Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Microbial Strains
2.2. Domain Analyses and Cloning of Linearized DNA
2.3. Recombinant Plasmid Construction and Purification of the Enzyme
2.4. Assay of Enzyme Activity
2.5. Bioinformatics Analysis, Structure Model Prediction and Molecular Docking
2.6. Function of PQQ-like Domain in Inclusion Body Formation
2.7. Characterization of the Purified OUC-CglA-DPQQ
2.8. Products’ Analyses of OUC-CglA-DPQQ
2.9. Hydrolysis Process Analysis
3. Results and Discussion
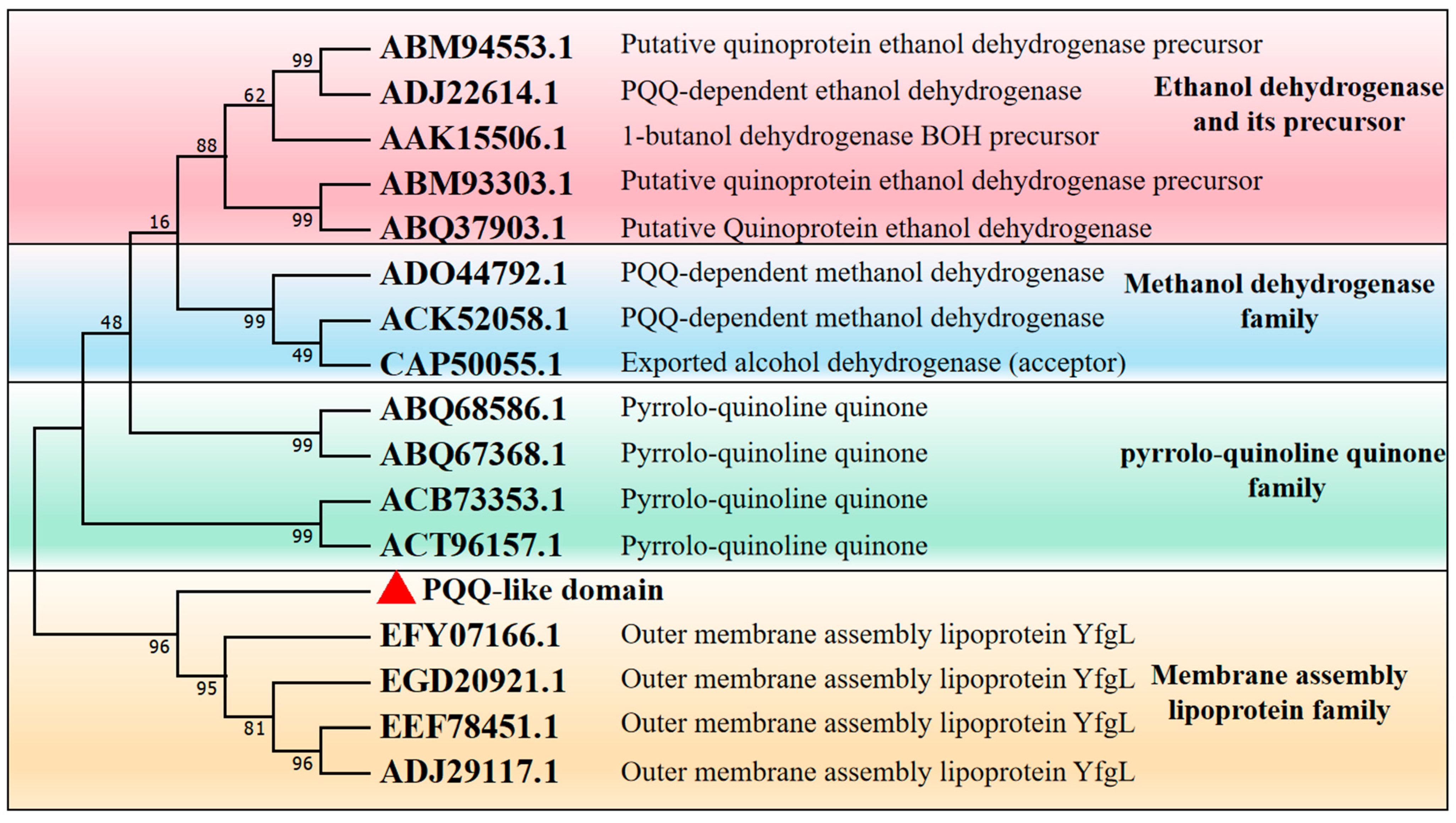
3.1. Sequence Analysis of λ-Carrageenase Mutant OUC-CglA-DPQQ
3.2. Heterologous Expression and Purification of OUC-CglA-DPQQ
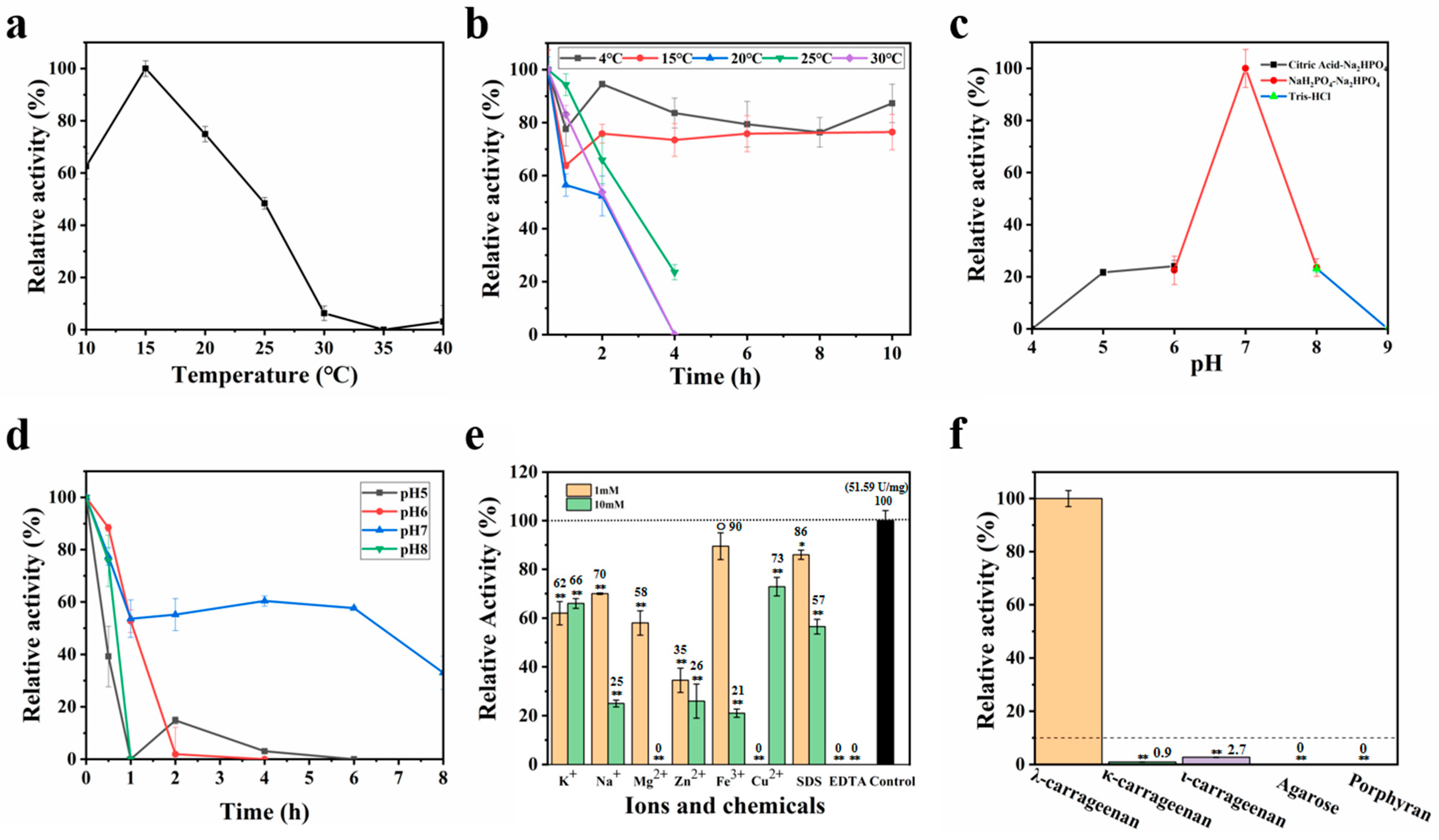
3.3. Characteristics of OUC-CglA-DPQQ
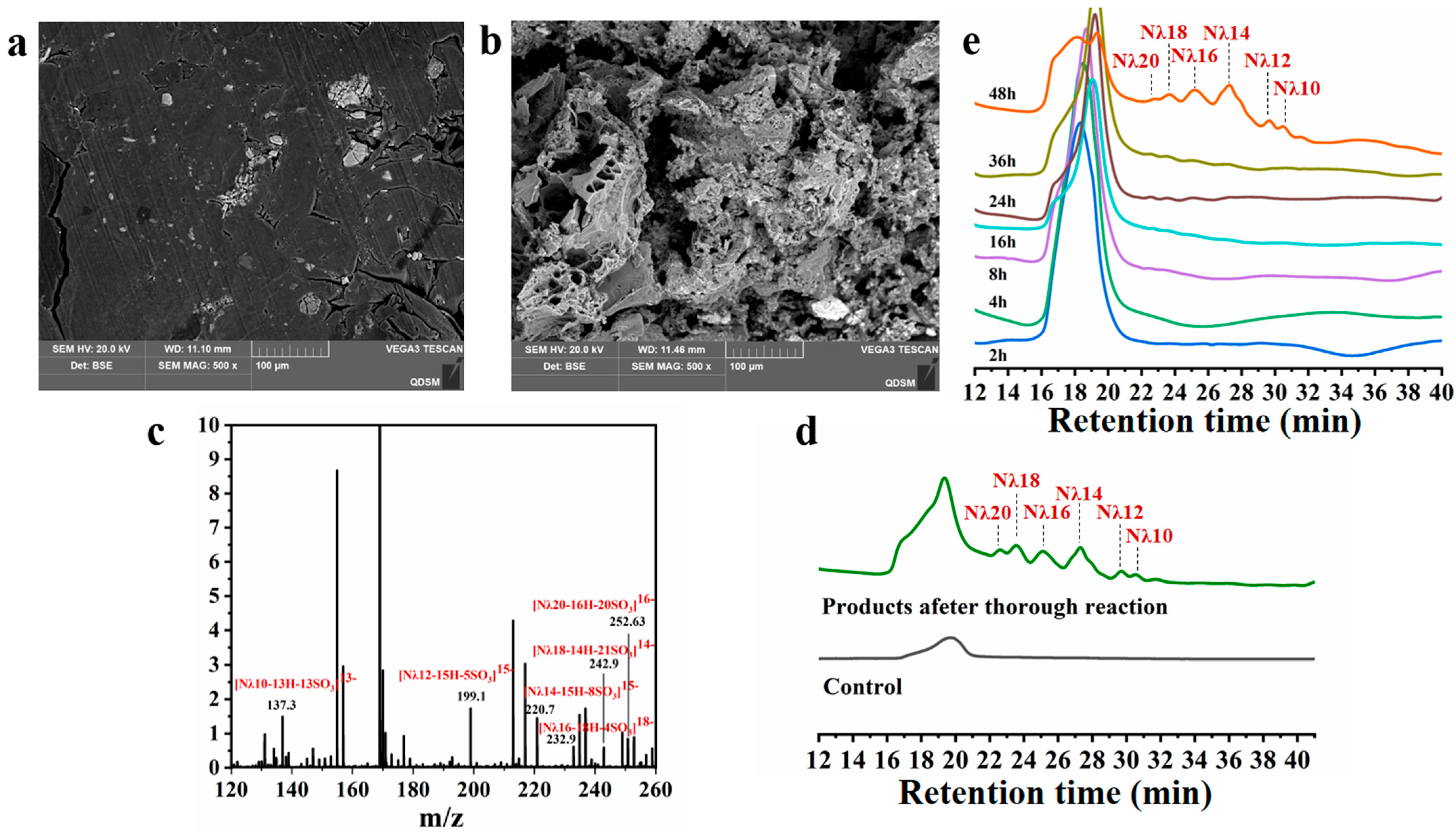
3.4. Product Analysis
3.5. Hydrolysis Process Analysis
3.6. Analysis of Catalytic Centre and Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campo, V.L. Carrageenans: Biological properties, chemical modifications and structural analysis-a review. Carbohydr. Polym. 2009, 77, 167–180. [Google Scholar] [CrossRef]
- Yu, G.; Guan, H.; Ioanoviciu, A.S.; Sikkander, S.A.; Thanawiroon, C.; Tobacman, J.K.; Toida, T.; Linhardt, R.J. Structural studies on κ-carrageenan derived oligosaccharides. Carbohydr. Res. 2002, 337, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wei, Y.; Zhang, Y.; Xu, Y.; Zheng, L.; Zhu, B.; Yao, Z. Carrageenan oligosaccharides: A comprehensive review of preparation, isolation, purification, structure, biological activities and applications. Algal Res. 2022, 61, 102593. [Google Scholar] [CrossRef]
- Zhu, B.; Ni, F.; Sun, Y.; Zhu, X.; Yin, H.; Yao, Z.; Du, Y. Insight into carrageenases: Major review of sources, category, property, purification method, structure, and applications. Crit. Rev. Biotechnol. 2018, 38, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, N.; Han, Q.; Li, X.; Jung, S.; Suk Lee, B.; Kumar Mongre, R.; Wang, Z.P.; Wang, L.; Lee, M.S. Production of a thermo-tolerant kappa-carrageenase via a food-grade host and anti-oxidant activity of its enzymatic hydrolysate. Food Chem. 2021, 339, 128027. [Google Scholar] [CrossRef] [PubMed]
- Kalitnik, A.A.; Anastyuk, S.D.; Sokolova, E.V.; Kravchenko, A.O.; Khasina, E.I.; Yermak, I.M. Oligosaccharides of κ/β-carrageenan from the red alga Tichocarpus crinitus and their ability to induce interleukin 10. J. Appl. Phycol. 2015, 28, 545–553. [Google Scholar] [CrossRef]
- Johnson, A.; Kong, F.; Miao, S.; Thomas, S.; Ansar, S.; Kong, Z.L. In-Vitro Antibacterial and Anti-Inflammatory Effects of Surfactin-Loaded Nanoparticles for Periodontitis Treatment. J. Nanomater. 2021, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Necas, J.; Bartosikova, L. Carrageenan: A review. Vet. Med. 2013, 58, 187–205. [Google Scholar] [CrossRef]
- Yuan, H.; Song, J. Preparation, structural characterization and in vitro anti-tumor activity of kappa-carrageenan oligosaccharide fraction from Kappaphycus striatum. J. Appl. Phycol. 2005, 17, 7–13. [Google Scholar] [CrossRef]
- Groult, H.; Cousin, R.; Chot-Plassot, C.; Maura, M.; Bridiau, N.; Piot, J.M.; Maugard, T.; Fruitier-Arnaudin, I. λ-Carrageenan Oligosaccharides of Distinct Anti-Heparanase and Anticoagulant Activities Inhibit MDA-MB-231 Breast Cancer Cell Migration. Mar. Drugs 2019, 17, 140. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Zhang, L.; Yan, Z.; Shen, J.; Chang, Y.; Wang, J. ι-Carrageenan Tetrasaccharide from ι-Carrageenan Inhibits Islet β Cell Apoptosis Via the Upregulation of GLP-1 to Inhibit the Mitochondrial Apoptosis Pathway. J. Agric. Food Chem. 2020, 69, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, Y.; Cai, W.; Wang, Q.; Chang, Y.; Sun, Y.; Dong, P.; Wang, J. Novel ι-Carrageenan Tetrasaccharide Alleviates Liver Lipid Accumulation via the Bile Acid−FXR−SHP/PXR Pathway to Regulate Cholesterol Conversion and Fatty Acid Metabolism in InsulinResistant Mice. J. Agric. Food Chem. 2021, 69, 9813–9821. [Google Scholar] [CrossRef] [PubMed]
- Masselin, A.; Rousseau, A.; Pradeau, S.; Fort, L.; Gueret, R.; Buon, L.; Armand, S.; Cottaz, S.; Choisnard, L.; Fort, S. Optimizing Chitin Depolymerization by Lysozyme to Long-Chain Oligosaccharides. Mar. Drugs 2021, 19, 320. [Google Scholar] [CrossRef] [PubMed]
- van Hoffen, E.; Ruiter, B.; Faber, J.; M’Rabet, L.; Knol, E.F.; Stahl, B.; Arslanoglu, S.; Moro, G.; Boehm, G.; Garssen, J. A specific mixture of short-chain galacto-oligosaccharides and long-chain fructo-oligosaccharides induces a beneficial immunoglobulin profile in infants at high risk for allergy. Allergy 2009, 64, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhu, B.; Yao, Z.; Jiang, J. Directed preparation, structure-activity relationship and applications of alginate oligosaccharides with specific structures: A systematic review. Food. Res. Int. 2023, 170, 112990. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Cheng, D.; Liu, Z.; Sun, J.; Mao, X. Advances in agaro-oligosaccharides preparation and bioactivities for revealing the structure-function relationship. Food. Res. Int. 2021, 145, 110408. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Hatada, Y. A novel enzyme, λ-carrageenase, isolated from a deep-sea bacterium. J. Biochem. 2006, 140, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, Q.; Seswita-Zilda, D. Purifification and characterization of a thermostable λ-carrageenase from a hot spring bacterium, Bacillus sp. Biotechnol. Lett. 2014, 36, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Ni, M. Study on λ-carrageenase of marine bacterium Cellulophaga sp. QY201. Master’s Thesis, Ocean University of China, Qingdao, China, 2008. [Google Scholar]
- Lu, Z.; Jiang, H.; Hamouda, H.I.; Wang, T.; Dong, Y.; Mao, X. Biochemical Characterization of a Cold-Adapted λ-Carrageenase OUC-CglA from Maribacter vaceletii: An Efficient Tool for λ-Carrageenan Degradation. J. Agric. Food Chem. 2022, 70, 12135–12142. [Google Scholar] [CrossRef] [PubMed]
- Guibet, M.; Colin, S.; Barbeyron, T.; Genicot, S.; Kloareg, B.; Michel, G.; Helbert, W. Degradation of lambda-carrageenan by Pseudoalteromonas carrageenovora lambda-carrageenase: A new family of glycoside hydrolases unrelated to kappa- and iota-carrageenases. Biochem. J. 2007, 404, 105–114. [Google Scholar] [CrossRef]
- Sun, Y.; Cao, S.; Zhang, Y.; Xue, C.; Xiao, H.; Chang, Y. Expression and Characterization of a Novel λ-Carrageenase Cgl150A_Wa from Wenyingzhuangia aestuarii. J. Ocean Univ. China. 2024, 23, 209–215. [Google Scholar] [CrossRef]
- Wei, Q.; Ran, T.; Ma, C.; He, J.; Xu, D.; Wang, W. Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway. J. Biol. Chem. 2016, 291, 15575–15587. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.E.; Jones, A.D.; Mercer, R.S.; Rucker, R.B. Characterization of pyrroloquinoline quinone amino acid derivatives by electrospray ionization mass spectrometry and detection in human milk. Anal. Biochem. 1999, 269, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Noji, N.; Nakamura, T.; Kitahata, N.; Taguchi, K.; Kudo, T.; Yoshida, S.; Tsujimoto, M.; Sugiyama, T.; Asami, T. Simple and sensitive method for pyrroloquinoline quinone (PQQ) analysis in various foods using liquid chromatography/electrospray-ionization tandem mass spectrometry. J. Agric. Food Chem. 2007, 55, 7258–7263. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Nukada, H.; Urakami, T.; Murphy, M.P. Antioxidant and pro-oxidant properties of pyrroloquinoline quinone (PQQ): Implications for its function in biological systems. Biochem. Pharmacol. 2003, 65, 67–74. [Google Scholar] [CrossRef]
- Misra, H.S.; Khairnar, N.P.; Barik, A.; Indira Priyadarsini, K.; Mohan, H.; Apte, S.K. Pyrroloquinoline-quinone: A reactive oxygen species scavenger in bacteria. FEBS Lett. 2004, 578, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, F.; Stites, T.E.; Anderson, P.; Storms, D.; Chan, I.; Eghbali, S.; Rucker, R. Pyrroloquinoline quinone improves growth and reproductive performance in mice fed chemically defined diets. Exp. Biol. Med. 2003, 228, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Kamimura, A.; Watanabe, F.; Kamiya, T.; Watanabe, D.; Yamamoto, E.; Fukagawa, M.; Hasumi, K.; Suzuki, E. Effects of orally administered pyrroloquinoline quinone disodium salt on dry skin conditions in mice and healthy female subjects. J. Nutr. Sci. Vitaminol. 2015, 61, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wen, C.; Lin, J.; Shen, G. Protective effect of pyrroloquinoline quinine on ultraviolet A irradiation-induced human dermalfibroblast senescence in vitro proceeds via the antiapoptotic sirtuin 1/nuclear factor derived erythroid 2-related factor 2/heme oxygenase 1 pathway. Mol. Med. Rep. 2015, 12, 4382–4388. [Google Scholar] [CrossRef][Green Version]
- Lumpe, H.; Daumann, L.J. Studies of Redox Cofactor Pyrroloquinoline Quinone and Its Interaction with Lanthanides (III) and Calcium (II). Inorg. Chem. 2019, 58, 8432–8441. [Google Scholar] [CrossRef] [PubMed]
- Heuck, A.; Schleiffer, A.; Clausen, T. Clausen, Augmenting β-Augmentation: Structural Basis of How BamB Binds BamA and May Support Folding of Outer Membrane Proteins. J. Mol. Biol. 2011, 406, 659–666. [Google Scholar] [CrossRef]
- Takeda, K.; Umezawa, K.; Várnai, A.; Eijsink, V.G.; Igarashi, K.; Yoshida, M.; Nakamura, N. Fungal PQQ-dependent dehydrogenases and their potential in biocatalysis. Curr. Opin. Chem. Biol. 2019, 49, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Xue, Y.T. Optimization of microwave assisted extraction, chemical characterization and antitumor activities of polysaccharides from Porphyra haitanensis. Carbohydr. Polym. 2019, 206, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Jiang, H.; Fu, L.; Ci, F.; Mao, X. Porphyran and oligoporphyran originating from red algae Porphyra: Preparation, biological activities, and potential applications. Food Chem. 2021, 349, 129209. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, X.; Zhang, Q.; Fu, L.; Tan, J.; Zhao, L. Biochemical Characterization of a Carrageenase, Car1383, Derived From Associated Bacteria of Antarctic macroalgae. Front. Microbiol. 2022, 13, 851182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Jiang, B.; Zhang, Y.; Sun, W.; Pu, Z.; Bao, Y. Purification and characterization of a cold-adapted κ-carrageenase from Pseudoalteromonas sp. ZDY3. Protein Expres Purif. 2021, 178, 105768. [Google Scholar] [CrossRef]
- Muzyed, S.; Howlader, M.M.; Tuvikene, R. Fermentation optimization, purification and biochemical characterization of ι-carrageenase from marine bacterium Cellulophaga baltica. Int. J. Biol. Macromol. 2021, 166, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Matard-Mann, M.; Bernard, T.; Leroux, C.; Barbeyron, T.; Larocque, R.; Préchoux, A.; Jeudy, A.; Jam, M.; Nyvall Collén, P.; Michel, G.; et al. Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases. J. Biol. Chem. 2017, 292, 19919–19934. [Google Scholar] [CrossRef] [PubMed]
- Michel, G.; Chantalat, L.; Fanchon, E.; Henrissat, B.; Kloareg, B.; Dideberg, O. The iota-carrageenase of Alteromonas fortis. A betahelix fold containing enzyme for the degradation of a highly polyanionic polysaccharide. J. Biol. Chem. 2001, 276, 40202–40209. [Google Scholar] [CrossRef] [PubMed]
- Michel, G.; Chantalat, L.; Duee, E.; Barbeyron, T.; Henrissat, B.; Kloareg, B.; Dideberg, O. The kappa-carrageenase of P. carrageenovora features a tunnel shaped active site: A novel insight in the evolution of Clan-B glycoside hydrolases. Structure 2001, 9, 513–525. [Google Scholar] [CrossRef] [PubMed]
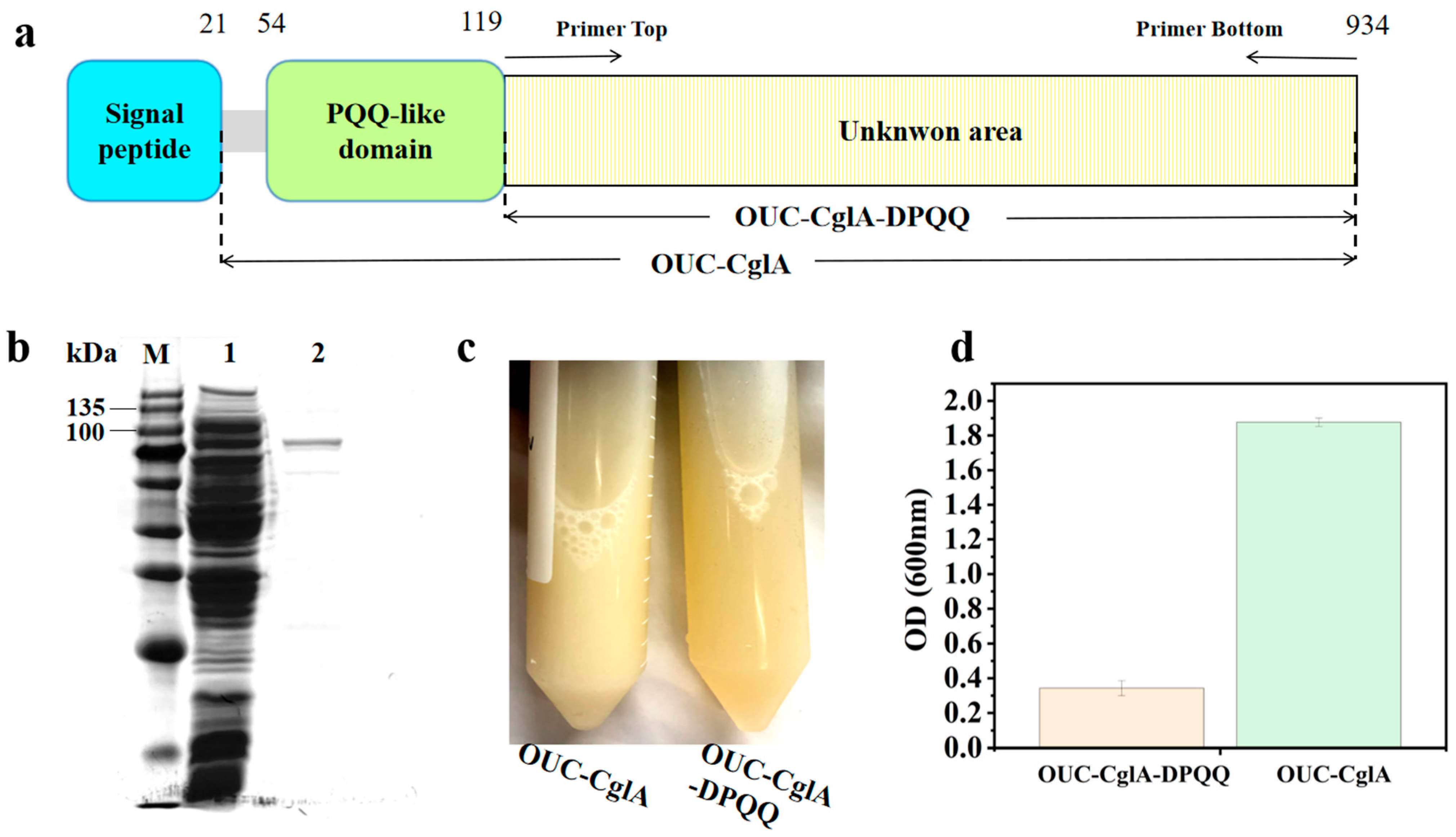

| Protein | Total Protein (mg) | Pure Enzyme (mg) | Pure Enzyme Ratio (%) | Specific Activity (U/mg) | Molecular Mass (kDa) |
|---|---|---|---|---|---|
| OUC-CglA-DPQQ | 84.438 | 3.28 | 3.9 | 51.59 | 93.7 |
| OUC-CglA | 59.248 | 0.68 | 1.1 | 418.68 | 105.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.; Jiang, H.; Yang, D.; Tang, H.; Hamouda, H.I.; Wang, T.; Mao, X. Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides. Foods 2024, 13, 1923. https://doi.org/10.3390/foods13121923
Lu Z, Jiang H, Yang D, Tang H, Hamouda HI, Wang T, Mao X. Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides. Foods. 2024; 13(12):1923. https://doi.org/10.3390/foods13121923
Chicago/Turabian StyleLu, Zewei, Hong Jiang, Dianqi Yang, Hengxin Tang, Hamed I. Hamouda, Tao Wang, and Xiangzhao Mao. 2024. "Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides" Foods 13, no. 12: 1923. https://doi.org/10.3390/foods13121923
APA StyleLu, Z., Jiang, H., Yang, D., Tang, H., Hamouda, H. I., Wang, T., & Mao, X. (2024). Characterization of a λ-Carrageenase Mutant with the Generation of Long-Chain λ-Neocarrageenan Oligosaccharides. Foods, 13(12), 1923. https://doi.org/10.3390/foods13121923








