Development of Predictive Modeling for Removal of Multispecies Biofilms of Salmonella Enteritidis, Escherichia coli, and Campylobacter jejuni from Poultry Slaughterhouse Surfaces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Inoculum Preparation
2.3. Surface Preparation
2.4. Compounds and Time of Contact
2.5. Adhesion Test
2.6. Removal of Formed Biofilms
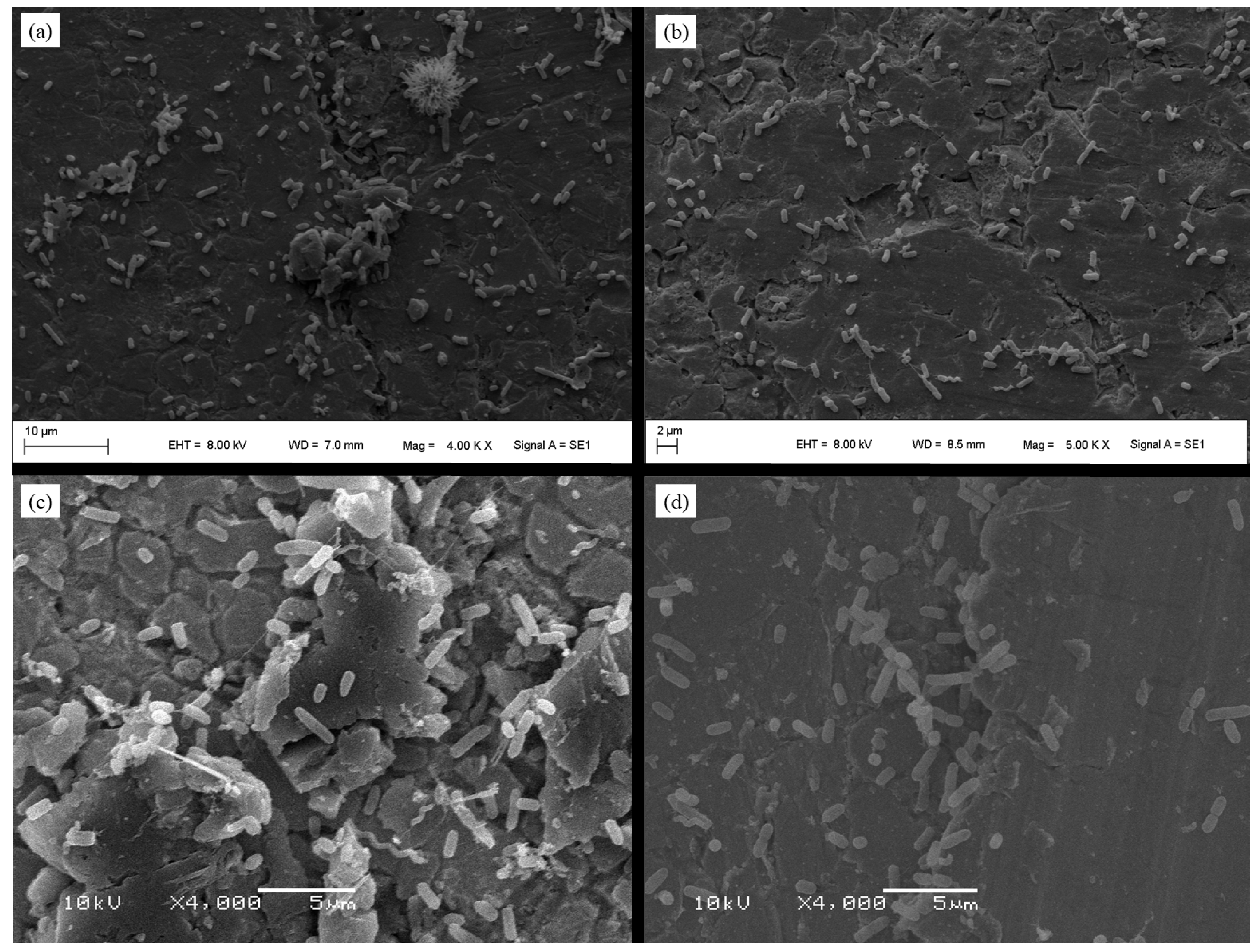
2.7. Scanning Electron Microscopy (SEM)
2.8. Development of Predictive Mathematical Models for Multispecies Biofilms
2.9. Statistical Analyses
3. Results
3.1. Adhesion
3.2. Biofilm Removal
3.3. Predictive Mathematical Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wessels, K.; Rip, D.; Gouws, P. Characteristics, Current Control Methods and the Potential of Bacteriophage Use. Foods 2021, 10, 1742. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 6 April 2024).
- Centers for Diseases Control and Prevention. Campylobacter. Available online: https://www.cdc.gov/campylobacter/about/index.html (accessed on 31 January 2024).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Making Food Safer to Eat Reducing Contamination from the Farm to the Table. Available online: http://www.cdc.gov/vitalsigns (accessed on 24 February 2024).
- Brazilian Ministry of Heath. Surtos de Doenças de Transmissão Hídrica e Alimentar. Informe. 2024. Available online: https://bvsms.saude.gov.br/bvs/publicacoes/manual_integrado_vigilancia_doencas_alimentos.pdf (accessed on 17 April 2024).
- Mitchell, N.M.; Johnson, J.R.; Johnston, B.; Curtiss, R.; Mellata, M. Zoonotic Potential of Escherichia coli Isolates from Retail Chicken Meat Products and Eggs. Appl. Environ. Microbiol. 2015, 81, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella Biofilms: An Overview on Occurrence, Structure, Regulation and Eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Carvalho, D.; Menezes, R.; Chitolina, G.Z.; Kunert-Filho, H.C.; Wilsmann, D.E.; Borges, K.A.; Furian, T.Q.; Salle, C.T.P.; de Souza Moraes, H.L.; do Nascimento, V.P. Antibiofilm Activity of the Biosurfactant and Organic Acids against Foodborne Pathogens at Different Temperatures, Times of Contact, and Concentrations. Braz. J. Microbiol. 2022, 53, 1051–1064. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Borges, K.A.; Furian, T.Q.; Souza, S.N.; Menezes, R.; Tondo, E.C.; Salle, C.T.P.; de Souza Moraes, H.L.; Nascimento, V.P. Biofilm Formation Capacity of Salmonella Serotypes at Different Conditions. Pesqui. Vet. Bras. 2018, 38, 71–76. [Google Scholar] [CrossRef]
- Yaron, S.; Römling, U. Biofilm Formation by Enteric Pathogens and Its Role in Plant Colonization and Persistence. Microb. Biotechnol. 2014, 7, 496–516. [Google Scholar] [CrossRef] [PubMed]
- Teh, A.H.T.; Lee, S.M.; Dykes, G.A. Does Campylobacter jejuni Form Biofilms in Food-Related Environments? Appl. Environ. Microbiol. 2014, 80, 5154–5160. [Google Scholar] [CrossRef]
- Skyberg, J.A.; Siek, K.E.; Doetkott, C.; Nolan, L.K. Biofilm Formation by Avian Escherichia coli in Relation to Media, Source and Phylogeny. J. Appl. Microbiol. 2007, 102, 548–554. [Google Scholar] [CrossRef]
- Tatchou-Nyamsi-König, J.A.; Dague, E.; Mullet, M.; Duval, J.F.L.; Gaboriaud, F.; Block, J.C. Adhesion of Campylobacter jejuni and Mycobacterium avium onto Polyethylene Terephtalate (PET) Used for Bottled Waters. Water Res. 2008, 42, 4751–4760. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-Species Biofilms: Living with Friendly Neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Mondal, D. Effect of Biofilm on Production of Poultry. In Focus on Bacterial Biofilms; Das, T., Ed.; IntechOpen: Rijeka, Croatia, 2022; Volume 1, pp. 1–21. [Google Scholar]
- Grigore-Gurgu, L.; Bucur, F.I.; Borda, D.; Alexa, E.-A.; Neagu, C.; Nicolau, A.I. Biofilms Formed by Pathogens in Food and Food Processing Environments. In Bacterial Biofilms; Dincer, S., Özdenefe, M., Arkut, A., Eds.; IntechOpen: Rijeka, Croatia, 2020; Volume 1, pp. 1–33. ISBN 978-1-78985-900-3. [Google Scholar]
- Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in Multispecies Biofilms: Do They Actually Matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Klančnik, A.; Gobin, I.; Jeršek, B.; Smole Možina, S.; Vučkovi, D.; Tušek Žnidarič, M.; Abram, M. Adhesion of Campylobacter Jejuni Is Increased in Association with Foodborne Bacteria. Microorganisms 2020, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.; Chitolina, G.Z.; Wilsmann, D.E.; Lucca, V.; Dias de Emery, B.; Borges, K.A.; Furian, T.Q.; Salle, C.T.P.; de Souza Moraes, H.L.; do Nascimento, V.P. Adhesion Capacity of Salmonella Enteritidis, Escherichia coli and Campylobacter jejuni on Polystyrene, Stainless Steel, and Polyethylene Surfaces. Food Microbiol. 2023, 114, 104280. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef]
- Jay, J.M. Microbiologia de Alimentos, 1st ed.; Rech, R., Geimba, M.P., Flôres, S.H., Frazzon, J., Carvalho, A.L.O., Frazzon, A.P.G., Oliveira, F.A., Oliveira, F.C., Bianchini, A., Silva, A.C.A., et al., Eds.; Artmed: Porto Alegre, Brazil, 2005; Volume 1, ISBN 853630507X. [Google Scholar]
- Ross, T.; Tienungoon, S. Predictive Modelling of the Growth and Survival of Listeria in Fishery Products. Int. J. Food Microbiol. 2000, 62, 231–245. [Google Scholar] [CrossRef]
- Mcdonald, K.; Sun, D.-W. Predictive Food Microbiology for the Meat Industry: A Review. Int. J. Food Microbiol. 1999, 52, 1–27. [Google Scholar] [CrossRef]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The Estimation of the Bactericidal Power of the Blood. Epidemiol. Infect. 1938, 38, 732–749. [Google Scholar] [CrossRef]
- ISO 18593:2018; Microbiology of Food and Animal Feeding Stuffs—Horizontal Methods for Sampling Techniques from Surfaces Using Contact Plates and Swabs. International Organization for Standardization: Geneva, Switzerland, 2018; pp. 1–11.
- Gibson, H.; Taylor, J.; Hall, K.; Holah, J. Effectiveness of Cleaning Techniques Used in the Food Industry in Terms of the Removal of Bacterial Biofilms. J. Appl. Microbiol. 1999, 87, 41–48. [Google Scholar] [CrossRef]
- Center for Microscopy and Microanalysis. Preparação de Amostras Biológicas Para MEV. Available online: https://www.ufrgs.br/cmm/wp-content/uploads/2019/11/PREPARA%C3%87%C3%83O-DE-AMOSTRAS-BIOL%C3%93GICAS-PARA-MEV.pdf (accessed on 6 February 2022).
- Winans, J.B.; Wucher, B.R.; Nadell, C.D. Multispecies Biofilm Architecture Determines Bacterial Exposure to Phages. PLoS Biol. 2022, 20, e3001913. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Wu, H.; Høiby, N.; Molin, S.; Song, Z.-J. Current Understanding of Multi-Species Biofilms. Int. J. Oral Sci. 2011, 3, 74–81. [Google Scholar] [CrossRef]
- Joshi, R.V.; Gunawan, C.; Mann, R. We Are One: Multispecies Metabolism of a Biofilm Consortium and Their Treatment Strategies. Front. Microbiol. 2021, 12, 635432. [Google Scholar] [CrossRef]
- Araújo, P.M.; Batista, E.; Fernandes, M.H.; Fernandes, M.J.; Gama, L.T.; Fraqueza, M.J. Assessment of Biofilm Formation by Campylobacter spp. Isolates Mimicking Poultry Slaughterhouse Conditions. Poult. Sci. 2022, 101, 101586. [Google Scholar] [CrossRef]
- do Valle Gomes, M.Z.; Nitschke, M. Evaluation of Rhamnolipid and Surfactin to Reduce the Adhesion and Remove Biofilms of Individual and Mixed Cultures of Food Pathogenic Bacteria. Food Control. 2012, 25, 441–447. [Google Scholar] [CrossRef]
- Rienzo, M.A.; Stevenson, P.; Marchant, R.; Banat, I.M. Antibacterial Properties of Biosurfactants against Selected Gram-Positive and-Negative Bacteria. FEMS Microbiol. Lett. 2016, 363, 224. [Google Scholar] [CrossRef]
- Nitschke, M.; Araújo, L.V.; Costa, S.G.V.A.O.; Pires, R.C.; Zeraik, A.E.; Fernandes, A.C.L.B.; Freire, D.M.G.; Contiero, J. Surfactin Reduces the Adhesion of Food-Borne Pathogenic Bacteria to Solid Surfaces. Lett. Appl. Microbiol. 2009, 49, 241–247. [Google Scholar] [CrossRef]
- Akbas, M.Y.; Kokumer, T. The Prevention and Removal of Biofilm Formation of Staphylococcus aureus Strains Isolated from Raw Milk Samples by Citric Acid Treatments. Int. J. Food Sci. Technol. 2015, 50, 1666–1672. [Google Scholar] [CrossRef]
- Piras, F.; Fois, F.; Consolati, S.G.; Mazza, R.; Mazzette, R. Influence of Temperature, Source, and Serotype on Biofilm Formation of Salmonella enterica Isolates from Pig Slaughterhouses. J. Food Prot. 2015, 78, 1875–1878. [Google Scholar] [CrossRef]
- Oliveira, D.C.V.; Fernandes Júnior, A.; Kaneno, R.; Silva, M.G.; Araújo Júnior, J.P.; Silva, N.C.C.; Rall, V.L.M. Ability of Salmonella spp. to Produce Biofilm Is Dependent on Temperature and Surface Material. Foodborne Pathog. Dis. 2014, 11, 478–483. [Google Scholar] [CrossRef]
- Valderrama, W.B.; Ostiguy, N.; Cutter, C.N. Multivariate Analysis Reveals Differences in Biofilm Formation Capacity among Listeria monocytogenes Lineages. Biofouling 2014, 30, 1199–1209. [Google Scholar] [CrossRef]
- Bernardes, P. Modelagem Da Adesão de Bacillus cereus Ao Aço Inoxidável Em Função Do Tempo e Da Temperatura e Influência Da Rugosidade e Da Hidrofobicidade Sobre a Adesão. Master’s Thesis, Universidade Federal de Viçosa, Viçosa, Brazil, 2008. [Google Scholar]
- Chmielewski, R.A.N.; Frank, J.F. A Predictive Model for Heat Inactivation of Listeria monocytogenes Biofilm on Buna-N Rubber. LWT Food Sci. Technol. 2006, 39, 11–19. [Google Scholar] [CrossRef]
- Zhang, T. Modeling Biofilms: From Genes to Communities. Processes 2017, 5, 5. [Google Scholar] [CrossRef]
- Salle, C.T.P.; Guahyba, A.S.; Wald, V.B.; Silva, A.B.; Salle, F.O.; Nascimento, V.P. Use of Artificial Neural Networks To Estimate Production Parameters Of Broiler Breeders in The Breeding Phase. Br. Poult. Sci. 2003, 44, 211–217. [Google Scholar] [CrossRef]
- Savegnago, R.P.; Nunes, B.N.; Caetano, S.L.; Ferraudo, A.S.; Schmidt, G.S.; Ledur, M.C.; Munari, D.P. Comparison Of Logistic And Neural Network Models To Fit To The Egg Production Curve of White Leghorn Hens. Poult. Sci. 2011, 90, 705–711. [Google Scholar] [CrossRef]
- Schneider, A.; Hommel, G.; Blettner, M. Linear Regression Analysis. Dtsch. Arztebl. 2010, 107, 776–782. [Google Scholar] [CrossRef]
| Salmonella Enteritidis + Escherichia coli | Salmonella Enteritidis + Campylobacter jejuni | Escherichia coli + Campylobacter jejuni | Salmonella Enteritidis + Escherichia coli + Campylobacter jejuni | |||||
|---|---|---|---|---|---|---|---|---|
| Stainless Steel | Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | Polyethylene | |
| Salmonella Enteritidis | 6.26 a | 5.43 a | 5.92 a | 5.62 a | - | - | 6.14 a | 5.78 a |
| Escherichia coli | 6.66 b | 6.15 b | - | - | 6.40 a | 6.09 a | 6.35 a | 6.13 b |
| Campylobacter jejuni | - | - | 4.29 b | 4.23 b | 4.32 b | 4.40 b | 4.40 b | 4.57 c |
| Treatment | Temperature (°C) | Salmonella Enteritidis | Escherichia coli | ||
|---|---|---|---|---|---|
| Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | ||
| Control | 4 | 5.30 ± 0.17 aA | 6.12 ± 0.20 bA | 5.65 ± 0.6 aA | 6.69 ± 0.22 bA |
| 12 | 5.26 ± 0.22 aA | 6.19 ± 0.32 bA | 6.25 ± 0.35 aA | 6.71 ± 0.28 bA | |
| 25 | 5.72 ± 0.07 aB | 6.46 ± 0.19 bB | 6.54 ± 0.35 aB | 6.57 ± 0.39 aA | |
| Rhamnolipid | 4 | 5.07 ± 0.22 aA | 5.76 ± 0.18 bA | 5.41 ± 0.67 aA | 6.18 ± 1.52 bA |
| 12 | 5.26 ± 0.31 aA | 6.07 ± 0.17 bB | 6.16 ± 0.30 aB | 6.23 ± 1.67 aA | |
| 25 | 5.33 ± 0.31 aA | 6.26 ± 0.14 bB | 6.33 ± 0.34 aB | 6.36 ± 1.55 aA | |
| Citric Acid | 4 | 0 aA | 0.44 ± 0.93 aA | 0 aA | 0 aA |
| 12 | 0 aA | 0 aA | 0 aA | 0 aA | |
| 25 | 0.29 ± 0.87 aA | 0.66 ± 1.32 aA | 0 aA | 1.32 ± 1.59 bB | |
| Benzalkonium Chloride | 4 | 3.10 ± 1.79 aA | 3.85 ± 0.73 aA | 3.23 ± 2.05 aA | 3.37 ± 1.52 aA |
| 12 | 1.59 ± 1.52 aA | 2.81 ± 1.67 bA | 1.03 ± 1.60 aB | 1.09 ± 1.67 aB | |
| 25 | 0.75 ± 1.49 aB | 4.20 ± 1.75 bA | 0.58 ± 1.15 aB | 0.78 ± 1.55 aB | |
| Treatment | Temperature (°C) | Salmonella Enteritidis | Campylobacter jejuni | ||
|---|---|---|---|---|---|
| Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | ||
| Control | 4 | 4.97 ± 0.23 aA | 6.07 ± 0.39 bA | 4.17 ± 0.32 aA | 4.43 ± 0.14 aA |
| 12 | 5.96 ± 0.64 aB | 5.76 ± 0.25 aA | 4.11 ± 0.28 aA | 4.28 ± 0.24 aA | |
| 25 | 5.93 ± 0.68 aB | 5.93 ± 0.41 aA | 4.41 ± 0.33 aA | 4.15 ± 0.24 aA | |
| Rhamnolipid | 4 | 4.13 ± 1.58 aA | 5.48 ± 0.26 bA | 3.97 ± 0.12 aA | 3.96 ± 0.15 aA |
| 12 | 5.74 ± 0.82 aB | 5.47 ± 0.23 aA | 4.38 ± 0.40 aA | 4.22 ± 0.12 aA | |
| 25 | 6.16 ± 0.63 aB | 5.77 ± 0.39 aA | 4.01 ± 0.26 aA | 3.86 ± 1.48 aA | |
| Citric Acid | 4 | 0 aA | 0 aA | 0 aA | 0 aA |
| 12 | 0 aA | 0 aA | 0 aA | 0 aA | |
| 25 | 0 aA | 0 aA | 0 aA | 0.74 ± 1.30 aA | |
| Benzalkonium Chloride | 4 | 2.79 ± 1.66 aA | 3.78 ± 2.19 aA | 0 aA | 0.34 ± 1.50 aA |
| 12 | 0.97 ± 1.47 aB | 1.86 ± 1.82 aA | 0 aA | 0.45 ± 1.03 aA | |
| 25 | 0.38 ± 1.13 aB | 1.38 ± 1.64 aB | 0.40 ± 1.19 aA | 0 aA | |
| Treatment | Temperature (°C) | Escherichia coli | Campylobacter jejuni | ||
|---|---|---|---|---|---|
| Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | ||
| Control | 4 | 5.73 ± 0.19 aA | 6.39 ± 0.09 bA | 4.34 ± 0.23 aA | 4.23 ± 0.21 aA |
| 12 | 6.31 ± 0.29 aB | 6.39 ± 0.13 aA | 4.31 ± 0.22 aA | 4.08 ± 0.43 aA | |
| 25 | 6.23 ± 0.22 aB | 6.43 ± 0.42 aA | 4.55 ± 0.20 aA | 4.66 ± 0.13 aA | |
| Rhamnolipid | 4 | 5.45 ± 0.38 aA | 6.20 ± 0.16 bA | 3.89 ± 0.35 aA | 3.84 ± 0.33 aA |
| 12 | 6.17 ± 0.37 aB | 6.26 ± 0.07 aA | 4.19 ± 0.39 aA | 3.73 ± 0.55 aA | |
| 25 | 6.12 ± 0.23 aB | 6.35 ± 0.23 aA | 4.40 ± 0.26 aA | 3.62 ± 1.40 aA | |
| Citric Acid | 4 | 0 aA | 0 aA | 0 aA | 0 aA |
| 12 | 0 aA | 0 aA | 0 aA | 0 aA | |
| 25 | 0.44 ± 1.31 aA | 0.41 ± 1.23 aA | 0 aA | 0.73 ± 1.45 aA | |
| Benzalkonium Chloride | 4 | 2.66 ± 1.58 aA | 3.59 ± 1.60 aA | 0.68 ± 1.36 aA | 0 aA |
| 12 | 0.44 ± 1.31 aB | 2.05 ± 2.00 bA | 0.41 ± 1.22 aA | 0 aA | |
| 25 | 0 aB | 1.34 ± 1.62 bB | 0 aA | 0 aA | |
| Treatment | Temperature (°C) | Salmonella Enteritidis | Escherichia coli | Campylobacter jejuni | |||
|---|---|---|---|---|---|---|---|
| Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | Polyethylene | Stainless Steel | ||
| Control | 4 | 5.51 ± 0.27 aA | 6.33 ± 0.38 bA | 5.91 ± 0.17 aA | 6.40 ± 0.39 bA | 4.47 ± 0.17 aA | 4.28 ± 0.64 aA |
| 12 | 5.88 ± 0.32 aB | 5.96 ± 0.13 aA | 6.37 ± 0.20 aB | 6.56 ± 0.19 aA | 4.53 ± 0.24 aB | 4.34 ± 0.35 bA | |
| 25 | 5.94 ± 0.42 aB | 6.13 ± 0.52 aA | 6.12 ± 0.07 aB | 6.10 ± 0.22 aB | 4.70 ± 0.09 aB | 4.58 ± 0.35 aA | |
| Rhamnolipid | 4 | 5.25 ± 0.39 aA | 6.02 ± 0.42 bA | 5.54 ± 0.22 | 6.21 ± 0.38 | 4.19 ± 0.33 aA | 4.17 ± 2.05 aA |
| 12 | 5.45 ± 0.22 aAB | 5.62 ± 0.36 aA | 5.93 ± 0.31 | 6.35 ± 0.24 | 4.44 ± 0.37 aB | 4.15 ± 1.60 aA | |
| 25 | 5.89 ± 0.50 aB | 5.75 ± 0.22 aA | 5.70 ± 0.31 | 5.60 ± 0.42 | 4.48 ± 0.12 aB | 3.99 ± 1.15 bA | |
| Citric Acid | 4 | 0 aA | 0 aA | 0 aA | 0 aA | 0 aA | 0 aA |
| 12 | 0 aA | 0 aA | 0 aA | 0 aA | 0 aA | 0 aA | |
| 25 | 0.32 ± 0.97 aA | 0. 39 ± 0.97 aA | 0.29 ± 0.87 aA | 0 aA | 0 aA | 0 aA | |
| Benzalkonium Chloride | 4 | 2.75 ± 1.58 aA | 3.96 ± 1.67 aA | 2.89 ± 1.80 aA | 3.19 ± 1.32 aA | 0 aA | 0 aA |
| 12 | 0.71 ± 1.44 aB | 3.54 ± 1.40 bA | 0.36 ± 1.52 aB | 1.54 ± 1.47 aA | 0 aA | 0 aA | |
| 25 | 1.11 ± 1.69 aB | 2.19 ± 1.70 aA | 0.29 ± 1.50 aB | 2.07 ± 1.97 bA | 0 aA | 0 aA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, D.; Chitolina, G.Z.; Wilsmann, D.E.; Lucca, V.; Emery, B.D.d.; Borges, K.A.; Furian, T.Q.; Santos, L.R.d.; Moraes, H.L.d.S.; Nascimento, V.P.d. Development of Predictive Modeling for Removal of Multispecies Biofilms of Salmonella Enteritidis, Escherichia coli, and Campylobacter jejuni from Poultry Slaughterhouse Surfaces. Foods 2024, 13, 1703. https://doi.org/10.3390/foods13111703
Carvalho D, Chitolina GZ, Wilsmann DE, Lucca V, Emery BDd, Borges KA, Furian TQ, Santos LRd, Moraes HLdS, Nascimento VPd. Development of Predictive Modeling for Removal of Multispecies Biofilms of Salmonella Enteritidis, Escherichia coli, and Campylobacter jejuni from Poultry Slaughterhouse Surfaces. Foods. 2024; 13(11):1703. https://doi.org/10.3390/foods13111703
Chicago/Turabian StyleCarvalho, Daiane, Gabriela Zottis Chitolina, Daiane Elisa Wilsmann, Vivian Lucca, Brunna Dias de Emery, Karen Apellanis Borges, Thales Quedi Furian, Luciana Ruschel dos Santos, Hamilton Luiz de Souza Moraes, and Vladimir Pinheiro do Nascimento. 2024. "Development of Predictive Modeling for Removal of Multispecies Biofilms of Salmonella Enteritidis, Escherichia coli, and Campylobacter jejuni from Poultry Slaughterhouse Surfaces" Foods 13, no. 11: 1703. https://doi.org/10.3390/foods13111703






