Investigation of Freezing and Freeze-Drying for Preserving and Re-Using a Whole Microbial Cheese Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Preparation of Microbial Inocula for Cheese Production
2.2.1. Starters and Ripening Yeast (Inoculation of Milk)
2.2.2. Ripening Bacteria (Inoculation through the Smearing Procedure of Cheese)
2.3. Experimental Cheese Production
2.3.1. Day 0 of Cheese Production
2.3.2. Day 1 of Cheese Production
2.3.3. Day 2 of Cheese Production
2.4. Experimental Design for Cheese Stabilization and Storage
2.5. Microbial Analysis (Culturability and pH Measurements)
2.6. Differential Scanning Calorimetry (DSC) of Frozen and Freeze-Dried Samples
2.7. Water Activity and Water Content Measurements of the Freeze-Dried Cheese Samples
2.8. Cheese Production with Frozen Ecosystems
2.9. Statistical Analyses
3. Results and Discussion
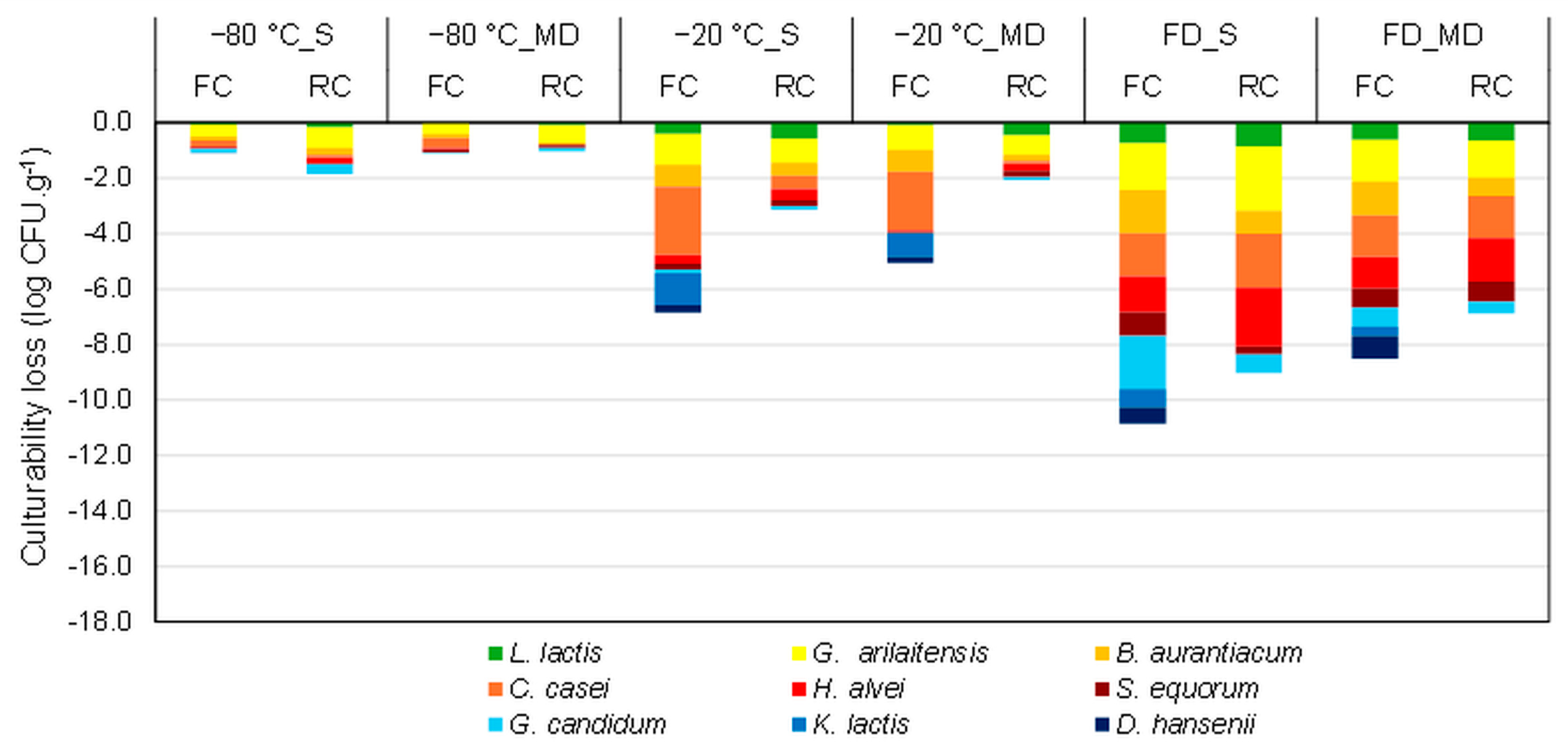
3.1. Effect of the Stabilization Process on the Cheese Microbial Community Composition
3.1.1. Stabilization Processes
3.1.2. Cheese Matrix and Protective Molecules
3.1.3. Specific Microorganism Sensitivity
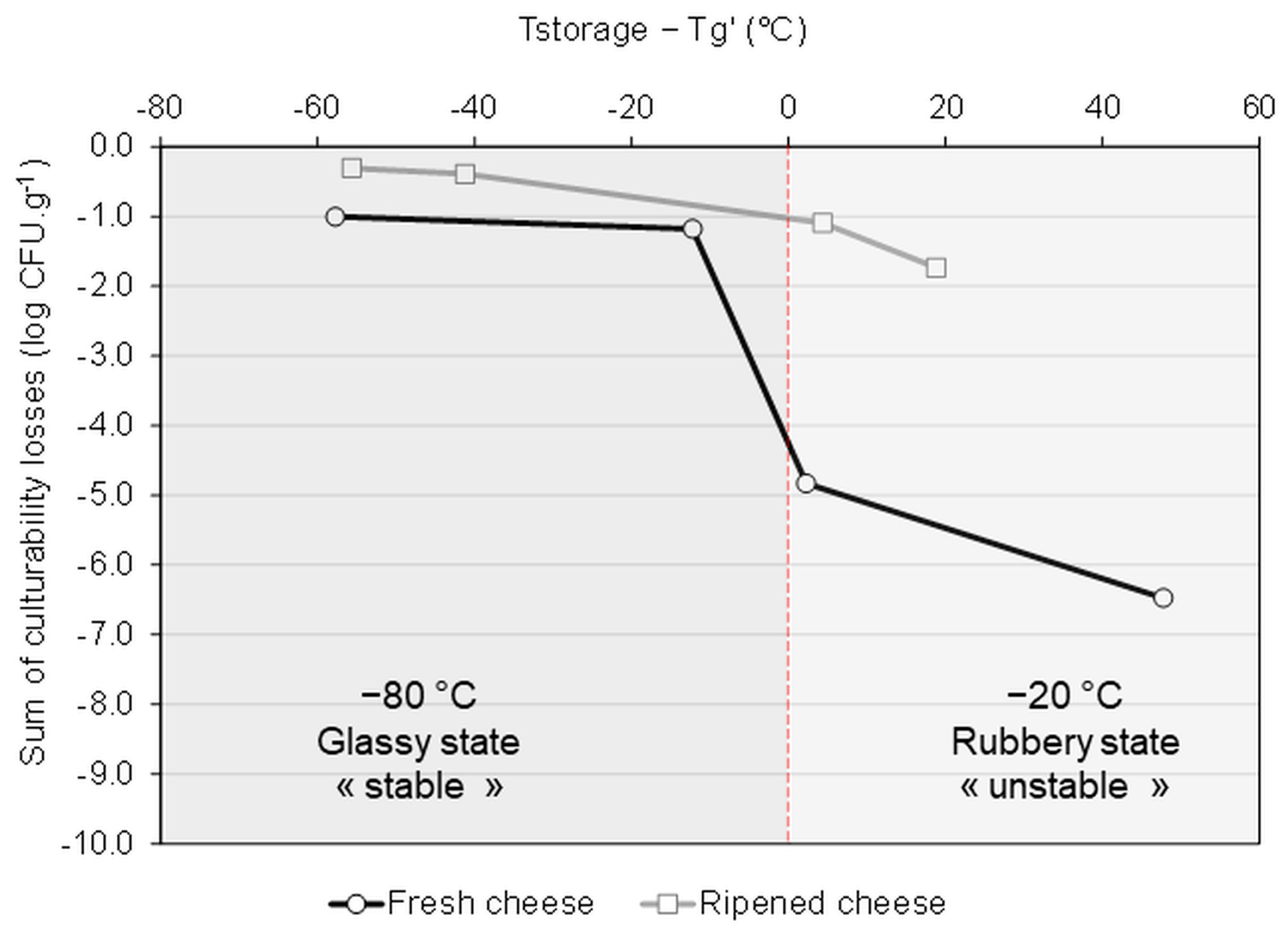
3.2. Evolution of the Composition of the Cheese Microbial Community during Storage in the Frozen and Dehydrated States
3.3. Correlation between the Storage Stability and the Physical Events Observed by DSC
3.4. Cheese-Making and Ripening with Frozen Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prakash, O.; Nimonkar, Y.; Desai, D. A Recent Overview of Microbes and Microbiome Preservation. Indian J. Microbiol. 2020, 60, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Esquinas-Alcázar, J. Protecting Crop Genetic Diversity for Food Security: Political, Ethical and Technical Challenges. Nat. Rev. Genet. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional Cheeses: Rich and Diverse Microbiota with Associated Benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Berland, M.; Cadiou, J.; Levenez, F.; Galleron, N.; Quinquis, B.; Thirion, F.; Gauthier, F.; Le Chatelier, E.; Plaza Oñate, F.; Schwintner, C.; et al. High Engraftment Capacity of Frozen Ready-to-Use Human Fecal Microbiota Transplants Assessed in Germ-Free Mice. Sci. Rep. 2021, 11, 4365. [Google Scholar] [CrossRef] [PubMed]
- Reygner, J.; Charrueau, C.; Delannoy, J.; Mayeur, C.; Robert, V.; Cuinat, C.; Meylheuc, T.; Mauras, A.; Augustin, J.; Nicolis, I.; et al. Freeze-Dried Fecal Samples Are Biologically Active after Long-Lasting Storage and Suited to Fecal Microbiota Transplantation in a Preclinical Murine Model of Clostridioides difficile Infection. Gut Microbes 2020, 11, 1405–1422. [Google Scholar] [CrossRef] [PubMed]
- Burz, S.D.; Abraham, A.-L.; Fonseca, F.; David, O.; Chapron, A.; Béguet-Crespel, F.; Cénard, S.; Le Roux, K.; Patrascu, O.; Levenez, F.; et al. A Guide for Ex Vivo Handling and Storage of Stool Samples Intended for Fecal Microbiota Transplantation. Sci. Rep. 2019, 9, 8897. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-D.; Alexander, A.; Ke, S.; Valilis, E.M.; Hu, S.; Li, B.; DuPont, H.L. Stability and Efficacy of Frozen and Lyophilized Fecal Microbiota Transplant (FMT) Product in a Mouse Model of Clostridium Difficile Infection (CDI). Anaerobe 2017, 48, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Aira, A.; Rubio, E.; Ruiz, A.; Vergara, A.; Casals-Pascual, C.; Rico, V.; Suñé-Negre, J.M.; Soriano, A. New Procedure to Maintain Fecal Microbiota in a Dry Matrix Ready to Encapsulate. Front. Cell. Infect. Microbiol. 2022, 12, 899257. [Google Scholar] [CrossRef]
- Tatangelo, V.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Ambrosini, R. Effect of Preservation Method on the Assessment of Bacterial Community Structure in Soil and Water Samples. FEMS Microbiol. Lett. 2014, 356, 32–38. [Google Scholar] [CrossRef]
- Rubin, B.E.R.; Gibbons, S.M.; Kennedy, S.; Hampton-Marcell, J.; Owens, S.; Gilbert, J.A. Investigating the Impact of Storage Conditions on Microbial Community Composition in Soil Samples. PLoS ONE 2013, 8, e70460. [Google Scholar] [CrossRef]
- Lauber, C.L.; Zhou, N.; Gordon, J.I.; Knight, R.; Fierer, N. Effect of Storage Conditions on the Assessment of Bacterial Community Structure in Soil and Human-Associated Samples: Influence of Short-Term Storage Conditions on Microbiota. FEMS Microbiol. Lett. 2010, 307, 80–86. [Google Scholar] [CrossRef]
- Laurin, V.; Labbé, N.; Juteau, P.; Parent, S.; Villemur, R. Long-Term Storage Conditions for Carriers with Denitrifying Biomass of the Fluidized, Methanol-Fed Denitrification Reactor of the Montreal Biodome, and the Impact on Denitrifying Activity and Bacterial Population. Water Res. 2006, 40, 1836–1840. [Google Scholar] [CrossRef]
- Kerckhof, F.M.; Courtens, E.N.; Geirnaert, A.; Hoefman, S.; Ho, A.; Vilchez-Vargas, R.; Pieper, D.H.; Jauregui, R.; Vlaeminck, S.E.; Van de Wiele, T.; et al. Optimized Cryopreservation of Mixed Microbial Communities for Conserved Functionality and Diversity. PLoS ONE 2014, 9, e99517. [Google Scholar] [CrossRef]
- Vlaeminck, S.E.; Geets, J.; Vervaeren, H.; Boon, N.; Verstraete, W. Reactivation of Aerobic and Anaerobic Ammonium Oxidizers in OLAND Biomass after Long-Term Storage. Appl. Microbiol. Biotechnol. 2007, 74, 1376–1384. [Google Scholar] [CrossRef]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Preservation of Kefir Grains, a Comparative Study. LWT-Food Sci. Technol. 1997, 30, 77–84. [Google Scholar] [CrossRef]
- Gökırmaklı, Ç.; Şatır, G.; Guzel-Seydim, Z.B. Microbial Viability and Nutritional Content of Water Kefir Grains under Different Storage Conditions. Food Sci. Nutr. 2024, 1–8. [Google Scholar] [CrossRef]
- Nikolaou, A.; Sgouros, G.; Mitropoulou, G.; Santarmaki, V.; Kourkoutas, Y. Freeze-Dried Immobilized Kefir Culture in Low Alcohol Winemaking. Foods 2020, 9, 115. [Google Scholar] [CrossRef]
- Béal, C.; Fonseca, F. Freezing of Probiotic Bacteria. In Advances in Probiotic Technology; Taylor & Francis Group: Boca Raton, FL, USA, 2015; pp. 179–212. ISBN 978-1-4987-3453-0. [Google Scholar]
- Velly, H.; Fonseca, F.; Passot, S.; Delacroix-Buchet, A.; Bouix, M. Cell Growth and Resistance of Lactococcus Lactis Subsp. Lactis TOMSC161 Following Freezing, Drying and Freeze-Dried Storage Are Differentially Affected by Fermentation Conditions. J. Appl. Microbiol. 2014, 117, 729–740. [Google Scholar] [CrossRef]
- Guerrero Sanchez, M.; Passot, S.; Campoy, S.; Olivares, M.; Fonseca, F. Effect of Protective Agents on the Storage Stability of Freeze-Dried Ligilactobacillus Salivarius CECT5713. Appl. Microbiol. Biotechnol. 2022, 106, 7235–7249. [Google Scholar] [CrossRef]
- Tovilla Coutiño, M.D.L.; Passot, S.; Trelea, I.-C.; Ropers, M.-H.; Gohon, Y.; Fonseca, F. Multiobjective Optimization of Frozen and Freeze-Dried Lactobacillus Delbrueckii Subsp. Bulgaricus CFL1 Production via the Modification of Fermentation Conditions. J. Appl. Microbiol. 2023, 134, lxad003. [Google Scholar] [CrossRef]
- Perdana, J.; Bereschenko, L.; Fox, M.B.; Kuperus, J.H.; Kleerebezem, M.; Boom, R.M.; Schutyser, M.A.I. Dehydration and Thermal Inactivation of Lactobacillus Plantarum WCFS1: Comparing Single Droplet Drying to Spray and Freeze Drying. Food Res. Int. 2013, 54, 1351–1359. [Google Scholar] [CrossRef]
- Broeckx, G.; Vandenheuvel, D.; Henkens, T.; Kiekens, S.; van den Broek, M.F.L.; Lebeer, S.; Kiekens, F. Enhancing the Viability of Lactobacillus Rhamnosus GG after Spray Drying and during Storage. Int. J. Pharm. 2017, 534, 35–41. [Google Scholar] [CrossRef]
- Mushtaq, M.; Gani, A.; Shetty, P.H.; Masoodi, F.A.; Ahmad, M. Himalayan Cheese (Kalari/Kradi): Effect of Different Storage Temperatures on Its Physicochemical, Microbiological and Antioxidant Properties. LWT-Food Sci. Tech. 2015, 63, 837–845. [Google Scholar] [CrossRef]
- Dugat-Bony, E.; Straub, C.; Teissandier, A.; Onésime, D.; Loux, V.; Monnet, C.; Irlinger, F.; Landaud, S.; Leclercq-Perlat, M.-N.; Bento, P.; et al. Overview of a Surface-Ripened Cheese Community Functioning by Meta-Omics Analyses. PLoS ONE 2015, 10, e0124360. [Google Scholar] [CrossRef]
- Leclercq-Perlat, M.-N.; Oumer, A.; Bergere, J.-L.; Spinnler, H.-E.; Corrieu, G. Growth of Debaryomyces Hansenii on a Bacterial Surface-Ripened Soft Cheese. J. Dairy. Res. 1999, 66, 271–281. [Google Scholar] [CrossRef]
- Wang, G.; Chen, Y.; Xia, Y.; Song, X.; Ai, L. Characteristics of Probiotic Preparations and Their Applications. Foods 2022, 11, 2472. [Google Scholar] [CrossRef]
- Fenster, K.; Freeburg, B.; Hollard, C.; Wong, C.; Rønhave Laursen, R.; Ouwehand, A. The Production and Delivery of Probiotics: A Review of a Practical Approach. Microorganisms 2019, 7, 83. [Google Scholar] [CrossRef]
- Sehrawat, R.; Abdullah, S.; Khatri, P.; Kumar, L.; Kumar, A.; Mujumdar, A.S. Role of Drying Technology in Probiotic Encapsulation and Impact on Food Safety. Dry. Technol. 2022, 40, 1562–1581. [Google Scholar] [CrossRef]
- Guerrero Sanchez, M.; Passot, S.; Ghorbal, S.; Campoy, S.; Olivares, M.; Fonseca, F. Insights into the Mechanisms of L. Salivarius CECT5713 Resistance to Freeze-Dried Storage. Cryobiology 2023, 112, 104556. [Google Scholar] [CrossRef]
- Costello, S.P.; Conlon, M.A.; Vuaran, M.S.; Roberts-Thomson, I.C.; Andrews, J.M. Faecal Microbiota Transplant for Recurrent Clostridium Difficile Infection Using Long-Term Frozen Stool Is Effective: Clinical Efficacy and Bacterial Viability Data. Aliment. Pharmacol. Ther. 2015, 42, 1011–1018. [Google Scholar] [CrossRef]
- Satokari, R.; Mattila, E.; Kainulainen, V.; Arkkila, P.E.T. Simple Faecal Preparation and Efficacy of Frozen Inoculum in Faecal Microbiota Transplantation for Recurrent C Clostridium Difficile Infection—An Observational Cohort Study. Aliment. Pharmacol. Ther. 2015, 41, 46–53. [Google Scholar] [CrossRef]
- Youngster, I.; Sauk, J.; Pindar, C.; Wilson, R.G.; Kaplan, J.L.; Smith, M.B.; Alm, E.J.; Gevers, D.; Russell, G.H.; Hohmann, E.L. Fecal Microbiota Transplant for Relapsing Clostridium Difficile Infection Using a Frozen Inoculum From Unrelated Donors: A Randomized, Open-Label, Controlled Pilot Study. Clin. Infect. Dis. 2014, 58, 1515–1522. [Google Scholar] [CrossRef]
- Jiang, Z.D.; Ajami, N.J.; Petrosino, J.F.; Jun, G.; Hanis, C.L.; Shah, M.; Hochman, L.; Ankoma-Sey, V.; DuPont, A.W.; Wong, M.C.; et al. Randomised Clinical Trial: Faecal Microbiota Transplantation for Recurrent Clostridum Difficile Infection—Fresh, or Frozen, or Lyophilised Microbiota from a Small Pool of Healthy Donors Delivered by Colonoscopy. Aliment. Pharmacol. Ther. 2017, 45, 899–908. [Google Scholar] [CrossRef]
- Bolla, P.A.; De Los Angeles Serradell, M.; De Urraza, P.J.; De Antoni, G.L. Effect of Freeze-Drying on Viability and in Vitro Probiotic Properties of a Mixture of Lactic Acid Bacteria and Yeasts Isolated from Kefir. J. Dairy. Res. 2011, 78, 15–22. [Google Scholar] [CrossRef]
- Guerrero Sanchez, M.; Passot, S.; Campoy, S.; Olivares, M.; Fonseca, F. Ligilactobacillus salivarius Functionalities, Applications, and Manufacturing Challenges. Appl. Microbiol. Biotechnol. 2022, 106, 57–80. [Google Scholar] [CrossRef]
- Jayaprakash, P.; Gaiani, C.; Edorh, J.-M.; Borges, F.; Beaupeux, E.; Maudhuit, A.; Desobry, S. Comparison of Electrostatic Spray Drying, Spray Drying, and Freeze Drying for Lacticaseibacillus Rhamnosus GG Dehydration. Foods 2023, 12, 3117. [Google Scholar] [CrossRef]
- Samedi, L.; Charles, A.L. Viability of 4 Probiotic Bacteria Microencapsulated with Arrowroot Starch in the Simulated Gastrointestinal Tract (GIT) and Yoghurt. Foods 2019, 8, 175. [Google Scholar] [CrossRef]
- Champagne, C.P.; Gardner, N.; Brochu, E.; Beaulieu, Y. The Freeze-Drying of Lactic Acid Bacteria. A Review. Can. Inst. Food Sci. Technol. J. 1991, 24, 118–128. [Google Scholar] [CrossRef]
- Bozoglu, T.F.; Ozilgen, M.; Bakir, U. Survival Kinetics of Lactic Acid Starter Cultures during and after Freeze Drying. Enzyme Microb. Technol. 1987, 9, 531–537. [Google Scholar] [CrossRef]
- Schoug, Å.; Olsson, J.; Carlfors, J.; Schnürer, J.; Håkansson, S. Freeze-Drying of Lactobacillus Coryniformis Si3—Effects of Sucrose Concentration, Cell Density, and Freezing Rate on Cell Survival and Thermophysical Properties. Cryobiology 2006, 53, 119–127. [Google Scholar] [CrossRef]
- Miyamoto-Shinohara, Y.; Imaizumi, T.; Sukenobe, J.; Murakami, Y.; Kawamura, S.; Komatsu, Y. Survival Rate of Microbes after Freeze-Drying and Long-Term Storage. Cryobiology 2000, 41, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Polo, L.; Mañes-Lázaro, R.; Olmeda, I.; Cruz-Pio, L.E.; Medina, Á.; Ferrer, S.; Pardo, I. Influence of Freezing Temperatures Prior to Freeze-Drying on Viability of Yeasts and Lactic Acid Bacteria Isolated from Wine. J. Appl. Microbiol. 2017, 122, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- To, C.S.B.; Etzel, M.R. Spray Drying, Freeze-Drying, or Freezing of Three Different Lactic Acid Bacteria Species. J. Food Sci. 1997, 62, 576–578. [Google Scholar] [CrossRef]
- Bashandy, E.Y.; Heider, L.E. The Effect of Freezing of Milk Samples on the Cultural Results. Zentralblatt Für Veterinärmedizin Reihe B 1979, 26, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Luedecke, L.O.; Forster, T.L.; Williams, K.; Hillers, J.K. Effect of Freezing and Storage at −20 °C on Survival of Mastitis Pathogens. J. Dairy. Sci. 1972, 55, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Schukken, Y.H.; Smit, J.A.H.; Grommers, F.J.; Vandegeer, D.; Brand, A. Effect of Freezing on Bacteriologic Culturing of Mastitis Milk Samples. J. Dairy. Sci. 1989, 72, 1900–1906. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jinxia, L.; Xiao, D.; Haibo, D.; Liu, B.; Liu, Y.; Hairong, H. Enhancement of Viability for Staphylococcus Aureus During Freeze-Drying Using the Response Surface Methodology. In 2nd International Conference on Biomedical and Biological Engineering 2017 (BBE 2017); Atlantis Press: Amsterdam, The Netherlands, 2017. [Google Scholar]
- To, B.C.S.; Etzel, M.R. Survival of Brevibacterium linens (ATCC 9174) after Spray Drying, Freeze Drying, or Freezing. J. Food Sci. 1997, 62, 167–170. [Google Scholar] [CrossRef]
- Bjerketorp, J.; Röling, W.F.M.; Feng, X.-M.; Garcia, A.H.; Heipieper, H.J.; Håkansson, S. Formulation and Stabilization of an Arthrobacter Strain with Good Storage Stability and 4-Chlorophenol-Degradation Activity for Bioremediation. Appl. Microbiol. Biotechnol. 2018, 102, 2031–2040. [Google Scholar] [CrossRef] [PubMed]
- Monnet, C.; Loux, V.; Gibrat, J.-F.; Spinnler, E.; Barbe, V.; Vacherie, B.; Gavory, F.; Gourbeyre, E.; Siguier, P.; Chandler, M.; et al. The Arthrobacter Arilaitensis Re117 Genome Sequence Reveals Its Genetic Adaptation to the Surface of Cheese. PLoS ONE 2010, 5, e15489. [Google Scholar] [CrossRef]
- Hamoudi, L.; Goulet, J.; Ratti, C. Effect of Protective Agents on the Viability of Geotrichum Candidum during Freeze-Drying and Storage. J. Food Sci. 2007, 72, M45–M49. [Google Scholar] [CrossRef]
- Miyamoto-Shinohara, Y.; Nozawa, F.; Sukenobe, J.; Imaizumi, T. Survival of Yeasts Stored after Freeze-Drying or Liquid-Drying. J. Gen. Appl. Microbiol. 2010, 56, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Tantratian, S.; Sae-ngow, A.; Pradistsuwan, C.; Prakitchaiwattana, C.; Pukahuta, C. Survival of Kluyveromyces Marxianus with Stigmasterol as Subjected to Freezing Stress. J. Biosci. Bioeng. 2019, 128, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, F.; Marin, M.; Morris, G.J. Stabilization of Frozen Lactobacillus delbrueckii Subsp. Bulgaricus in Glycerol Suspensions: Freezing Kinetics and Storage Temperature Effects. Appl. Environ. Microbiol. 2006, 72, 6474–6482. [Google Scholar] [CrossRef] [PubMed]
- Passot, S.; Cenard, S.; Douania, I.; Trelea, I.C.; Fonseca, F. Critical Water Activity and Amorphous State for Optimal Preservation of Lyophilized Lactic Acid Bacteria. Food Chem 2012, 132, 1699–1705. [Google Scholar] [CrossRef]
- Lopez, C.; Briard-Bion, V.; Camier, B.; Gassi, J.Y. Milk Fat Thermal Properties and Solid Fat Content in Emmental Cheese: A Differential Scanning Calorimetry Study. J. Dairy. Sci. 2006, 89, 2894–2910. [Google Scholar] [CrossRef]
- Tunick, M.H. Effects of Homogenization and Proteolysis on Free Oil in Mozzarella Cheese. J. Dairy. Sci. 1994, 77, 2487–2493. [Google Scholar] [CrossRef]
- Kohl, W.; Achenbach, H.; Reichenbach, H. The Pigments of Brevibacterium Linens: Aromatic Carotenoids. Phytochemistry 1983, 22, 207–210. [Google Scholar] [CrossRef]
- Sutthiwong, N.; Dufossé, L. Production of Carotenoids by Arthrobacter arilaitensis Strains Isolated from Smear-Ripened Cheeses. FEMS Microbiol. Lett. 2014, 360, 174–181. [Google Scholar] [CrossRef]
- Leclercq-Perlat, M.-N.; Corrieu, G.; Spinnler, H.-E. The Color of Brevibacterium Linens Depends on the Yeast Used for Cheese Deacidification. J. Dairy. Sci. 2004, 87, 1536–1544. [Google Scholar] [CrossRef]



| Group | Species | Strain | Origin |
|---|---|---|---|
| Starter | Lactococcus lactis subsp. lactis | S3+ | SayFood collection, INRAE, Palaiseau, France |
| Lactococcus lactis subsp. lactis | S3- | SayFood collection, INRAE, Palaiseau, France | |
| Ripening Bacteria | Glutamicibacter arilaitensis * | Re117 | Institut Pasteur collection, Paris, France |
| Brevibacterium aurantiacum | ATCC 9174 | American Type Culture Collection, Rockville, MD, USA | |
| Corynebacterium casei | 2M01 | ABTE collection, UNICAEN, Caen, France | |
| Hafnia alvei | GB001 | SayFood collection, INRAE, Palaiseau, France | |
| Staphylococcus equorum | Mu2 | SayFood collection, INRAE, Palaiseau, France | |
| Ripening Yeasts | Kluyveromyces lactis | CLIB210 | SayFood collection, INRAE, Palaiseau, France |
| Debaryomyces hansenii | 304 | SayFood collection, INRAE, Palaiseau, France | |
| Geotrichum candidum | ATCC 204307 | American Type Culture Collection, Rockville, MD, USA |
| Test Production (Frozen Ecosystems) | Reference Production | |||||||
|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 1 | Day 2 | Day 22 | Day 0 | Day 1 | Day 2 | Day 22 | |
| pH | 6.57 | 4.55 | 4.57 | 7.09 | 6.69 | 4.66 | 4.45 | 6.99 |
| G. arilaitensis | - | - | 6.0 × 102 | 1.9 × 105 | - | - | 6.9 × 105 | 5.5 × 107 |
| B. aurantiacum | - | - | 1.1 × 104 | 1.4 × 106 | - | - | 3.4 × 105 | 4.5 × 107 |
| C. casei | - | - | 1.4 × 106 | 1.6 × 109 | - | - | 1.0 × 106 | 1.5 × 109 |
| H. alvei | - | - | 5.5 × 105 | 1.8 × 108 | - | - | 1.0 × 106 | 1.1 × 108 |
| S. equorum | - | - | 3.2 × 105 | 6.3 × 107 | - | - | 1.4 × 106 | 1.3 × 108 |
| K. lactis | 1.8 × 104 | 5.9 × 106 | 1.4 × 107 | ND | 5.2 × 104 | 3.0 × 106 | 8.3 × 106 | ND |
| D. hansenii | 8.8 × 103 | 4.3 × 104 | 3.5 × 105 | 2.0 × 105 | 2.8 × 105 | 3.06 × 106 | 1.3 × 107 | 1.0 × 106 |
| G. candidum | 1.0 × 101 | 2.9 × 103 | 2.0 × 106 | 8.6 × 107 | 3.0 × 102 | 1.1 × 105 | 8.8 × 104 | 1.1 × 108 |
| L. lactis S3+/S3- | 1.4 × 107 | 5.6 × 109 | 2.1 × 109 | 1.0 × 108 | 1.1 × 107 | 7.5 × 109 | 5.5 × 109 | 4.2 × 108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, W.; Passot, S.; Irlinger, F.; Fonseca, F. Investigation of Freezing and Freeze-Drying for Preserving and Re-Using a Whole Microbial Cheese Community. Foods 2024, 13, 1809. https://doi.org/10.3390/foods13121809
Cao W, Passot S, Irlinger F, Fonseca F. Investigation of Freezing and Freeze-Drying for Preserving and Re-Using a Whole Microbial Cheese Community. Foods. 2024; 13(12):1809. https://doi.org/10.3390/foods13121809
Chicago/Turabian StyleCao, Wenfan, Stéphanie Passot, Françoise Irlinger, and Fernanda Fonseca. 2024. "Investigation of Freezing and Freeze-Drying for Preserving and Re-Using a Whole Microbial Cheese Community" Foods 13, no. 12: 1809. https://doi.org/10.3390/foods13121809
APA StyleCao, W., Passot, S., Irlinger, F., & Fonseca, F. (2024). Investigation of Freezing and Freeze-Drying for Preserving and Re-Using a Whole Microbial Cheese Community. Foods, 13(12), 1809. https://doi.org/10.3390/foods13121809






