Polymorphisms within the PRKG1 Gene of Gannan Yaks and Their Association with Milk Quality Characteristics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Experimental Animals and Milk Composition Analysis
2.3. DNA Extraction
2.4. Genotyping
2.5. Statistical Analysis
3. Results
3.1. Analysis of PRKG1 Genotyping Results and Locus Genetic Parameters in Gannan Yak
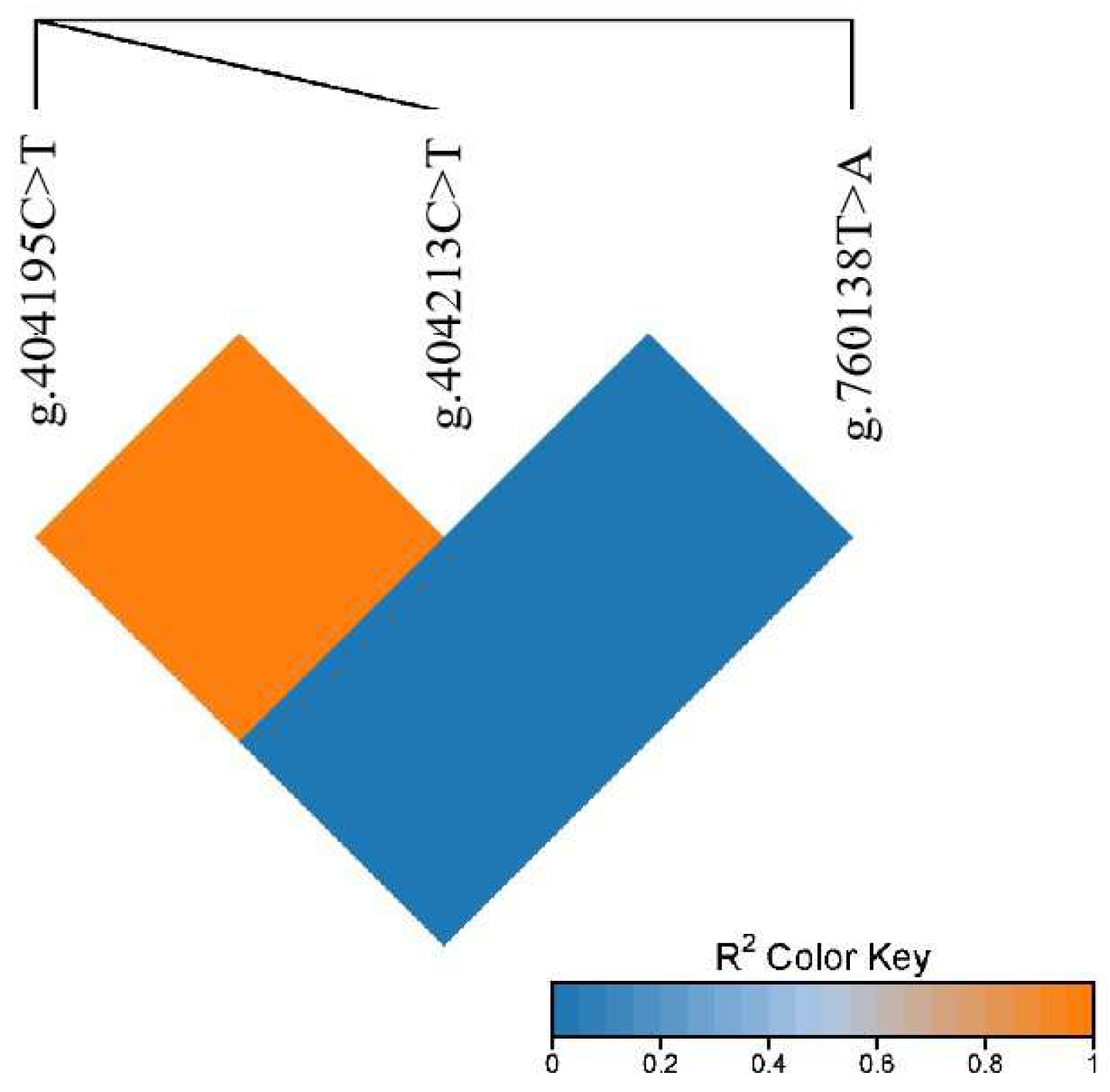
3.2. Linkage Disequilibrium Analysis of PRKG1 Gene SNPs in Gannan Yak
3.3. Association Analysis of Milk Traits and Genotypes of SNPs in Gannan Yak
4. Discussion
4.1. Genetic Polymorphism Analysis of the PRKG1 Gene in Gannan Yak
4.2. Correlation Analysis of PRKG1 Gene Polymorphism and Milk Quality Traits in Gannan Yak
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R.; et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; He, S.; Yu, Z.; Lan, D.; Xiong, X.; Li, Z. Transcriptomic study of yak mammary gland tissue during lactation. Anim. Biotechnol. 2022, 33, 672–679. [Google Scholar] [CrossRef]
- Cui, G.X.; Yuan, F.; Degen, A.A.; Liu, S.M.; Zhou, J.W.; Shang, Z.H.; Ding, L.M.; Mi, J.D.; Wei, X.H.; Long, R.J. Composition of the milk of yaks raised at different altitudes on the Qinghai-Tibetan Plateau. Int. Dairy J. 2016, 59, 29–35. [Google Scholar] [CrossRef]
- Li, H.; Yang, X.; Tang, D.; Xi, B.; Li, W.; Chen, Z.; Bao, Y.; Dingkao, R.; Gao, Y.; Wang, P.; et al. Exploring the link between microbial community structure and flavour compounds of traditional fermented yak milk in Gannan region. Food Chem. 2024, 435, 137553. [Google Scholar] [CrossRef]
- Li, F.; Cai, C.; Qu, K.; Liu, J.; Jia, Y.; Hanif, Q.; Chen, N.; Zhang, J.; Chen, H.; Huang, B.; et al. DGAT1 K232A polymorphism is associated with milk production traits in Chinese cattle. Anim. Biotechnol. 2021, 32, 427–431. [Google Scholar] [CrossRef]
- Čítek, J.; Brzáková, M.; Hanusová, L.; Hanuš, O.; Večerek, L.; Samková, E.; Křížová, Z.; Hoštičková, I.; Kávová, T.; Straková, K.; et al. Gene polymorphisms influencing yield, composition and technological properties of milk from Czech Simmental and Holstein cows. Anim. Biosci. 2021, 34, 2–11. [Google Scholar] [CrossRef]
- Li, C.; Sun, D.; Zhang, S.; Wang, S.; Wu, X.; Zhang, Q.; Liu, L.; Li, Y.; Qiao, L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS ONE 2014, 9, e96186. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Lv, X.; Liu, L.; Yang, Y.; Ma, Z.; Han, B.; Sun, D. A post-GWAS confirming effects of PRKG1 gene on milk fatty acids in a Chinese Holstein dairy population. BMC Genet. 2019, 20, 53. [Google Scholar] [CrossRef]
- Ni, J.; Xian, M.; Ren, Y.; Yang, L.; Li, Y.; Guo, S.; Ran, B.; Hu, J. Whole-genome resequencing reveals candidate genes associated with milk production trait in Guanzhong dairy goats. Anim. Genet. 2024, 55, 168–172. [Google Scholar] [CrossRef]
- Taye, M.; Kim, J.; Yoon, S.H.; Lee, W.; Hanotte, O.; Dessie, T.; Kemp, S.; Mwai, O.A.; Caetano-Anolles, K.; Cho, S.; et al. Whole genome scan reveals the genetic signature of African Ankole cattle breed and potential for higher quality beef. BMC Genet. 2017, 18, 11. [Google Scholar] [CrossRef]
- Ren, Y.; Chen, X.; Zheng, X.; Wang, F.; Sun, R.; Wei, L.; Zhang, Y.; Liu, H.; Lin, Y.; Hong, L.; et al. Diverse WGBS profiles of longissimus dorsi muscle in Hainan black goats and hybrid goats. BMC Genom. Data 2023, 24, 77. [Google Scholar] [CrossRef] [PubMed]
- Fishe, J.N.; Labilloy, G.; Higley, R.; Casey, D.; Ginn, A.; Baskovich, B.; Blake, K.V. Single Nucleotide Polymorphisms (SNPs) in PRKG1 & SPATA13-AS1 are associated with bronchodilator response: A pilot study during acute asthma exacerbations in African American children. Pharmacogenet Genom. 2021, 31, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Gago-Díaz, M.; Blanco-Verea, A.; Teixidó, G.; Huguet, F.; Gut, M.; Laurie, S.; Gut, I.; Carracedo, Á.; Evangelista, A.; Brion, M. PRKG1 and genetic diagnosis of early-onset thoracic aortic disease. Eur. J. Clin. Investig. 2016, 46, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, G.; Todeschini, P.; Paracchini, L.; Calura, E.; Fruscio, R.; Romani, C.; Beltrame, L.; Martini, P.; Ravaggi, A.; Ceppi, L.; et al. Expression profiles of PRKG1, SDF2L1 and PPP1R12A are predictive and prognostic factors for therapy response and survival in high-grade serous ovarian cancer. Int. J. Cancer 2020, 147, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.L.; Wong, J.C.; Johlfs, M.G.; Tsang, B.K.; Fiscus, R.R. Protein kinase G type Ialpha activity in human ovarian cancer cells significantly contributes to enhanced Src activation and DNA synthesis/cell proliferation. Mol. Cancer Res. 2010, 8, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.L.; Fraser, M.; Fiscus, R.R.; Tsang, B.K. Cisplatin alters nitric oxide synthase levels in human ovarian cancer cells: Involvement in p53 regulation and cisplatin resistance. Br. J. Cancer 2008, 98, 1803–1809. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Li, X.; Ma, Y.; Li, D. Differences in proteomic profiles of milk fat globule membrane in yak and cow milk. Food Chem. 2017, 221, 1822–1827. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, J.; Dai, R.; Ma, X.; Huang, C.; Ren, W.; Ma, X.; Lu, J.; Zhao, X.; Renqing, J.; et al. Comparative Study on Nutritional Characteristics and Volatile Flavor Substances of Yak Milk in Different Regions of Gannan. Foods 2023, 12, 2172. [Google Scholar] [CrossRef]
- Kalwar, Q.; Ma, X.; Xi, B.; Korejo, R.A.; Bhuptani, D.K.; Chu, M.; Yan, P. Yak milk and its health benefits: A comprehensive review. Front. Vet. Sci. 2023, 10, 1213039. [Google Scholar] [CrossRef]
- Abeygunawardana, D.I.; Ranasinghe, R.; De Silva, S.; Deshapriya, R.; Gamika, P.A.; Rajapakse, J. Effect of LHCGR and FSHR gene polymorphisms on fertility traits and milk yield of cross-bred dairy cows in Sri Lanka. Anim. Biotechnol. 2023, 34, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wen, Y.Y.; Liu, H.H.; Cao, H.Y.; Dong, X.Y.; Mao, H.G.; Yin, Z.Z. POMC gene expression, polymorphism, and the association with reproduction traits in chickens. Poult. Sci. 2020, 99, 2895–2901. [Google Scholar] [CrossRef]
- Li, M.; Sun, X.; Hua, L.; Lai, X.; Lan, X.; Lei, C.; Zhang, C.; Qi, X.; Chen, H. SIRT1 gene polymorphisms are associated with growth traits in Nanyang cattle. Mol. Cell. Probes 2013, 27, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, J.; Ma, X.; Ma, R.; Shen, J.; Liu, M.; Yu, D.; Feng, F.; Huang, C.; Ma, X.; et al. Polymorphisms of CCSER1 Gene and Their Correlation with Milk Quality Traits in Gannan Yak (Bos grunniens). Foods 2023, 12, 4318. [Google Scholar] [CrossRef]
- Yang, Y.; Han, L.; Yu, Q.; Gao, Y.; Song, R.; Zhao, S. Phosphoproteomic analysis of longissimus lumborum of different altitude yaks. Meat Sci. 2020, 162, 108019. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Raza, S.; Sun, Y.; Sabek, A.; Abbas, S.Q.; Shah, M.A.; Khan, R.; Abdelnour, S.A. Molecular characterization and analysis of the association of growth hormone 1 gene with growth traits in Chinese indigenous yak (Bos grunniens). Trop. Anim. Health Prod. 2021, 53, 221. [Google Scholar] [CrossRef] [PubMed]
- Serrote, C.; Reiniger, L.; Silva, K.B.; Rabaiolli, S.; Stefanel, C.M. Determining the Polymorphism Information Content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, F.; Gallais, A. Partitioning heterozygosity in subdivided populations: Some misuses of Nei’s decomposition and an alternative probabilistic approach. Mol. Ecol. 2020, 29, 2957–2962. [Google Scholar] [CrossRef]
- Zintzaras, E. Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. Eur. J. Epidemiol. 2010, 25, 553–560. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Hao, W.; Yin, W.; Ai, S.; Zhao, Y.; Duan, Z. Novel Single nucleotide polymorphisms and haplotype of MYF5 gene are associated with body measurements and ultrasound traits in grassland short-tailed sheep. Genes 2022, 13, 483. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Miladi, M.; Dukare, S.; Boulay, K.; Caudron-Herger, M.; Groß, M.; Backofen, R.; Diederichs, S. A pan-cancer analysis of synonymous mutations. Nat. Commun. 2019, 10, 2569. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Carmi, S.; Waldman, B.H.; Tkacz, I.D.; Naboishchikov, I.; Michaeli, S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J. Biol. Chem. 2013, 288, 4991–5006. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Guan, L.; Xuan, J.; Wang, H.; Yuan, Z.; Wu, M.; Liu, R.; Zhu, C.; Wei, C.; Zhao, F.; et al. Effect of polymorphisms in the CAMKMT gene on growth traits in Ujumqin sheep. Anim. Genet. 2016, 47, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Wang, W.; Zhang, D.; Li, C.; Huang, Y.; Ma, Z.; Wang, X.; Zhao, L.; Zhang, Y.; Yang, X.; et al. Identification of TRAPPC9 and BAIAP2 gene polymorphisms and their association with fat deposition-related traits in hu sheep. Front. Vet. Sci. 2022, 9, 928375. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, W.; Zhang, D.; Zhang, Y.; Zhao, Y.; Li, X.; Zhao, L.; Cheng, J.; Xu, D.; Yang, X.; et al. Polymorphisms of PLIN1 and MOGAT1 genes and their association with feed efficiency in Hu sheep. Gene 2024, 897, 148072. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhou, Y.; Zheng, X.; Guo, J.; Duan, H.; Zhou, S.; Yan, W. Yak Milk: Nutritional Value, Functional Activity, and Current Applications. Foods 2023, 12, 2090. [Google Scholar] [CrossRef] [PubMed]
- Or-Rashid, M.M.; Odongo, N.E.; Subedi, B.; Karki, P.; McBride, B.W. Fatty acid composition of yak (Bos grunniens) cheese including conjugated linoleic acid and trans-18:1 fatty acids. J. Agric. Food Chem. 2008, 56, 1654–1660. [Google Scholar] [CrossRef]
- Guo, X.; Long, R.; Kreuzer, M.; Ding, L.; Shang, Z.; Zhang, Y.; Yang, Y.; Cui, G. Importance of functional ingredients in yak milk-derived food on health of Tibetan nomads living under high-altitude stress: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 292–302. [Google Scholar] [CrossRef]
- Sheng, Q.; Li, J.; Alam, M.S.; Fang, X.; Guo, M. Gross composition and nutrient profiles of Chinese yak (Maiwa) milk. Int. J. Food Sci. Technol. 2008, 43, 568–572. [Google Scholar] [CrossRef]
- Lin, K.; Zhang, L.W.; Han, X.; Xin, L.; Meng, Z.X.; Gong, P.M.; Cheng, D.Y. Yak milk casein as potential precursor of angiotensin I-converting enzyme inhibitory peptides based on in silico proteolysis. Food Chem. 2018, 254, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Ma, Z.; Ramachandran, M.; De Souza, C.; Han, X.; Zhang, L. ACE inhibitory peptide KYIPIQ derived from yak milk casein induces nitric oxide production in HUVECs and diffuses via a transcellular mechanism in Caco-2 monolayers. Process Biochem. 2020, 99, 103–111. [Google Scholar] [CrossRef]
- Mao, X.; Cheng, X.; Wang, X.; Wu, S. Free-radical-scavenging and anti-inflammatory effect of yak milk casein before and after enzymatic hydrolysis. Food Chem. 2011, 126, 484–490. [Google Scholar] [CrossRef]
- Pei, J.; Jiang, H.; Li, X.; Jin, W.; Tao, Y. Antimicrobial peptides sourced from post-butter processing waste yak milk protein hydrolysates. AMB Express 2017, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- Garattini, S. Glutamic acid, twenty years later. J. Nutr. 2000, 130, 901S–909S. [Google Scholar] [CrossRef]
- Alim, M.A.; Wang, P.; Wu, X.P.; Li, C.; Cui, X.G.; Zhang, S.L.; Zhang, Q.; Zhang, Y.; Sun, D.X. Effect of FASN gene on milk yield and milk composition in the Chinese Holstein dairy population. Anim. Genet. 2014, 45, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, R.; Xu, L.; Su, D.; Li, Y.; Liu, L.; Ma, Z.; Sun, D.; Han, B. Fatty acid desaturase 2 affects the milk-production traits in Chinese Holsteins. Anim. Genet. 2022, 53, 422–426. [Google Scholar] [CrossRef]
- Khan, M.Z.; Wang, D.; Liu, L.; Usman, T.; Wen, H.; Zhang, R.; Liu, S.; Shi, L.; Mi, S.; Xiao, W.; et al. Significant genetic effects of JAK2 and DGAT1 mutations on milk fat content and mastitis resistance in Holsteins. J. Dairy Res. 2019, 86, 388–393. [Google Scholar] [CrossRef]
- Khan, M.Z.; Dari, G.; Khan, A.; Yu, Y. Genetic polymorphisms of TRAPPC9 and CD4 genes and their association with milk production and mastitis resistance phenotypic traits in Chinese Holstein. Front. Vet. Sci. 2022, 9, 1008497. [Google Scholar] [CrossRef]
| SNPs | Position | Genotypic Frequencies | Allelic Frequencies | He | Ne | PIC | p Value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| g.404195C>T | Intron | CC | CT | TT | C | T | 0.500 | 1.999 | 0.375 | 0.757 |
| 0.247 | 0.488 | 0.265 | 0.491 | 0.509 | ||||||
| g.404213C>T | Intron | CC | CT | TT | C | T | 0.500 | 1.999 | 0.375 | 0.757 |
| 0.247 | 0.488 | 0.265 | 0.491 | 0.509 | ||||||
| g.760138T>C | Intron | TT | TC | CC | T | C | 0.362 | 1.568 | 0.297 | 0.351 |
| 0.568 | 0.389 | 0.044 | 0.762 | 0.238 | ||||||
| SNPs g.404195C>T, SNPs g.404213C>T | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Casein % | Protein % | Fat % | SNFs % | Lactose % | Acidity °T | TSs % |
| CC | 4.10 ± 0.25 | 4.91 ± 0.33 | 6.07 ± 2.49 | 11.21 ± 0.43 | 4.95 ± 0.13 | 12.36 ± 1.20 | 17.2 ± 2.45 |
| CT | 4.02 ± 0.31 | 4.81 ± 0.41 | 5.47 ± 2.71 | 11.18 ± 0.48 | 4.99 ± 0.16 | 12.18 ± 1.28 | 16.54 ± 2.68 |
| TT | 4.18 ± 0.25 | 5.04 ± 0.37 | 5.24 ± 2.65 | 11.45 ± 0.44 | 4.98 ± 0.15 | 12.79 ± 1.30 | 16.55 ± 2.52 |
| p-Value | p = 0.012 | p = 0.006 | p = 0.34 | p = 0.007 | p = 0.358 | p = 0.044 | p = 0.383 |
| SNPs g.760138T>C | |||||||
| Genotype | Casein % | Protein % | Fat % | SNFs % | Lactose % | Acidity °T | TSs % |
| TT | 3.97 ± 0.27 | 4.78 ± 0.37 | 4.87 ± 2.24 | 11.17 ± 0.41 | 4.99 ± 0.13 | 12.18 ± 1.17 | 15.91 ± 2.18 |
| TC | 4.11 ± 0.29 | 4.90 ± 0.41 | 5.87 ± 2.64 | 11.26 ± 0.48 | 4.97 ± 0.16 | 12.40 ± 1.39 | 17.02 ± 2.58 |
| CC | 4.25 ± 0.22 | 5.09 ± 0.31 | 6.08 ± 3.20 | 11.46 ± 0.51 | 4.98 ± 0.15 | 12.79 ± 1.12 | 17.43 ± 3.04 |
| p-Value | p = 0.000 | p = 0.003 | p = 0.052 | p = 0.037 | p = 0.619 | p = 0.137 | p = 0.013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, F.; Yang, G.; Ma, X.; Zhang, J.; Huang, C.; Ma, X.; La, Y.; Yan, P.; Zhandui, P.; Liang, C. Polymorphisms within the PRKG1 Gene of Gannan Yaks and Their Association with Milk Quality Characteristics. Foods 2024, 13, 1913. https://doi.org/10.3390/foods13121913
Feng F, Yang G, Ma X, Zhang J, Huang C, Ma X, La Y, Yan P, Zhandui P, Liang C. Polymorphisms within the PRKG1 Gene of Gannan Yaks and Their Association with Milk Quality Characteristics. Foods. 2024; 13(12):1913. https://doi.org/10.3390/foods13121913
Chicago/Turabian StyleFeng, Fen, Guowu Yang, Xiaoyong Ma, Juanxiang Zhang, Chun Huang, Xiaoming Ma, Yongfu La, Ping Yan, Pingcuo Zhandui, and Chunnian Liang. 2024. "Polymorphisms within the PRKG1 Gene of Gannan Yaks and Their Association with Milk Quality Characteristics" Foods 13, no. 12: 1913. https://doi.org/10.3390/foods13121913






