Isolation and Identification of Lichen Photobionts Collected from Different Environments in North of Portugal and Evaluation of Bioactivities of Their Extracts
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Lichen Sampling
2.3. Isolation and Purification of Photobiont
2.4. Photobiont Identification
2.4.1. Microalgae Culture Conditions
2.4.2. Light Microscopy and Morphological Characterization
2.4.3. DNA Extraction, Amplification (PCR) and Sequencing
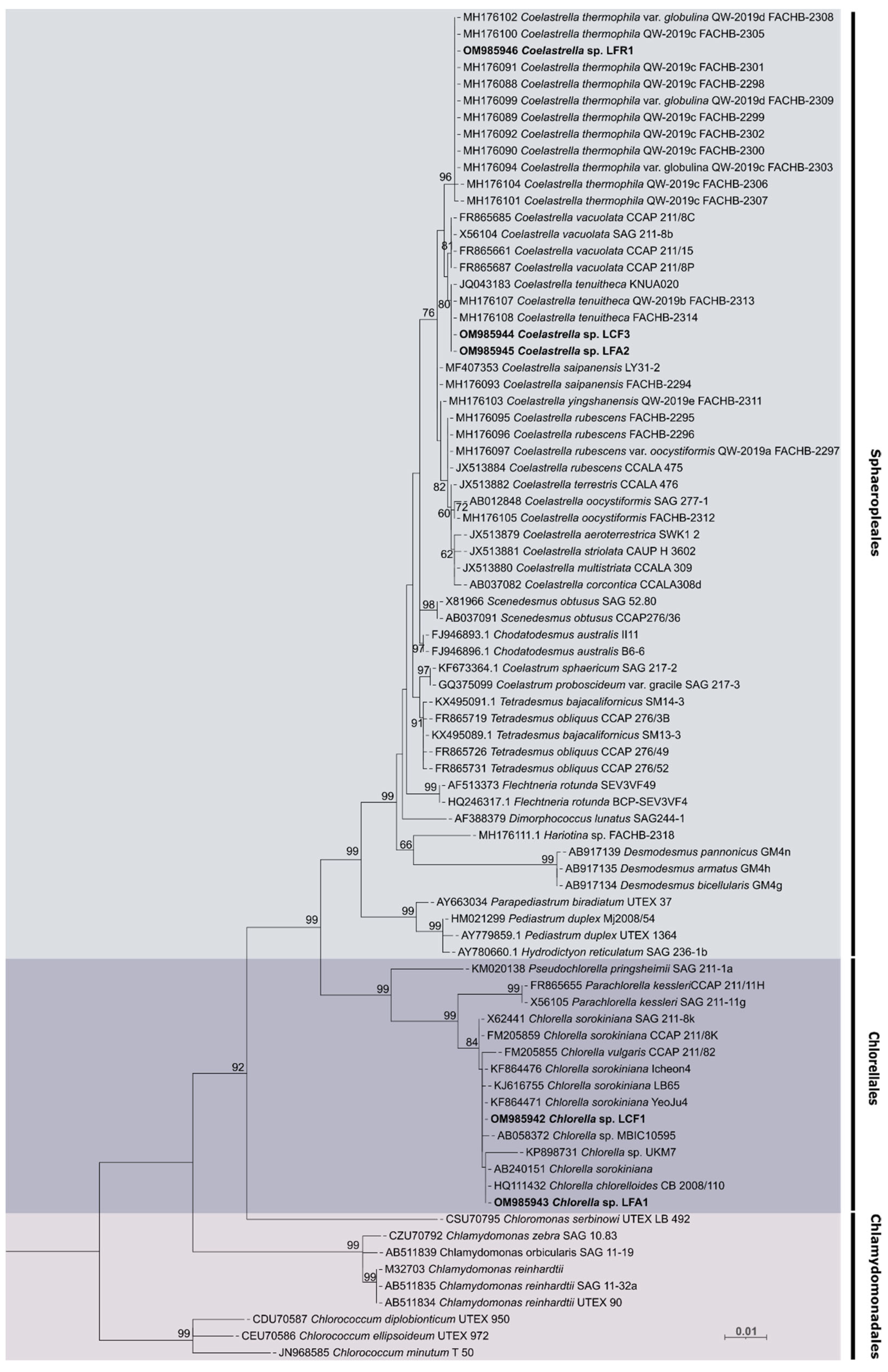
2.4.4. Phylogenetic Analysis
2.5. Ethanolic Extracts Production
2.6. Assessment of Bioactivities
2.6.1. Antioxidant Activity
2,2-Diphenyl-1-Picrylhydrazyl (DPPH) Assay
Ferric Reducing Antioxidant Power (FRAP) Assay
2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic Acid) (ABTS) Assay
Phenolics Content
2.6.2. Antibacterial Activity
2.6.3. Anti-Inflammatory and Anti-Aging Potential: Inhibition of Hyaluronidase Activity
2.6.4. Antidiabetic Activity
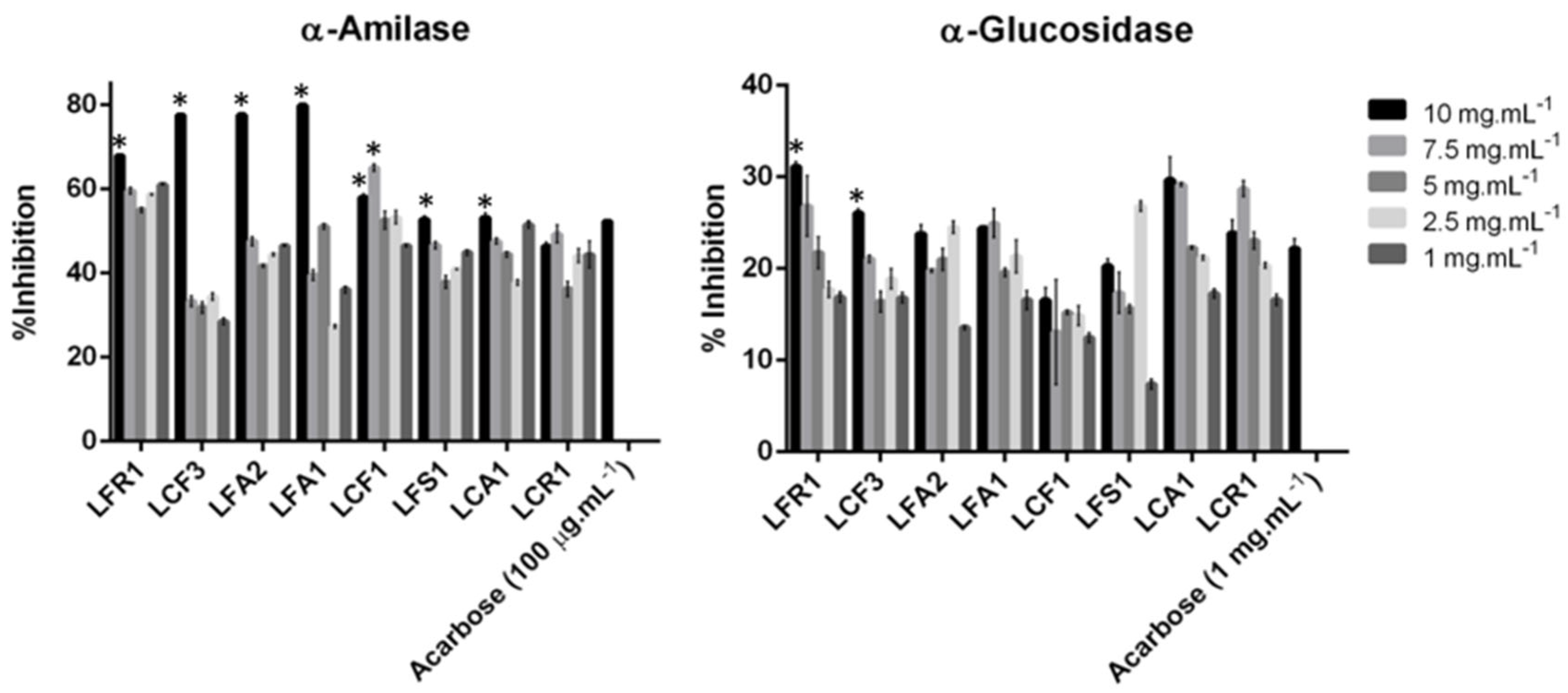
α–Amylase Inhibition Assay
α–Glucosidase Inhibition Assay
2.6.5. Cytotoxicity
3. Results and Discussion
3.1. Isolation and Identification of Photobionts
3.2. Bioactivities
3.2.1. Phenolic Content and Antioxidant Activity
3.2.2. Antimicrobial Activity
3.2.3. Anti-Inflammatory and Anti-Aging Potential
3.2.4. Antidiabetic Activity
3.2.5. Cytotoxicity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Domínguez-Morueco, N.; Moreno, H.; Barreno, E.; Catalá, M. Preliminary assessment of terrestrial microalgae isolated from lichens as testing species for environmental monitoring: Lichen phycobionts present high sensitivity to environmental micropollutants. Ecotoxicol. Environ. Saf. 2014, 99, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, R.; Del Hoyo, A.; Díaz-Rodríguez, C.; Coello, A.J.; Del Campo, E.M.; Barreno, E.; Catalá, M.; Casano, L.M. Lichen Rehydration in Heavy Metal-Polluted Environments: Pb Modulates the Oxidative Response of Both Ramalina farinacea Thalli and Its Isolated Microalgae. Microb. Ecol. 2015, 69, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Sanders, W.B. Lichens: The Interface between Mycology and Plant Morphology. BioScience 2001, 51, 1025–1035. [Google Scholar] [CrossRef]
- Catalá, S.; del Campo, E.M.; Barreno, E.; García-Breijo, F.J.; Reig-Armiñana, J.; Casano, L.M. Coordinated ultrastructural and phylogenomic analyses shed light on the hidden phycobiont diversity of Trebouxia microalgae in Ramalina fraxinea. Mol. Phylogenetics Evol. 2016, 94, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Mendoza, F.; Domaschke, S.; García, M.A.; Jordan, P.; Martín, M.P.; Printzen, C. Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Mol. Ecol. 2011, 20, 1208–1232. [Google Scholar] [CrossRef] [PubMed]
- Goga, M.; Elečko, J.; Marcinčinová, M.; Ručová, D.; Bačkorová, M.; Bačkor, M. Lichen Metabolites: An Overview of Some Secondary Metabolites and Their Biological Potential. In Co-Evolution of Secondary Metabolites; Merillon, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–36. [Google Scholar]
- Sadowsky, A.; Meeßee, J. The Resistance of Lichen Photobionts to Extreme Abiotic Stressors on Earth, in Space and in Simulations. In Algal and Cyanobacteria Symbioses; World Scientific: London, UK, 2015; pp. 575–609. [Google Scholar]
- Calcott, M.J.; Ackerley, D.F.; Knight, A.; Keyzers, R.A.; Owen, J.G. Secondary metabolism in the lichen symbiosis. Chem. Soc. Rev. 2018, 47, 1730–1760. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, B. (Ed.) Lichen Secondary Metabolites—Bioactive Properties and Pharmaceutical Potential; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Oksanen, I. Ecological and biotechnological aspects of lichens. Appl. Microbiol. Biotechnol. 2006, 73, 723–734. [Google Scholar] [CrossRef]
- Zambare, V.P.; Christopher, L.P. Biopharmaceutical potential of lichens. Pharm. Biol. 2012, 50, 778–798. [Google Scholar] [CrossRef]
- Beckett, R.P.; Minibayeva, F.; Solhaug, K.A.; Roach, T. Photoprotection in lichens: Adaptations of photobionts to high light. Lichenol. 2021, 53, 21–33. [Google Scholar] [CrossRef]
- Niedermeyer, T.; Brönstrup, M. 10 Natural Product Drug Discovery from Microalgae; Posten, C., Walter, C., Eds.; De Gruyter: Berlin, Germany, 2012; pp. 169–202. [Google Scholar]
- Varshney, P.; Mikulic, P.; Vonshak, A.; Beardall, J.; Wangikar, P.P. Extremophilic micro-algae and their potential contribution in biotechnology. Bioresour. Technol. 2015, 184, 363–372. [Google Scholar] [CrossRef]
- Francenia Santos Sánchez, N.; Salas-Coronado, R.; Villanueva-Cañongo, C.; Hernández-Carlos, B. Antioxidant Compounds and Their Antioxidant Mechanism. In Antioxidants; Shalaby, E., Ed.; Intechopen: London, UK, 2019; Chapter 2. [Google Scholar]
- Pirian, K.; Moein, S.; Sohrabipour, J.; Rabiei, R.; Blomster, J. Antidiabetic and antioxidant activities of brown and red macroalgae from the Persian Gulf. J. Appl. Phycol. 2017, 29, 3151–3159. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; El-Shenody, R.A.E.K.; Bases, E.A.; El-Shafay, S.M. Comparative assessment of antioxidant activity and biochemical composition of four seaweeds, Rocky Bay of Abu Qir in Alexandria, Egypt. Food Sci. Technol. 2020, 41, 29–40. [Google Scholar] [CrossRef]
- Stojkovic, D.; Smiljkovic, M.; Ciric, A.; Glamoclija, J.; Van Griensven, L.; Ferreira, I.C.F.R.; Sokovic, M. An insight into antidiabetic properties of six medicinal and edible mushrooms: Inhibition of α-amylase and α-glucosidase linked to type-2 diabetes. South Afr. J. Bot. 2019, 120, 100–103. [Google Scholar] [CrossRef]
- EL Shafay, S.; El-Sheekh, M.; Bases, E.; El-Shenody, R. Antioxidant, antidiabetic, anti-inflammatory and anticancer potential of some seaweed extracts. Food Sci. Technol. 2022, 42, e20521. [Google Scholar] [CrossRef]
- Zandi, K.; Tajbakhsh, S.; Nabipour, I.; Rastian, Z.; Yousefi, F.; Sharafian, S.; Sartavi, K. In vitro antitumor activity of Gracilaria corticata (a red alga) against Jurkat and molt-4 human cancer cell lines. Afr. J. Biotechnol. 2010, 9, 6787–6790. [Google Scholar]
- Buhren, B.A.; Schrumpf, H.; Hoff, N.-P.; Bölke, E.; Hilton, S.; Gerber, P.A. Hyaluronidase: From clinical applications to molecular and cellular mechanisms. Eur. J. Med. Res. 2016, 21, 5. [Google Scholar] [CrossRef] [PubMed]
- Boukhris, S.; Athmouni, K.; Hamza-Mnif, I.; Siala-Elleuch, R.; Ayadi, H.; Nasri, M.; Sellami-Kamoun, A. The Potential of a Brown Microalga Cultivated in High Salt Medium for the Production of High-Value Compounds. BioMed Res. Int. 2017, 2017, 4018562. [Google Scholar] [CrossRef] [PubMed]
- Calatayud, A.; Guera, A.; Fos, S.; Barreno, E. A new method to isolate lichen algae by using percoll® gradient centrifugation. Lichenol. 2001, 33, 361–366. [Google Scholar] [CrossRef]
- Gasulla, F.; Guéra, A.; Barreno, E. A simple and rapid method for isolating lichen photobionts. Symbiosis 2010, 51, 175–179. [Google Scholar] [CrossRef]
- Yoshimura, I.; Yamamoto, Y.; Nakano, T.; Finnie, J. Isolation and Culture of Lichen Photobionts and Mycobionts. In Protocols in Lichenology: Culturing, Biochemistry, Ecophysiology and Use in Biomonitoring; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef]
- Kotai, J. Instructions for preparation of modified nutrient solution Z8 for algae. Nor. Inst. Water Res. 1972, 11, 5. [Google Scholar]
- van der Staay, G.W.M.; Guillou, L.; Vaulot, D.; Claustre, H.; Medlin, L.K.; der Staay, S.Y.M.-V. Abundance and diversity of prymnesiophytes in the picoplankton coμmunity from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 2000, 45, 98–109. [Google Scholar] [CrossRef]
- Katana, A.; Kwiatowski, J.; Spalik, K.; Zakryś, B.; Szalacha, E.; Szymańska, H. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J. Phycol. 2001, 37, 443–451. [Google Scholar] [CrossRef]
- Maltsev, Y.; Krivova, Z.; Maltseva, S.; Maltseva, K.; Gorshkova, E.; Kulikovskiy, M. Lipid accumulation by Coelastrella multistriata (Scenedesmaceae, Sphaeropleales) during nitrogen and phosphorus starvation. Sci. Rep. 2021, 11, 19818. [Google Scholar] [CrossRef] [PubMed]
- Goecke, F.; Noda, J.; Paliocha, M.; Gislerød, H.R. Revision of Coelastrella (Scenedesmaceae, Chlorophyta) and first register of this green coccoid microalga for continental Norway. World J. Microbiol. Biotechnol. 2020, 36, 149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Song, H.; Liu, X.; Liu, B.; Hu, Z.; Liu, G. Morphology and molecular phylogeny of coccoid green algae Coelastrella sensu lato (Scenedesmaceae, Sphaeropeales), including the description of three new species and two new varieties. J. Phycol. 2019, 55, 1290–1305. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Yoshida, R.; Ohkoshi, K.; Toyoshima, H. Coelastrella astaxanthina sp. nov. (Sphaeropleales, Chlorophyceae), a novel microalga isolated from an asphalt surface in midsummer in Japan. Phycol. Res. 2019, 68, 107–114. [Google Scholar] [CrossRef]
- Suarez-Montes, D.; Borrell, Y.J.; Gonzalez, J.M.; Rico, J.M. Isolation and identification of microalgal strains with potential as carotenoids producers from a municipal solid waste landfill. Sci. Total Environ. 2022, 802, 149755. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Youn, J.S.; Kim, Y.-J.; Na, H.J.; Jung, H.R.; Song, C.K.; Kang, S.Y.; Kim, J.Y. Antioxidant activity and contents of leaf extracts obtained from Dendropanax morbifera LEV are dependent on the collecting season and extraction conditions. Food Sci. Biotechnol. 2018, 28, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. Available online: http://linkinghub.elsevier.com/retrieve/pii/S0003269796902924 (accessed on 12 January 2024). [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Saleh, B.; Al-Mariri, A. Antimicrobial activity of the marine algal extracts against selected pathogens. J. Agric. Sci. Technol. 2017, 19, 1067–1077. [Google Scholar]
- Sozmen, A.B.; Canbay, E.; Sozmen, E.Y.; Ovez, B. The effect of temperature and light intensity during cultivation of Chlorella miniata on antioxidant, anti-inflammatory potentials and phenolic compound accumulation. Biocatal. Agric. Biotechnol. 2018, 14, 366–374. [Google Scholar] [CrossRef]
- Ferreira-Santos, P.M.; Genisheva, Z.; Botelho, C.; Santos, J.; Ramos, C.; Teixeira, J.A.; Rocha, C.M. Unravelling the Biological Potential of Pinus pinaster Bark Extracts. Antioxidants 2020, 9, 334. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, A.; Nogueira, E.; Azoia, N.G.; Sárria, M.P.; Abreu, A.S.; Shimanovich, U.; Rollett, A.; Härmark, J.; Hebert, H.; Guebitz, G.; et al. Size controlled protein nanoemulsions for active targeting of folate receptor positive cells. Colloids Surf. B Biointerfaces 2015, 135, 90–98. [Google Scholar] [CrossRef]
- Gonçalves, R.F.S.; Martins, J.T.; Abrunhosa, L.; Vicente, A.A.; Pinheiro, A.C. Nanoemulsions for Enhancement of Curcumin Bioavailability and Their Safety Evaluation: Effect of Emulsifier Type. Nanomaterials 2021, 11, 815. [Google Scholar] [CrossRef]
- Zhong, X.; Zhu, Y.; Wang, Y.; Zhao, Q.; Huang, H. Effects of three antibiotics on growth and antioxidant response of Chlorella pyrenoidosa and Anabaena cylindrica. Ecotoxicol. Environ. Saf. 2021, 211, 111954. [Google Scholar] [CrossRef]
- Hajimahmoodi, M.; Faramarzi, M.A.; Mohammadi, N.; Soltani, N.; Oveisi, M.R.; Nafissi-Varcheh, N. Evaluation of antioxidant properties and total phenolic contents of some strains of microalgae. J. Appl. Phycol. 2010, 22, 43–50. [Google Scholar] [CrossRef]
- Monteiro, M.; Santos, R.A.; Iglesias, P.; Couto, A.; Serra, C.R.; Gouvinhas, I.; Barros, A.; Oliva-Teles, A.; Enes, P.; Díaz-Rosales, P. Effect of extraction method and solvent system on the phenolic content and antioxidant activity of selected macro- and microalgae extracts. J. Appl. Phycol. 2020, 32, 349–362. [Google Scholar] [CrossRef]
- Minhas, A.K.; Hodgson, P.; Barrow, C.J.; Adholeya, A. A Review on the Assessment of Stress Conditions for Simultaneous Production of Microalgal Lipids and Carotenoids. Front. Microbiol. 2016, 7, 546. [Google Scholar] [CrossRef] [PubMed]
- Goiris, K.; Muylaert, K.; Fraeye, I.; Foubert, I.; De Brabanter, J.; De Cooman, L. Antioxidant potential of microalgae in relation to their phenolic and carotenoid content. J. Appl. Phycol. 2012, 24, 1477–1486. [Google Scholar] [CrossRef]
- Maadane, A.; Merghoub, N.; Ainane, T.; El Arroussi, H.; Benhima, R.; Amzazi, S.; Bakri, Y.; Wahby, I. Antioxidant activity of some Moroccan marine microalgae: Pufa profiles, carotenoids and phenolic content. J. Biotechnol. 2015, 215, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Wootton-Beard, P.C.; Moran, A.; Ryan, L. Stability of the total antioxidant capacity and total polyphenol content of 23 commercially available vegetable juices before and after in vitro digestion measured by FRAP, DPPH, ABTS and Folin–Ciocalteu methods. Food Res. Int. 2011, 44, 217–224. [Google Scholar] [CrossRef]
- Elfalleh, W.; Hannachi, H.; Tlili, N.; Yahia, Y.; Nasri, N.; Ferchichi, A. Total phenolic contents and antioxidant activities of pomegranate peel, seed, leaf and flower. J. Med. Plants Res. 2011, 6, 4724–4730. [Google Scholar] [CrossRef]
- Machado, A. Antioxidant capacity of phenolic acids extracted from the biomass from spirulina sp. LEB-18. Brazilian J. Dev. 2021, 7, 25956–25970. [Google Scholar] [CrossRef]
- Singh, S.; Kate, B.N.; Banerjee, U.C. Bioactive Compounds from Cyanobacteria and Microalgae: An Overview. Crit. Rev. Biotechnol. 2005, 25, 73–95. [Google Scholar] [CrossRef]
- Yao, S.; Lyu, S.; An, Y.; Lu, J.; Gjermansen, C.; Schramm, A. Microalgae-bacteria symbiosis in microalgal growth and biofuel production: A review. J. Appl. Microbiol. 2019, 126, 359–368. [Google Scholar] [CrossRef]
- Santos, C.A.; Reis, A. Microalgal symbiosis in biotechnology. Appl. Microbiol. Biotechnol. 2014, 98, 5839–5846. [Google Scholar] [CrossRef] [PubMed]
- Tabarzad, M.; Atabaki, V.; Hosseinabadi, T. Anti-inflammatory Activity of Bioactive Compounds from Microalgae and Cyanobacteria by Focusing on the Mechanisms of Action. Mol. Biol. Rep. 2020, 47, 6193–6205. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, N.; Sakaki, S.; Yamaguchi, Y.; Takenaka, H. Inhibitory effects of microalgae on the activation of hyaluronidase. J. Appl. Phycol. 2001, 13, 489–492. [Google Scholar] [CrossRef]
- Kanno, T.; Shinpo, K.; Masada, M.; Tamura, G. Growth-promoting factor for yeast from an extract of Chlorella vulgaris CK-5. J. Ferment. Bioeng. 1996, 81, 577. [Google Scholar] [CrossRef]
- Lauritano, C.; Ianora, A. Marine Organisms with Anti-Diabetes Properties. Mar. Drugs 2016, 14, 220. [Google Scholar] [CrossRef] [PubMed]
- Lebovitz, H.E. Oral Antidiabetic Agents. Drugs 1992, 44, 21–28. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, F. Evaluation of the green alga Chlorella pyrenoidosa for management of diabetes. J. Food Drug Anal. 2012, 20, 28. [Google Scholar] [CrossRef]
- Priatni, S.; Ratnaningrum, D.; Kosasih, W. The Screening of Antidiabetic Activity and The Cultivation Study of Local Marine Microalgae. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1011, 012066. [Google Scholar] [CrossRef]
- Talero, E.; García-Mauriño, S.; Ávila-Román, J.; Rodríguez-Luna, A.; Alcaide, A.; Motilva, V. Bioactive Compounds Isolated from Microalgae in Chronic Inflammation and Cancer. Mar. Drugs 2015, 13, 6152–6209. [Google Scholar] [CrossRef] [PubMed]
- Murugan, K.; Iyer, V.V. Differential growth inhibition of cancer cell lines and antioxidant activity of extracts of red, brown, and green marine algae. Vitr. Cell. Dev. Biol. Anim. 2013, 49, 324–334. Available online: http://www.jstor.org/stable/26588114 (accessed on 6 May 2024). [CrossRef]
- Czerwonka, A.; Kalawaj, K.; Slawinska-Brych, A.; Lemieszek, M.K.; Bartnik, M.; Wojtanowski, K.K.; Zdzisinska, B.; Rzeski, W. Anticancer effect of the water extract of a commercial Spirulina (Arthrospira platensis) product on the human lung cancer A549 cell line. Biomed. Pharmacother. 2018, 106, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Reyna-Martinez, R.; Gomez-Flores, R.; Lopez-Chuken, U.; Quintanilla-Licea, R.; Caballero-Hernandez, D.; Rodriguez-Padilla, C.; Beltran-Rocha, J.C.; Tamez-Guerra, P. Antitumor activity of Chlorella sorokiniana and Scenedesmus sp. microalgae native of Nuevo Leon State, Mexico. PeerJ 2018, 6, e4358. [Google Scholar] [CrossRef] [PubMed]



| No | Primer Name | Sequence (5′-3′) | Reference |
|---|---|---|---|
| 1 | 18SF | ACCTGGTTGATCCTGCCAG | [27] |
| 2 | 18S402F | GCTACCACATCCAAGGAAGGCA | [28] |
| 3 | 18S895F | GTCAGAGGTGAAATTCTTGGAT | [28] |
| 4 | 18S919R | TAAATCCAAGAATTTCACCTCT | [28] |
| 5 | 18S1339R | CTCGTTCGTTAACGGAATTAACC | [28] |
| 6 | 18SR | TGATCCTTCYGCAGGTTCAC | [27] |
| Strain Code | Lichen Morphology | Substrate | Sampling Site | Collection Data | Lat. (N) Long. (W) | Alt. (m) |
|---|---|---|---|---|---|---|
| LFE3 | Fruticose | Wood | Esposende, Braga, PT | 16 June 2017 | 41°31′49.3″ 8°46′58.9″ | 1 |
| LCF2 | Crustose | Tree | Barcelos, Braga, PT | 16 June 2017 | 41°29′40.4″ 8°38′45.1″ | 283 |
| LCE2 | Crustose | Wood | Esposende, Braga, PT | 16 June 2017 | 41°31′41.9″ 8°46′53.7″ | 1 |
| LFE1 | Foliose | Tree | Esposende, Braga, PT | 16 June 2017 | 41°32′07.2″ 8°47′06.1″ | 4 |
| LFF1 | Foliose | Rock | Barcelos, Braga, PT | 16 June 2017 | 41°29′40.4″ 8°38′45.1″ | 288 |
| LFA1 | Fruticose | Tree | Braga, PT | 18 October 2017 | 41°33′42.9″ 8°23′46.4″ | 202 |
| LCA2 | Crustose | Rock | Barcelos, Braga, PT | 19 October 2017 | 41°33′19.2″ 8°38′22.1″ | 121 |
| LFS1 | Fruticose | Sand | Esposende, Braga, PT | 19 October 2017 | 41°33′20.0″ 8°47′33.5″ | 2 |
| LCA1 | Foliose | Tree | Barcelos, Braga, PT | 19 October 2017 | 41°33′19.5″ 8°38′23.9″ | 116 |
| LCF1 | Fruticose | Tree | Barcelos, Braga, PT | 19 October 2017 | 41°34′42.8″ 8°33′38.4″ | 302 |
| LFV1 | Fruticose | Tree | Viana do Castelo, PT | 15 January 2018 | 41°48′02.1″ 8°43′57.6″ | 374 |
| LCF3 | Foliose | Tree | Barcelos, Braga, PT | 08 February 2018 | 41°34′41.7″ 8°33′41.5″ | 291 |
| LFR1 | Foliose | Rock | Esposende, Braga, PT | 23 April 2018 | 41°35′27.6″ 8°46′40.9″ | 201 |
| LFA2 | Fruticose | Tree | Esposende, Braga, PT | 23 April 2018 | 41°33′23.8″ 8°45′40.5″ | 192 |
| LCR1 | Crustose | Rock | Esposende, Braga, PT | 23 April 2018 | 41°33′22.2″ 8°45′40.4″ | 192 |
| LFP1 | Foliose | Tree | Póvoa de Varzim, Porto, PT | 23 April 2018 | 41°23′54.7″ 8°45′14.0″ | 18 |
| LFP2 | Crustose | Rock | Póvoa de Varzim, Porto, PT | 23 April 2018 | 41°23′55.0″ 8°45′21.3″ | 18 |
| LFR2 | Fruticose | Rock | Esposende, Braga, PT | 01 May 2018 | 41°33′30.3″ 8°45′34.9″ | 187 |
| Strain Identification | Cell Dimensions Ø (µm) | Cell Shape | Cell Color |
|---|---|---|---|
| Coelastrella sp. LFR1 | 8.6 ± 0.7 µm | spheroidal to ellipsoidal | Green |
| Coelastrella sp. LCF3 | 6.4 ± 1.4 µm | spheroidal to ellipsoidal | Green |
| Coelastrella sp. LFA2 | 6.0 ± 0.9 µm | spheroidal to ellipsoidal | Green |
| Chlorella sp. LFA1 | 4.5 ± 0.6 µm | spheroidal to ellipsoidal | Green |
| Chlorella sp. LCF1 | 4.5 ± 0.4 µm | spheroidal to ellipsoidal | Green |
| Minimum Inhibitory Concentration (MIC) | ||||||||
|---|---|---|---|---|---|---|---|---|
| LFR1 | LCF3 | LFA2 | LFA1 | LCF1 | LFS1 | LCA1 | LCR1 | |
| Microorganisms | mg.mL−1 | |||||||
| E. coli | NA * | NA | NA | NA | >5 | >5 | NA | NA |
| Listeria | >5 | NA | NA | NA | >5 | >5 | >5 | NA |
| Salmonella | >5 | NA | NA | NA | >5 | >5 | >5 | NA |
| (a) | |||||
| Viability of Caco-2 Cell Line (%) | |||||
| Photobiont Extracts Concentration | |||||
| 0.5 mg.mL−1 | 1.25 mg.mL−1 | 2.5 mg.mL−1 | 3.75 mg.mL−1 | 5 mg.mL−1 | |
| Control * | 100.00 | ||||
| LFR1 | 105.96 ± 1.46 | 112.03 ± 2.06 | 86.53 ± 6.63 | 62.40 ± 7.87 | 24.02 ± 1.82 |
| LFA1 | 98.44 ± 1.78 | 91.72 ± 1.01 | 88.69 ± 2.14 | 92.22 ± 1.94 | 13.22 ± 1.49 |
| LCF3 | 110.95 ± 1.65 | 76.23 ± 3.41 | 78.66 ± 6.50 | 85.99 ±3.97 | 96.28 ± 8.26 |
| LFA2 | 104.01 ± 2.61 | 89.15 ± 7.26 | 89.98 ± 5.88 | 93.53 ± 4.71 | 85.29 ± 5.21 |
| LFS1 | 117.11 ± 4.37 | 86.29 ± 1.68 | 79.42 ± 2.91 | 60.24 ± 6.21 | 13.41 ± 1.82 |
| LCA1 | 98.44 ± 5.77 | 88.26 ± 4.65 | 71.77 ± 2.53 | 51.74 ± 4.65 | 10.24 ± 0.93 |
| LCF1 | 117.79 ± 4.29 | 78.60 ± 4.01 | 68.21 ± 2.10 | 34.73 ± 6.83 | 13.97 ± 2.19 |
| LCR1 | 117.20 ± 4.94 | 96.65 ± 7.13 | 84.48 ± 3.47 | 67.66 ± 7.56 | 28.31 ± 6.33 |
| (b) | |||||
| Viability of Bj5ta cell line (%) | |||||
| Photobiont extracts concentration | |||||
| 0.5 mg.mL−1 | 1.25 mg.mL−1 | 2.5 mg.mL−1 | 3.75 mg.mL−1 | 5 mg.mL−1 | |
| Control * | 100.00 | ||||
| LFR1 | 98.27 ± 3.15 | 89.03 ± 5.89 | 63.52 ± 5.29 | 103.71 ± 6.08 | 129.78 ± 3.50 |
| LFA1 | 82.10 ± 9.62 | 10.17 ± 1.62 | 10.96 ± 0.77 | 27.61 ± 1.45 | 48.91 ± 0.58 |
| LCF3 | 102.23 ± 2.50 | 91.26 ± 2.72 | 73.88 ± 2.97 | 63.59 ± 4.15 | 125.82 ± 3.20 |
| LFA2 | 80.65 ± 5.77 | 79.10 ± 2.99 | 81.23 ± 853 | 91.21 ± 10.20 | 101.10 ± 4.88 |
| LFS1 | 106.53 ± 6.38 | 109.42 ± 4.52 | 29.39 ± 2.28 | 24.59 ± 2.24 | 39.67 ± 0.63 |
| LCA1 | 98.44 ± 3.44 | 77.44 ± 4.93 | 20.14 ± 1.61 | 30.63 ± 0.68 | 40.49 ± 0.48 |
| LCF1 | 117.12 ± 1.33 | 106.34 ± 1.39 | 27.82 ± 0.84 | 35.30 ± 2.15 | 67.12 ± 1.93 |
| LCR1 | 97.32 ± 1.77 | 84.35 ± 7.00 | 46.52 ± 3.09 | 54.81 ± 3.51 | 62.23 ± 3.32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loureiro, L.; Morais, J.; Silva, R.; Martins, J.T.; Geada, P.; Vasconcelos, V.; Vicente, A.A. Isolation and Identification of Lichen Photobionts Collected from Different Environments in North of Portugal and Evaluation of Bioactivities of Their Extracts. Foods 2024, 13, 1759. https://doi.org/10.3390/foods13111759
Loureiro L, Morais J, Silva R, Martins JT, Geada P, Vasconcelos V, Vicente AA. Isolation and Identification of Lichen Photobionts Collected from Different Environments in North of Portugal and Evaluation of Bioactivities of Their Extracts. Foods. 2024; 13(11):1759. https://doi.org/10.3390/foods13111759
Chicago/Turabian StyleLoureiro, Luís, João Morais, Raquel Silva, Joana T. Martins, Pedro Geada, Vítor Vasconcelos, and António A. Vicente. 2024. "Isolation and Identification of Lichen Photobionts Collected from Different Environments in North of Portugal and Evaluation of Bioactivities of Their Extracts" Foods 13, no. 11: 1759. https://doi.org/10.3390/foods13111759
APA StyleLoureiro, L., Morais, J., Silva, R., Martins, J. T., Geada, P., Vasconcelos, V., & Vicente, A. A. (2024). Isolation and Identification of Lichen Photobionts Collected from Different Environments in North of Portugal and Evaluation of Bioactivities of Their Extracts. Foods, 13(11), 1759. https://doi.org/10.3390/foods13111759








