang, F.Y.; Zhou, H.Y.; Du, L. Optimization of Methyl Anthranilate Synthesis Process by Response Surface Methodology and Its Reaction Mechanism. Synthesis 2022, 54, 5261–5272. [Google Scholar] [CrossRef]
- Wang, B.; Wang, C.; Du, L.; Zhao, Q.; Yin, J. Optimization of Process and Improvement in Continuous Production of Synthesis of Methyl Anthranilate. Chem. Eng. Trans. 2020, 81, 139–144. [Google Scholar]
- Choi, K.R.; Lee, S.Y. Systems metabolic engineering of microorganisms for food and cosmetics production. Nat. Rev. Bioeng. 2023, 1, 832–857. [Google Scholar] [CrossRef]
- Johnson, A.; Deshmukh, P.; Kaushik, S.; Sharma, V. Microbial Bio-production of Proteins and Valuable Metabolites. In Microbial Interventions in Agriculture and Environment: Volume 1: Research Trends, Priorities and Prospects; Singh, D.P., Gupta, V.K., Prabha, R., Eds.; Springer: Singapore, 2019; pp. 381–418. [Google Scholar]
- Kuivanen, J.; Kannisto, M.; Mojzita, D.; Rischer, H.; Toivari, M.; Jäntti, J. Engineering of Saccharomyces cerevisiae for anthranilate and methyl anthranilate production. Microb. Cell Factories 2021, 20, 34. [Google Scholar] [CrossRef]
- Gregory, P.; Bonita, S.; Mohamad, F. A method for the preparation of “natural” methyl anthranilate. WO8900203A, 1 December 1989. [Google Scholar]
- Chandrasekaran, S.D.; Vaithilingam, M.; Shanker, R.; Kumar, S.; Thiyur, S.; Babu, V.; Selvakumar, J.N.; Prakash, S. Exploring the In Vitro Thrombolytic Activity of Nattokinase From a New Strain Pseudomonas aeruginosa CMSS. Jundishapur J. Microbiol. 2015, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Nozal, M.J.; Bernal, J.L.; Toribio, L.; Jiménez, J.J.; Martín, M.T. High-performance liquid chromatographic determination of methyl anthranilate, hydroxymethylfurfural and related compounds in honey. J. Chromatogr. A 2001, 917, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357-U121. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Kim, H.; Choi, H.S. Fungi in salterns. J. Microbiol. 2019, 57, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Czachura, P.; Owczarek-Koscielniak, M.; Piatek, M. Salinomyces polonicus: A moderately halophilic kin of the most extremely halotolerant fungus Hortaea werneckii. Fungal Biol. 2021, 125, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Taupp, M.; Harmsen, D.; Heckel, F.; Schreier, P. Production of Natural Methyl Anthranilate by Microbial N-Demethylation of N-Methyl Methyl Anthranilate by the Topsoil-Isolated Bacterium Bacillus megaterium. J. Agric. Food Chem. 2005, 53, 9586–9589. [Google Scholar] [CrossRef]
- Zhu, M.; Zheng, J.; Xie, J.; Zhao, D.; Qiao, Z.W.; Huang, D.; Luo, H.B. Effects of environmental factors on the microbial community changes during medium-high temperature Daqu manufacturing. Food Res. Int. 2022, 153, 11. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Huang, H.; Tang, P.; Liang, J.; Liao, Q.; Chen, J.; Pang, L.; Yang, K.; Wei, H.; Chen, M.; et al. Associations between maternal exposure to multiple metals and metalloids and blood pressure in preschool children: A mixture-based approach. J. Trace Elem. Med. Biol. Organ Soc. Miner. Trace Elem. (GMS) 2024, 84, 127460. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Zhang, L. Decorrelated empirical likelihood for generalized linear models with high-dimensional longitudinal data. Stat. Probab. Lett. 2024, 211, 110135. [Google Scholar] [CrossRef]
- Lee, M.A.; Choi, Y.J.; Kim, Y.S.; Chon, S.Y.; Chung, Y.B.; Park, S.H.; Yun, Y.R.; Min, S.G.; Yang, H.C.; Seo, H.Y. Effects of salt type on the metabolites and microbial community in kimchi fermentation. Heliyon 2022, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Mi, T.; Wang, D.K.; Yao, S.J.; Yang, H.; Che, Y.L.; Wu, C.D. Effects of salt concentration on the quality and microbial diversity of spontaneously fermented radish paocai. Food Res. Int. 2022, 160, 14. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Li, S.; Wang, J.; Zhao, L.; Yan, Y.; Cao, Q.; Wu, T.T.; Liu, L.Y.; Wang, C.T. Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth. Mar. Drugs 2018, 16, 511. [Google Scholar] [CrossRef]
- Yao, Z.Y.; Gong, J.S.; Liu, Y.R.; Jiang, J.Y.; Zhang, Y.S.; Su, C.; Li, H.; Kang, C.L.; Liu, L.; Xu, Z.H.; et al. Genetic variation reveals the enhanced microbial hyaluronan biosynthesis via atmospheric and room temperature plasma. Carbohydr. Polym. 2023, 312, 13. [Google Scholar] [CrossRef]
- Zeng, Z.; Liu, Y.; Feng, X.Y.; Li, S.X.; Jiang, X.M.; Chen, J.Q.; Shao, Z.Q. The RNAome landscape of tomato during arbuscular mycorrhizal symbiosis reveals an evolving RNA layer symbiotic regulatory network. Plant Commun. 2023, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Nie, C.Y.; Li, Z.; Kang, J.; Wang, X.L.; Cui, Y.N. Physiological and Transcriptional Analyses Provide Insight into Maintaining Ion Homeostasis of Sweet Sorghum under Salt Stress. Int. J. Mol. Sci. 2023, 24, 11045. [Google Scholar] [CrossRef]
- Banik, S.; Dutta, D. Membrane Proteins in Plant Salinity Stress Perception, Sensing, and Response. J. Membr. Biol. 2023, 256, 109–124. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V.J.H. The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance. Horticulturae 2023, 9, 230. [Google Scholar] [CrossRef]
- Li, J.L.; Pang, Q.Y.; Yan, X.F. Unique Features of the m6A Methylome and Its Response to Salt Stress in the Roots of Sugar Beet (Beta vulgaris). Int. J. Mol. Sci. 2023, 24, 11659. [Google Scholar] [CrossRef]
- Liu, J.G.; Han, X.; Yang, T.; Cui, W.H.; Wu, A.M.; Fu, C.X.; Wang, B.C.; Liu, L.J. Genome-wide transcriptional adaptation to salt stress in Populus. BMC Plant Biol. 2019, 19, 367. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.A.; Ahmar, S.; Ali, B.; Saleem, M.H.; Khan, M.U.; Zhou, W.; Liu, S. The Role of Membrane Transporters in Plant Growth and Development, and Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 12792. [Google Scholar] [CrossRef] [PubMed]
- Danaeifar, A.; Khaleghi, E.; Zivdar, S.; Mehdikhanlou, K. Physiological, biochemical, and gene expression of Sour Orange (Citrus aurantium L.) to Iron(II)-Arginine Chelate under salinity, alkalinity, and salt-alkali combined stresses. Sci. Hortic. 2023, 319, 112146. [Google Scholar] [CrossRef]
- Heo, L.; Cho, Y.M.; Choi, J.; Lee, J.; Han, Y.; Han, S.W. Proteomic and Phenotypic Analyses of a Putative YggS Family Pyridoxal Phosphate-Dependent Enzyme in Acidovorax citrulli. Plant Pathol. J. 2023, 39, 235–244. [Google Scholar] [PubMed]
- Chen, H.; Xiong, L.M. Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 2005, 44, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Y.; Li, Q.X.; Zhang, Y.S.; Zhang, M.C.; Li, Z.H. Glycine betaine increases salt tolerance in maize (Zea mays L.) by regulating Na+ homeostasis. Front. Plant Sci. 2022, 13, 978304. [Google Scholar] [CrossRef] [PubMed]
- Daraz, U.; Ahmad, I.; Li, Q.S.; Zhu, B.; Saeed, M.F.; Li, Y.; Ma, J.; Wang, X.-B. Plant growth promoting rhizobacteria induced metal and salt stress tolerance in Brassica juncea through ion homeostasis. Ecotox. Environ. Safe. 2023, 267, 115657. [Google Scholar] [CrossRef]
- Mount, D.B.; Romero, M.F. The SLC26 gene family of multifunctional anion exchangers. Pflügers Arch. Eur. J. Physiol. 2004, 447, 710–721. [Google Scholar] [CrossRef]
- Rahmati, N.; Kunzelmann, K.; Xu, J.; Barone, S.; Sirianant, L.; De Zeeuw, C.I.; Soleimani, M. Slc26a11 is prominently expressed in the brain and functions as a chloride channel: Expression in Purkinje cells and stimulation of V H+-ATPase. Pflügers Arch. Eur. J. Physiol. 2013, 465, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Chattopadhyay, D. Adaptation of plants to salt stress: The role of the ion transporters. J. Plant Biochem. Biotechnol. 2021, 30, 668–683. [Google Scholar] [CrossRef]
- Diédhiou, C.; Golldack, D. Salt-dependent regulation of chloride channel transcripts in rice. Plant Sci. 2006, 170, 793–800. [Google Scholar] [CrossRef]
- Nakamura, A.; Fukuda, A.; Sakai, S.; Tanaka, Y. Molecular Cloning, Functional Expression and Subcellular Localization of Two Putative Vacuolar Voltage-gated Chloride Channels in Rice (Oryza sativa L.). Plant Cell Physiol. 2006, 47, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Zhang, H.D.; Dong, F.Y.; Zou, J.; Gao, C.B.; Zhu, Z.W.; Liu, Y.K. Multiple roles of wheat calmodulin genes during stress treatment and TaCAM2-D as a positive regulator in response to drought and salt tolerance. Int. J. Biol. Macromol. 2022, 220, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Steimle, B.L.; Bailey, D.K.; Smith, F.M.; Rosenblum, S.L.; Kosman, D.J.; Hanson, P. Calcium and the Ca-ATPase SPCA1 modulate plasma membrane abundance of ZIP8 and ZIP14 to regulate Mn(II) uptake in brain microvascular endothelial cells. J. Biol. Chem. 2022, 298, 102211. [Google Scholar] [CrossRef] [PubMed]
- Wojtunik-Kulesza, K.; Oniszczuk, A.; Waksmundzka-Hajnos, M. An attempt to elucidate the role of iron and zinc ions in development of Alzheimer’s and Parkinson’s diseases. Biomed. Pharmacother. 2019, 111, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, A.Q. Copper-instigated modulatory cell mortality mechanisms and progress in oncological treatment investigations. Front. Immunol. 2023, 14, 1236063. [Google Scholar] [CrossRef]
- Yin, Y.L.; Xu, Y.N.; Li, X.N.; Fan, S.G.; Wang, G.Y.; Fu, J.M. Physiological integration between Bermudagrass ramets improves overall salt resistance under heterogeneous salt stress. Physiol. Plant. 2022, 174, e13655. [Google Scholar] [CrossRef]
- Ma, R.; Liu, B.W.; Geng, X.; Ding, X.; Yan, N.; Sun, X.; Wang, W.L.; Sun, X.Z.; Zheng, C.S. Biological Function and Stress Response Mechanism of MYB Transcription Factor Family Genes. J. Plant Growth Regul. 2023, 42, 83–95. [Google Scholar] [CrossRef]
- Tate, J.J.; Marsikova, J.; Vachova, L.; Palkova, Z.; Cooper, T.G. Effects of Abolishing Whi2 on Nitrogen Catabolite Repression-Sensitive GATA-Factor Localization and Protein Production. Faseb J. 2022, 36. [Google Scholar] [CrossRef]
- Crespo, J.L.; Daicho, K.; Ushimaru, T.; Hall, M.N. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 34441–34444. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Escoda, B.; Ivanova, T.; Calvo, I.A. Yox1 links MBF-dependent transcription to completion of DNA synthesis. EMBO Rep. 2011, 12, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.M.; Ghizzoni, J.M.; Gil-Bona, A.; Wang, W.; Costanzo, M.; Li, R.; Flynn, M.J.; Zhu, L.J.; Myers, C.L.; Boone, C.; et al. Repression of essential cell cycle genes increases cellular fitness. PLoS Genet. 2022, 18, e1010349. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Du, Z.D.; Lu, S.R.; Wang, Z.; He, X. Regulation of Cat8 in energy metabolic balance and glucose tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2023, 107, 4605–4619. [Google Scholar] [CrossRef]
- MacLean, A.; Legendre, F.; Appanna, V.D. The tricarboxylic acid (TCA) cycle: A malleable metabolic network to counter cellular stress. Crit. Rev. Biochem. Mol. Biol. 2023, 17, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.; Edwards, R.M.; Coggins, J.R. The pentafunctional arom enzyme of Saccharomyces cerevisiae is a mosaic of monofunctional domains. Biochem. J. 1987, 246, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. PlantBiol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Ingrisano, R.; Tosato, E.; Trost, P.; Gurrieri, L.; Sparla, F. Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae. Plants 2023, 12, 3410. [Google Scholar] [CrossRef]
- Chen, Y.L.; Huang, W.X.; Zhang, F.T. Metabolomic Profiling of Dongxiang Wild Rice Under Salinity Demonstrates the Significant Role of Amino Acids in Rice Salt Stress. Front. Plant Sci. 2021, 12, 729004. [Google Scholar] [CrossRef]
- Xie, E.; Wei, X.J.; Ding, A.Z.; Zheng, L.; Wu, X.N.; Anderson, B. Short-Term Effects of Salt Stress on the Amino Acids of Phragmites australis Root Exudates in Constructed Wetlands. Water 2020, 12, 569. [Google Scholar] [CrossRef]
- Blaszczyk, A.J.; Wang, B.; Silakov, A.; Ho, J.V.; Booker, S.J. Efficient methylation of C2 in L-tryptophan by the cobalamin-dependent radical S-adenosylmethionine methylase TsrM requires an unmodified N1 amine. J. Biol. Chem. 2017, 292, 15456–15467. [Google Scholar] [CrossRef] [PubMed]
- Köllner, T.G.; Lenk, C.; Zhao, N.; Seidl-Adams, I.; Gershenzon, J.; Chen, F.; Degenhardt, J. Herbivore-Induced SABATH Methyltransferases of Maize That Methylate Anthranilic Acid Using S-Adenosyl-L-Methionine. Plant Physiol. 2010, 153, 1795–1807. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xian, F.; Yang, L.; Ye, H.; Xu, J.; Yue, X.; Wang, X. Revealing the Mechanism of Aroma Production Driven by High Salt Stress in Trichomonascus ciferrii WLW. Foods 2024, 13, 1593. https://doi.org/10.3390/foods13111593
Xian F, Yang L, Ye H, Xu J, Yue X, Wang X. Revealing the Mechanism of Aroma Production Driven by High Salt Stress in Trichomonascus ciferrii WLW. Foods. 2024; 13(11):1593. https://doi.org/10.3390/foods13111593
Chicago/Turabian StyleXian, Fangying, Lin Yang, Huaqing Ye, Jinlin Xu, Xiaoping Yue, and Xiaolan Wang. 2024. "Revealing the Mechanism of Aroma Production Driven by High Salt Stress in Trichomonascus ciferrii WLW" Foods 13, no. 11: 1593. https://doi.org/10.3390/foods13111593
APA StyleXian, F., Yang, L., Ye, H., Xu, J., Yue, X., & Wang, X. (2024). Revealing the Mechanism of Aroma Production Driven by High Salt Stress in Trichomonascus ciferrii WLW. Foods, 13(11), 1593. https://doi.org/10.3390/foods13111593






