Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
2.2. Isolation of AFB1-Degrading Strains
2.3. Aflatoxin Degradation Assay
2.4. Extraction and Detection of Residual AFB1
2.5. Identification of the HB2-2 Strain
2.6. Impact of Various Fermentation Conditions on the Degradation of AFB1
2.7. Degradation of AFB1 by Strain HB2-2 in Peanut Meal
2.8. The Different Cellular Fractions of Strain HB2-2 on AFB1 Degradation
2.9. The Impact of Heat Inactivation, Proteinase K, and Proteinase K Combined with Sodium Dodecyl Sulfate (SDS) Treatments on the AFB1 Degradation Activity of the Fermentation Supernatant
2.10. Analysis and Identification of Degraded Products
2.11. Cytotoxicity Assay of the Degradation Products of AFB1
2.12. Data Analysis
3. Results and Discussions
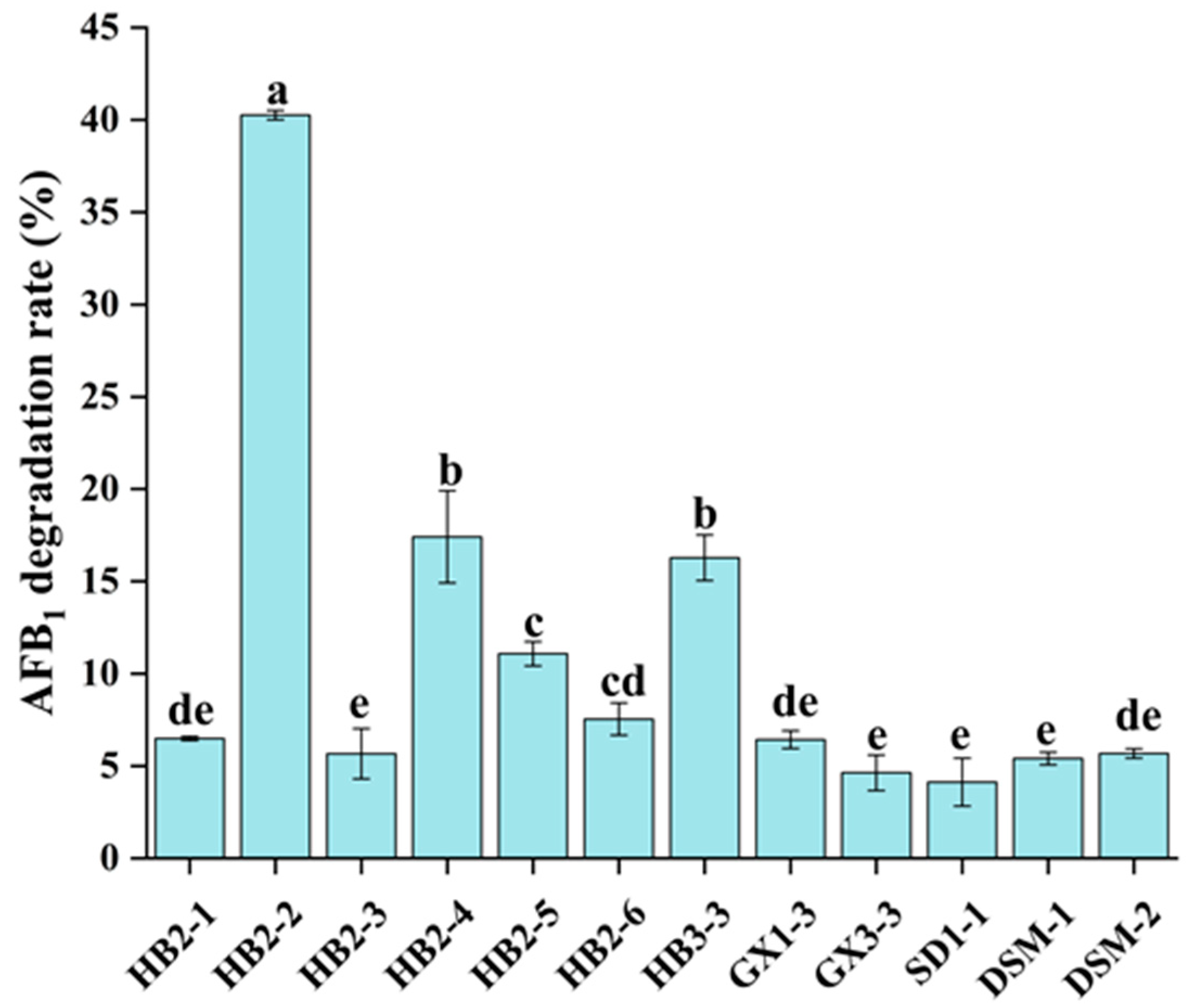
3.1. Screening of AFB1-Degrading Strains
3.2. Physiological, Morphological and 16s rRNA Characterization of Strain HB2-2
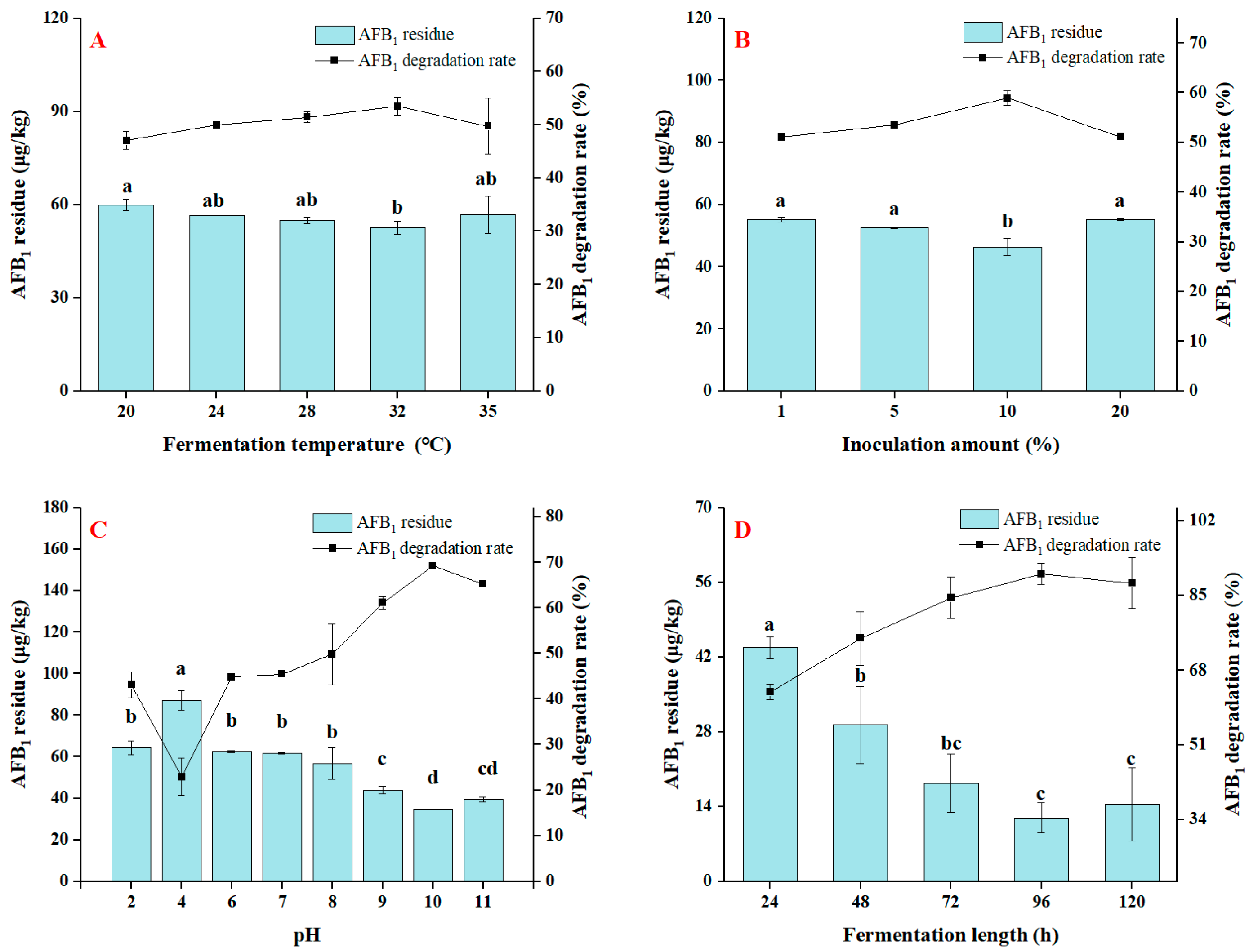
3.3. Effect of Fermentation Conditions on the Degradation of AFB1
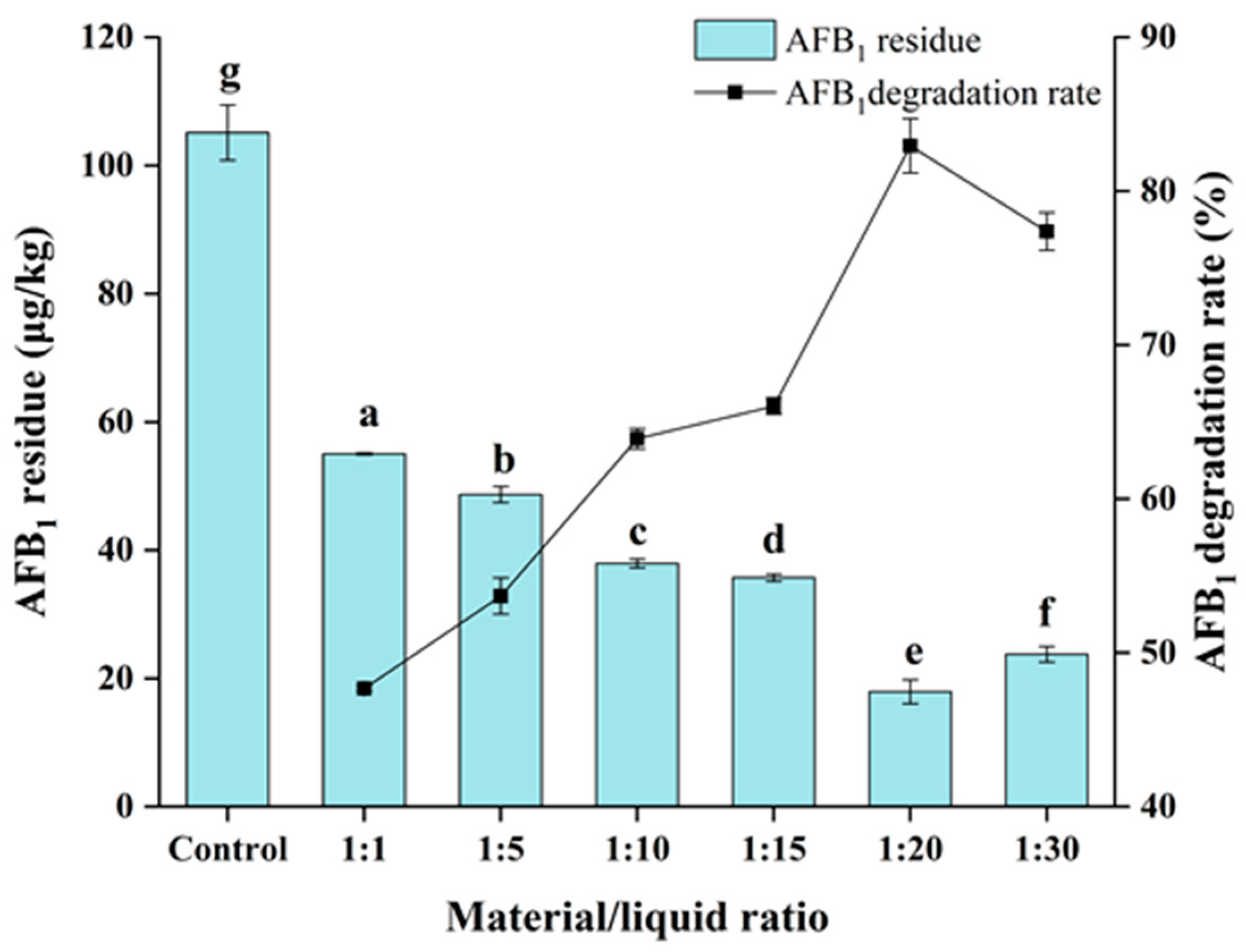
3.4. Degradation of AFB1 by E. faecium HB2-2 in Peanut Meal
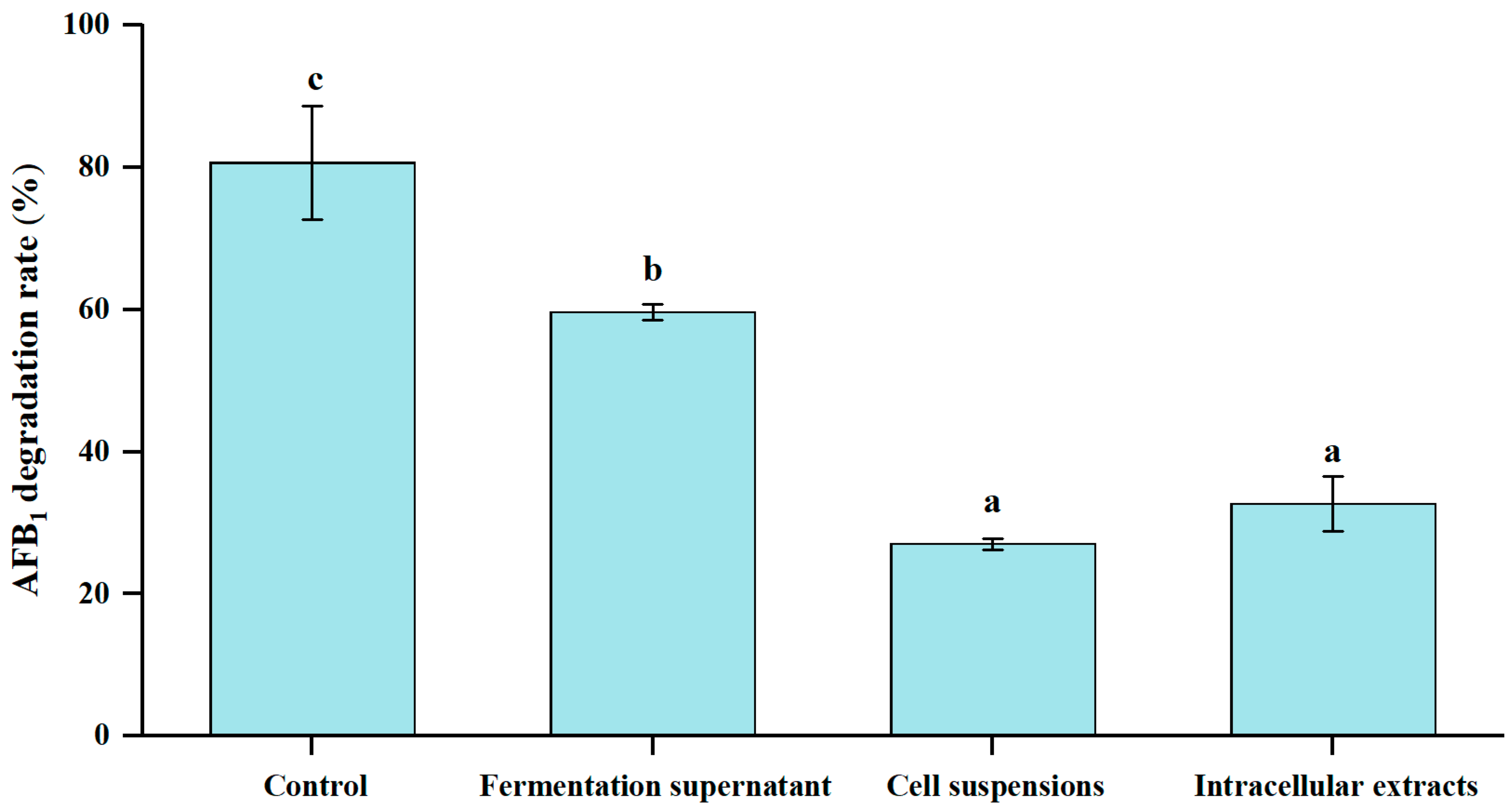
3.5. Different Cellular Fractions of E. faecium HB2-2 on AFB1 Degradation
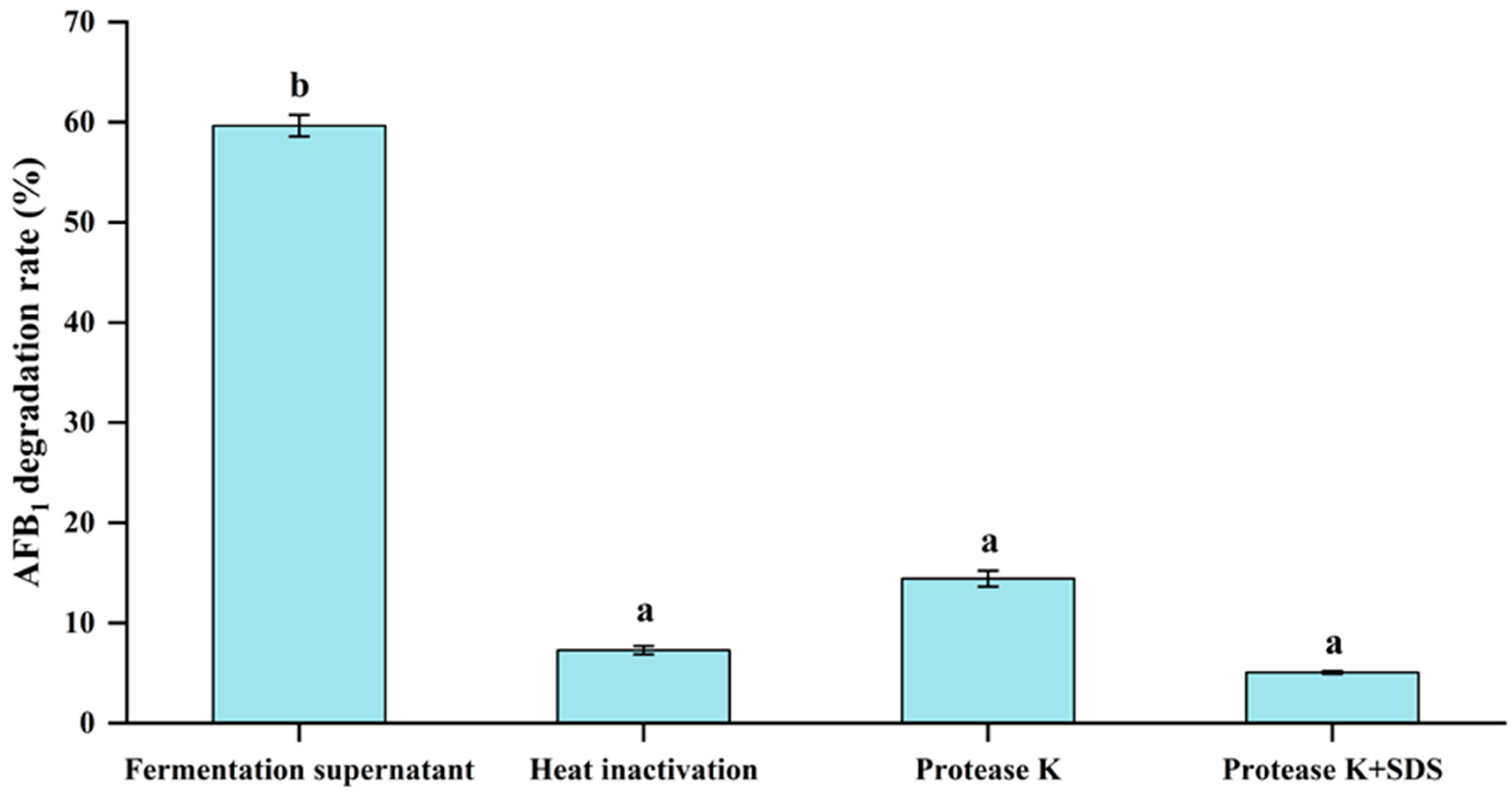
3.6. Effect of Heat Inactivation, Proteinase K and Proteinase K + SDS Treatments of Fermentation Supernatant on the AFB1 Degradation Activity
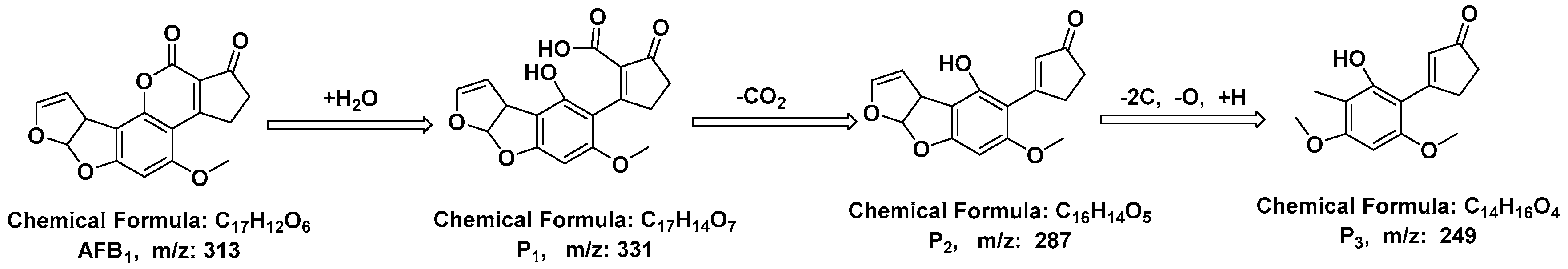
3.7. Identification of the AFB1 Degradation Products
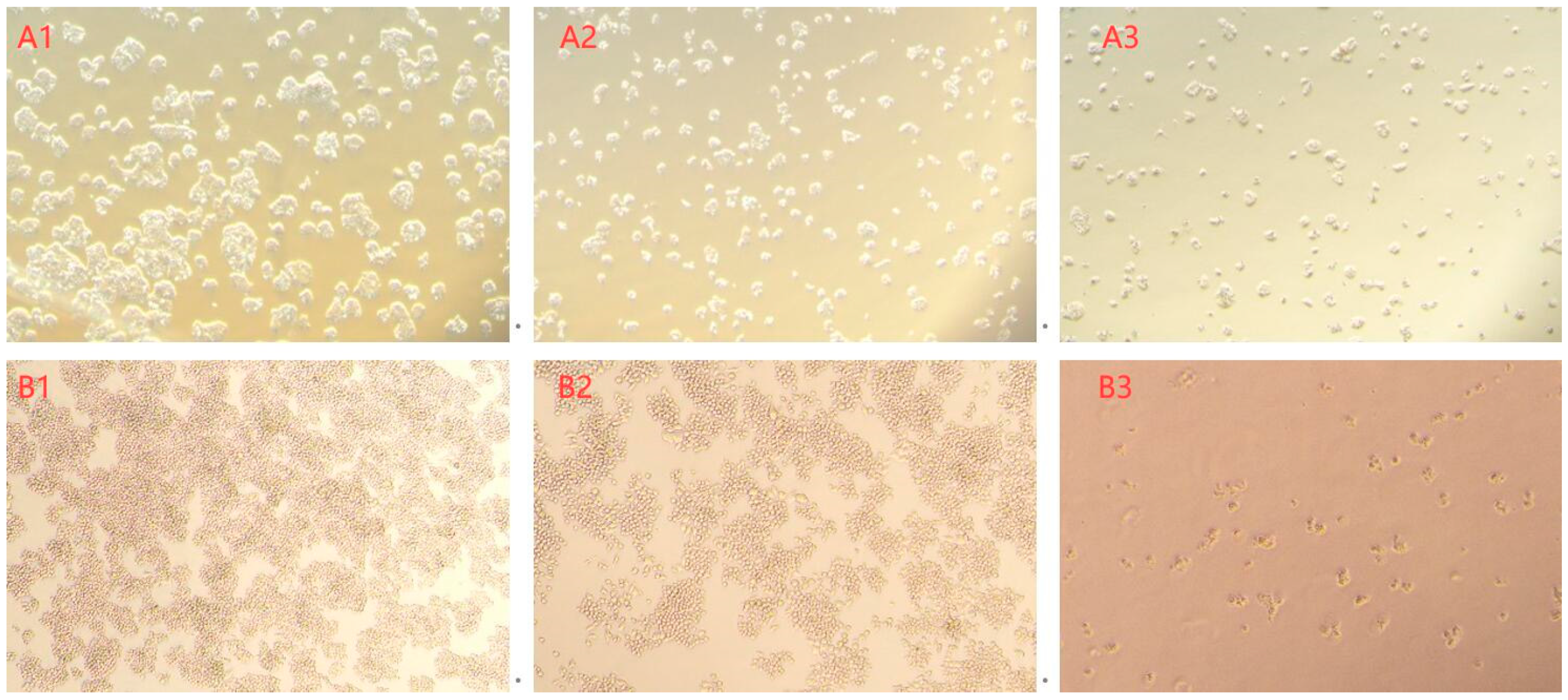
3.8. Cytotoxicity Assay of the AFB1 Degradation Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edite Bezerra Da Rocha, M.; Freire, F.D.C.O.; Erlan Feitosa Maia, F.; Izabel Florindo Guedes, M.; Rondina, D. Mycotoxins and Their Effects on Human and Animal Health. Food Control 2014, 36, 159–165. [Google Scholar] [CrossRef]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T.; et al. Impact of Food Processing and Detoxification Treatments on Mycotoxin Contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Kostić, A.; Milinčić, D.; Petrović, T.; Krnjaja, V.; Stanojević, S.; Barać, M.; Tešić, Ž.; Pešić, M. Mycotoxins and Mycotoxin Producing Fungi in Pollen: Review. Toxins 2019, 11, 64. [Google Scholar] [CrossRef]
- Dai, C.; Das Gupta, S.; Wang, Z.; Jiang, H.; Velkov, T.; Shen, J. T-2 Toxin and Its Cardiotoxicity: New Insights on the Molecular Mechanisms and Therapeutic Implications. Food Chem. Toxicol. 2022, 167, 113262. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Tian, E.; Hao, Z.; Tang, S.; Wang, Z.; Sharma, G.; Jiang, H.; Shen, J. Aflatoxin B1 Toxicity and Protective Effects of Curcumin: Molecular Mechanisms and Clinical Implications. Antioxidants 2022, 11, 2031. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; He, Z.; Xiong, D.; Long, M. Mechanisms by Which Microbial Enzymes Degrade Four Mycotoxins and Application in Animal Production: A Review. Anim. Nutr. 2023, 15, 256–274. [Google Scholar] [CrossRef] [PubMed]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide Contamination of Food-Crops with Mycotoxins: Validity of the Widely Cited ‘FAO Estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Omara, T.; Nassazi, W.; Omute, T.; Awath, A.; Laker, F.; Kalukusu, R.; Musau, B.; Nakabuye, B.V.; Kagoya, S.; Otim, G.; et al. Aflatoxins in Uganda: An Encyclopedic Review of the Etiology, Epidemiology, Detection, Quantification, Exposure Assessment, Reduction, and Control. Int. J. Microbiol. 2020, 2020, 4723612. [Google Scholar] [CrossRef] [PubMed]
- Raksha Rao, K.; Vipin, A.V.; Hariprasad, P.; Anu Appaiah, K.A.; Venkateswaran, G. Biological Detoxification of Aflatoxin B1 by Bacillus Licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar] [CrossRef]
- Haque, M.A.; Wang, Y.; Shen, Z.; Li, X.; Saleemi, M.K.; He, C. Mycotoxin Contamination and Control Strategy in Human, Domestic Animal and Poultry: A Review. Microb. Pathog. 2020, 142, 104095. [Google Scholar] [CrossRef]
- Li, S.; Li, C.; Chen, S.; Wang, X.; Liu, J.; Deng, X.; Cai, H.; Liu, G. Effects of Solid-State Fermentation on the Standardized Ileal Digestibility of Amino Acids and Apparent Metabolizable Energy in Peanut Meal Fed to Broiler Chickens. Fermentation 2023, 9, 346. [Google Scholar] [CrossRef]
- Liu, R.; Lu, M.; Wang, R.; Wang, S.; Chang, M.; Jin, Q.; Wang, X. Degradation of Aflatoxin B1 in Peanut Meal by Electron Beam Irradiation. Int. J. Food Prop. 2018, 21, 892–901. [Google Scholar] [CrossRef]
- Saalia, F.K.; Phillips, R.D. Reduction of Aflatoxins in Peanut Meal by Extrusion Cooking in the Presence of Nucleophiles. LWT-Food Sci. Technol. 2011, 44, 1511–1516. [Google Scholar] [CrossRef]
- Xia, Y.; He, R.; Sun, Y.; Zhou, H.; Gao, M.; Hu, X.; Cui, X.; Cheng, Q.; Wang, Z. Food-Grade Expression of Manganese Peroxidases in Recombinant Kluyveromyces Lactis and Degradation of Aflatoxin B1 Using Fermentation Supernatants. Front. Microbiol. 2022, 12, 821230. [Google Scholar] [CrossRef] [PubMed]
- Farzaneh, M.; Shi, Z.-Q.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 Degradation by Bacillus Subtilis UTBSP1 Isolated from Pistachio Nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Fernández-Juri, M.G.; Muzzolón, J.A.; Dalcero, A.M.; Magnoli, C.E. Effect of Acid Lactic Bacteria Isolated from Faeces of Healthy Dogs on Growth Parameters and Aflatoxin B1 Production by Aspergillus Species In Vitro. Mycotoxin Res. 2011, 27, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Wang, R.; Wang, L.; Li, Y.; Wang, Y.; Chen, Z. Detoxification of Aflatoxin in Corn Flour by Ozone: Detoxification of Aflatoxin in Corn Flour by Ozone. J. Sci. Food Agric. 2014, 94, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Sidu, S.; Tlou, M.G.; Mavumengwana, V. Aflatoxin B 1 Degradation by Liquid Cultures and Lysates of Three Bacterial Strains. Int. J. Food Microbiol. 2016, 233, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kong, Q.; Chi, C.; Shan, S.; Guan, B. Biotransformation of Aflatoxin B1 and Aflatoxin G1 in Peanut Meal by Anaerobic Solid Fermentation of Streptococcus thermophilus and Lactobacillus delbrueckii Subsp. bulgaricus. Int. J. Food Microbiol. 2015, 211, 1–5. [Google Scholar] [CrossRef]
- Zaccaria, M.; Dawson, W.; Russel Kish, D.; Reverberi, M.; Bonaccorsi Di Patti, M.C.; Domin, M.; Cristiglio, V.; Chan, B.; Dellafiora, L.; Gabel, F.; et al. Experimental–Theoretical Study of Laccase as a Detoxifier of Aflatoxins. Sci. Rep. 2023, 13, 860. [Google Scholar] [CrossRef]
- Cserháti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Háhn, J.; Tóth, S.; Nagy, I.; Kukolya, J. Mycotoxin-Degradation Profile of Rhodococcus Strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.H.; Guan, S.; Gao, X.; Ma, Q.G.; Lei, Y.P.; Bai, X.M.; Ji, C. Preparation, Purification and Characteristics of an Aflatoxin Degradation Enzyme from Myxococcus Fulvus ANSM068: An Aflatoxin Degradation Enzyme. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Q.; Chen, Z.-Y.; Liu, H.; Li, P. Investigation of Pseudomonas Fluorescens Strain 3JW1 on Preventing and Reducing Aflatoxin Contaminations in Peanuts. PLoS ONE 2017, 12, e0178810. [Google Scholar] [CrossRef] [PubMed]
- Witzler, J.J.P.; Pinto, R.A.; Font De Valdez, G.; De Castro, A.D.; Cavallini, D.C.U. Development of a Potential Probiotic Lozenge Containing Enterococcus Faecium CRL 183. LWT 2017, 77, 193–199. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Y.; Meng, Q.; Zhang, X.; Wang, X. Progress in the Application of Enterococcus Faecium in Animal Husbandry. Front. Cell. Infect. Microbiol. 2023, 13, 1168189. [Google Scholar] [CrossRef]
- Topcu, A.; Bulat, T.; Wishah, R.; Boyacı, I.H. Detoxification of Aflatoxin B1 and Patulin by Enterococcus Faecium Strains. Int. J. Food Microbiol. 2010, 139, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Z.; Wang, H.; Qiu, H.; Zhang, M.; Li, S.; Luo, X.; Song, Y.; Zhou, H.; Ma, W.; et al. Rapid Biodegradation of Aflatoxin B1 by Metabolites of Fusarium Sp. WCQ3361 with Broad Working Temperature Range and Excellent Thermostability. J. Sci. Food Agric. 2017, 97, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Shafi, S.; Shehata, S.; Shindia, A.; El-Meligy, K.; Khidr, A. Biodegradation of Aflatoxins by Bacteria. Egypt. J. Microbiol. 2018, 53, 241–254. [Google Scholar] [CrossRef]
- GB 5009.22—2016; Determination of Aflatoxins B and G in Food of National Food Safety Standard. National Health and Family Planning Commission, People’s Republic of China, State Food and Drug Administration: Beijing, China, 2016.
- Zhao, Y.; Wang, N.; Gao, H.-L.; Guo, Z.-X.; Lu, A.-X.; Guo, X.-J.; Lu, J.-H.; Luan, Y.-X. Determination of Aflatoxin B1 in Lotus Seed by High Performance Liquid Chromatography with Aptamer Affinity Column for Purification and Enrichment. Chin. J. Anal. Chem. 2020, 48, 662–669. [Google Scholar] [CrossRef]
- Schaad, N.W.; Jones, J.B.; Chun, W. Laboratory Guide for Identification of Plant Pathogenic Bacteria, 3rd ed.; APS Press: St. Paul, MN, USA, 2001. [Google Scholar]
- Karcz, J.; Bernas, T.; Nowak, A.; Talik, E.; Woznica, A. Application of Lyophilization to Prepare the Nitrifying Bacterial Biofilm for Imaging with Scanning Electron Microscopy. Scanning 2012, 34, 26–36. [Google Scholar] [CrossRef]
- Yue, X.; Li, Q.; Wang, H.; Sun, Y.; Wang, A.; Zhang, Q.; Zhang, C. Scanning Electron Microscopy of the Nail Plate in Onychomycosis Patients with Negative Fungal Culture. Scanning 2016, 38, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Blainski, J.M.L.; Da Rocha Neto, A.C.; Schimidt, E.C.; Voltolini, J.A.; Rossi, M.J.; Di Piero, R.M. Exopolysaccharides from Lactobacillus Plantarum Induce Biochemical and Physiological Alterations in Tomato Plant against Bacterial Spot. Appl. Microbiol. Biotechnol. 2018, 102, 4741–4753. [Google Scholar] [CrossRef] [PubMed]
- Galach’yants, A.D.; Bel’kova, N.L.; Sukhanova, E.V.; Romanovskaya, V.A.; Gladka, G.V.; Bedoshvili, E.D.; Parfenova, V.V. Diversity and Physiological and Biochemical Properties of Heterotrophic Bacteria Isolated from Lake Baikal Neuston. Microbiology 2016, 85, 604–613. [Google Scholar] [CrossRef]
- Berry, D.; Gutierrez, T. Evaluating the Detection of Hydrocarbon-Degrading Bacteria in 16S rRNA Gene Sequencing Surveys. Front. Microbiol. 2017, 8, 896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.F.; Nan, Z.B. First Report of Erwinia persicinus Causing Wilting of Medicago sativa Sprouts in China. Plant Dis. 2012, 96, 454. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.K.; Singh, Y.S.; Tripathy, P.S.; Kumar, R.; Abujam, S.K.; Parhi, J. DNA Barcoding and Phylogenetics of Freshwater Fish Fauna of Ranganadi River, Arunachal Pradesh. Gene 2020, 754, 144860. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Wu, T.; Zhang, H.; Sun, Z.; Mwabulili, F.; Xie, Y.; Sun, S.; Ma, W.; Li, Q.; Yang, Y.; et al. Mining Lactonase Gene from Aflatoxin B1-Degrading Strain Bacillus Megaterium and Degrading Properties of the Recombinant Enzyme. J. Agric. Food Chem. 2023, 71, 20762–20771. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, X.; Cui, Y.; Yuan, W. Ultrasound for Microalgal Cell Disruption and Product Extraction: A Review. Ultrason. Sonochem. 2022, 87, 106054. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.S.; Vashist, A.; Kumar, M.; Taneja, N.K.; Gautam, U.S.; Dwivedi, S.; Tyagi, J.S.; Gupta, R.K. Anti-Mycobacterial Activity of Heat and pH Stable High Molecular Weight Protein(s) Secreted by a Bacterial Laboratory Contaminant. Microb. Cell Fact. 2022, 21, 15. [Google Scholar] [CrossRef]
- Rigon, F.; Pereira, D.A.B.; Loregian, K.E.; Magnani, E.; Marcondes, M.I.; Branco, R.H.; Benedeti, P.D.B.; Paula, E.M. Use of Heating Methods and Xylose to Increase Rumen Undegradable Protein of Alternative Protein Sources: 1) Peanut Meal. Animals 2022, 13, 23. [Google Scholar] [CrossRef]
- Winogradoff, D.; John, S.; Aksimentiev, A. Protein Unfolding by SDS: The Microscopic Mechanisms and the Properties of the SDS-Protein Assembly. Nanoscale 2020, 12, 5422–5434. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Bahuguna, A.; Lee, J.S.; Sood, A.; Han, S.S.; Chun, H.S.; Kim, M. Degradation Mechanism of Aflatoxin B1 and Aflatoxin G1 by Salt Tolerant Bacillus Albus YUN5 Isolated from ‘Doenjang’, a Traditional Korean Food. Food Res. Int. 2023, 165, 112479. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Liu, Y.; Zhang, G. Interaction of Aflatoxin B 1 and Fumonisin B 1 in HepG2 Cell Apoptosis. Food Biosci. 2017, 20, 131–140. [Google Scholar] [CrossRef]
- Tadee, A.; Mahakunakorn, P.; Porasuphatana, S. Oxidative Stress and Genotoxicity of Co-Exposure to Chlorpyrifos and Aflatoxin B1 in HepG2 Cells. Toxicol. Ind. Health 2020, 36, 336–345. [Google Scholar] [CrossRef]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 Degradation by Stenotrophomonas Maltophilia and Other Microbes Selected Using Coumarin Medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef]
- Ali, S.; Hassan, M.; Essam, T.; Ibrahim, M.A.; Al-Amry, K. Biodegradation of Aflatoxin by Bacterial Species Isolated from Poultry Farms. Toxicon 2021, 195, 7–16. [Google Scholar] [CrossRef]
- Xu, L.; Eisa Ahmed, M.; Sangare, L.; Zhao, Y.; Selvaraj, J.; Xing, F.; Wang, Y.; Yang, H.; Liu, Y. Novel Aflatoxin-Degrading Enzyme from Bacillus Shackletonii L7. Toxins 2017, 9, 36. [Google Scholar] [CrossRef]
- Suresh, G.; Cabezudo, I.; Pulicharla, R.; Cuprys, A.; Rouissi, T.; Brar, S.K. Biodegradation of Aflatoxin B1 with Cell-Free Extracts of Trametes Versicolor and Bacillus Subtilis. Res. Vet. Sci. 2020, 133, 85–91. [Google Scholar] [CrossRef]
- Yue, X.; Ren, X.; Fu, J.; Wei, N.; Altomare, C.; Haidukowski, M.; Logrieco, A.F.; Zhang, Q.; Li, P. Characterization and Mechanism of Aflatoxin Degradation by a Novel Strain of Trichoderma Reesei CGMCC3.5218. Front. Microbiol. 2022, 13, 1003039. [Google Scholar] [CrossRef]
- Shu, X.; Wang, Y.; Zhou, Q.; Li, M.; Hu, H.; Ma, Y.; Chen, X.; Ni, J.; Zhao, W.; Huang, S.; et al. Biological Degradation of Aflatoxin B1 by Cell-Free Extracts of Bacillus Velezensis DY3108 with Broad PH Stability and Excellent Thermostability. Toxins 2018, 10, 330. [Google Scholar] [CrossRef]
- Teniola, O.; Addo, P.; Brost, I.; Farber, P.; Jany, K.; Alberts, J.; Vanzyl, W.; Steyn, P.; Holzapfel, W. Degradation of Aflatoxin B1 by Cell-Free Extracts of Rhodococcus Erythropolis and Mycobacterium Fluoranthenivorans Sp. Nov. DSM44556T. Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Eshelli, M.; Harvey, L.; Edrada-Ebel, R.; McNeil, B. Metabolomics of the Bio-Degradation Process of Aflatoxin B1 by Actinomycetes at an Initial pH of 6.0. Toxins 2015, 7, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, J.; Liu, Z.; Shi, Y.; Liu, J.; Xu, X.; Hao, S.; Mu, P.; Deng, F.; Deng, Y. Aflatoxin B1 Degradation and Detoxification by Escherichia Coli CG1061 Isolated From Chicken Cecum. Front. Pharmacol. 2019, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Afsharmanesh, H.; Perez-Garcia, A.; Zeriouh, H.; Ahmadzadeh, M.; Romero, D. Aflatoxin Degradation by Bacillus Subtilis UTB1 Is Based on Production of an Oxidoreductase Involved in Bacilysin Biosynthesis. Food Control 2018, 94, 48–55. [Google Scholar] [CrossRef]
- González Pereyra, M.L.; Martínez, M.P.; Cavaglieri, L.R. Presence of aiiA Homologue Genes Encoding for N-Acyl Homoserine Lactone-Degrading Enzyme in Aflatoxin B1-Decontaminating Bacillus Strains with Potential Use as Feed Additives. Food Chem. Toxicol. 2019, 124, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, J.; Jin, Y.; Wu, C.; Shen, D.; Zhang, S.; Zhou, R. Mechanism and Kinetics of Degrading Aflatoxin B1 by Salt Tolerant Candida Versatilis CGMCC 3790. J. Hazard. Mater. 2018, 359, 382–387. [Google Scholar] [CrossRef] [PubMed]
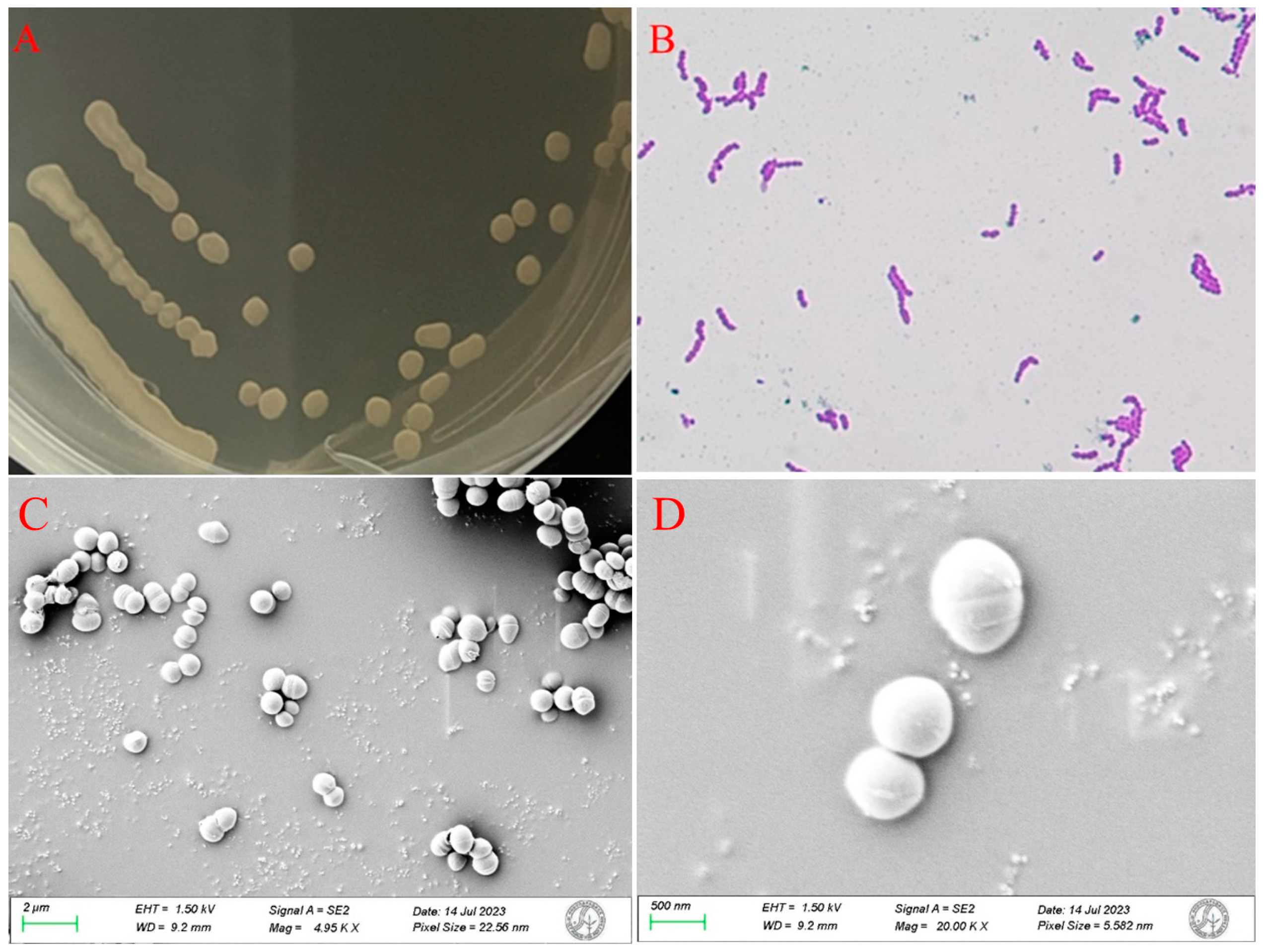



| Experimental Items | Results | Experimental Items | Results |
|---|---|---|---|
| Gram stain | + a | Oxidase | - |
| Methyl red test | - | Starch hydrolysis | + |
| Anaerobic growth | - | Tween 20 | + |
| Citrate utilization | - | Sucrose utilization | + |
| Propionate utilization | + | Glucose utilization | + |
| Lactose utilization | + | 2% NaCl | + |
| 7% NaCl | + | Growth at 4 °C | - |
| Growth at 28 °C | + | 37 °C growth | - |
| V-P test | - | pH6 | + |
| Contact enzymes | + | pH10 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, J.; Cao, L.; Du, X.; Zhang, Y.; Cong, Y.; He, J.; Zhang, W. Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2. Foods 2024, 13, 1887. https://doi.org/10.3390/foods13121887
Feng J, Cao L, Du X, Zhang Y, Cong Y, He J, Zhang W. Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2. Foods. 2024; 13(12):1887. https://doi.org/10.3390/foods13121887
Chicago/Turabian StyleFeng, Jiangtao, Ling Cao, Xiaoyan Du, Yvying Zhang, Yanxia Cong, Junbo He, and Weinong Zhang. 2024. "Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2" Foods 13, no. 12: 1887. https://doi.org/10.3390/foods13121887
APA StyleFeng, J., Cao, L., Du, X., Zhang, Y., Cong, Y., He, J., & Zhang, W. (2024). Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2. Foods, 13(12), 1887. https://doi.org/10.3390/foods13121887







