Effect of a New Fermentation Strain Combination on the Fermentation Process and Quality of Highland Barley Yellow Wine
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains
2.2. Fermentation Process of Highland Barley Yellow Wine
2.3. Determination of Enzyme Activity during Fermentation
2.3.1. Determination of α-Amylase Activity
2.3.2. Determination of Glucoamylase Activity
2.3.3. Determination of Acid Protease Activity
2.4. Analysis of the Physicochemical Parameters of Highland Barley Yellow Wine
2.5. Measurements of the Electronic Tongue
2.6. HS-SPME–GC–MS Detection
2.6.1. HS-SPME Conditions
2.6.2. GC–MS Conditions
2.7. Amino Acid Determination
2.7.1. Sample Pretreatment with Hydrolyzed Amino Acids (Minimum Protein Content Requirement: 1 mg/mL)
2.7.2. Preparation of Standards
2.7.3. Buffer
2.7.4. Experimental Conditions of the Amino Acid Analyzer
2.8. Statistical Analysis
3. Results and Discussion
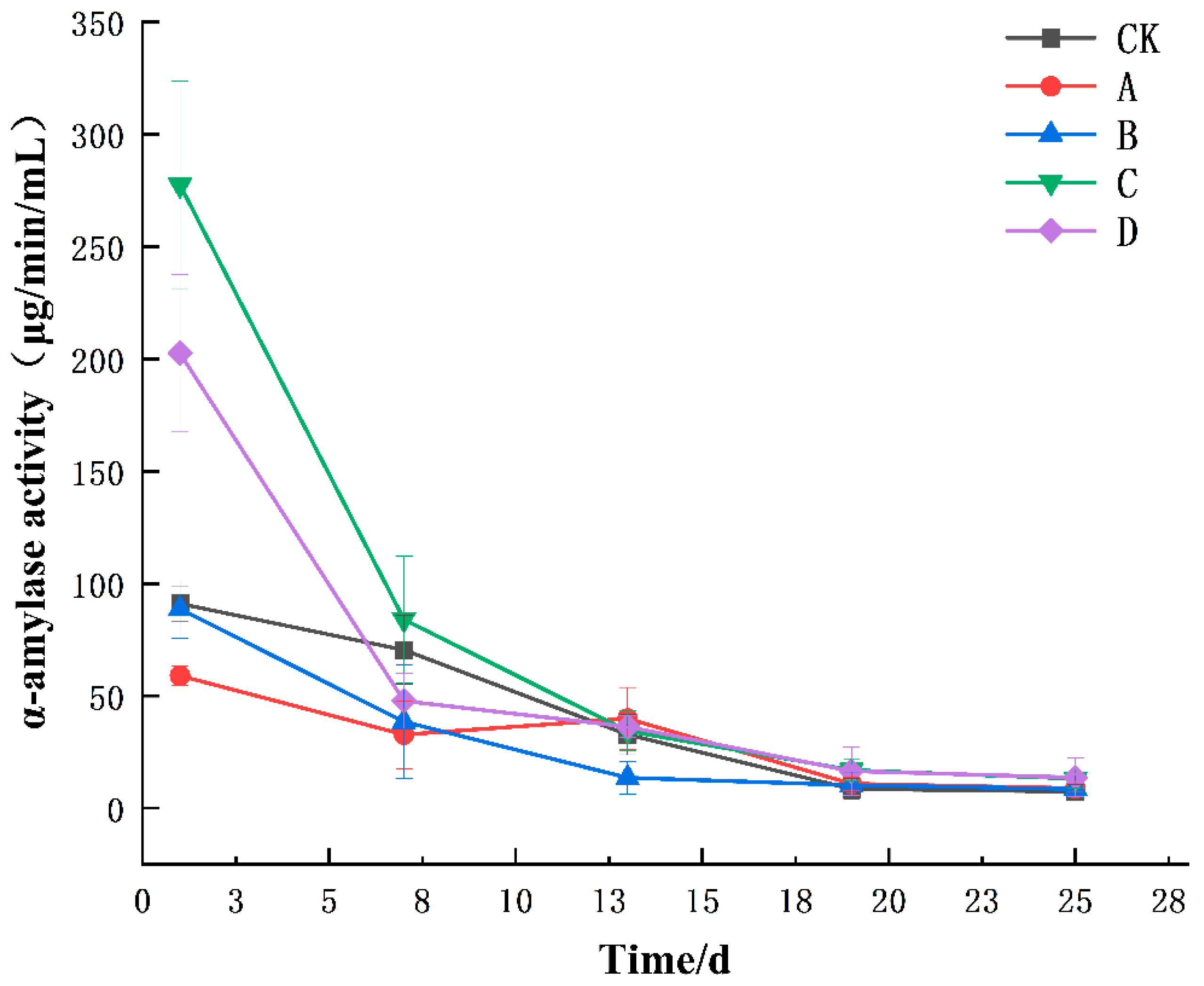
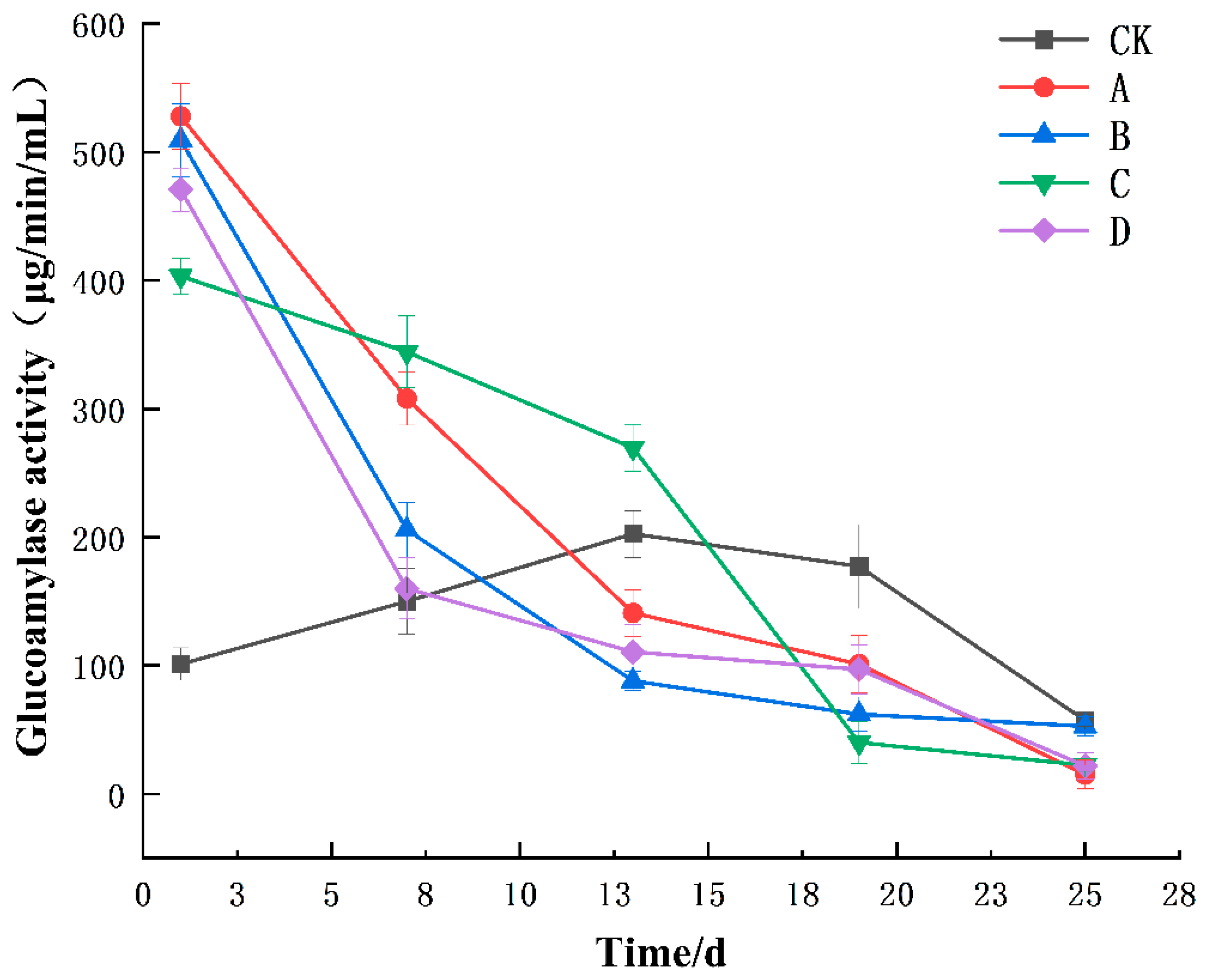
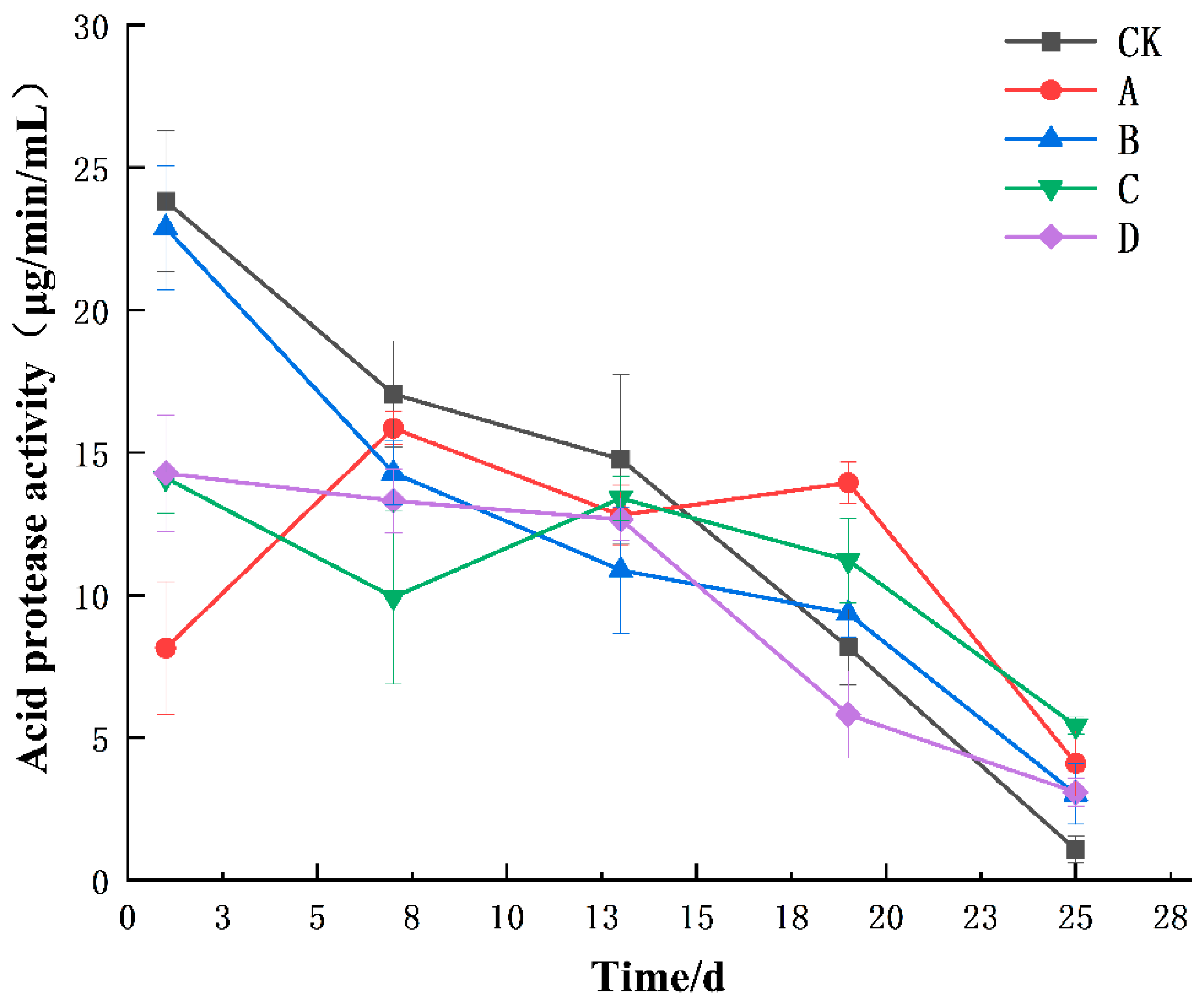
3.1. Changes in Enzyme Activities during the Fermentation of Yellow Wine
3.2. Physicochemical Properties of Yellow Wine
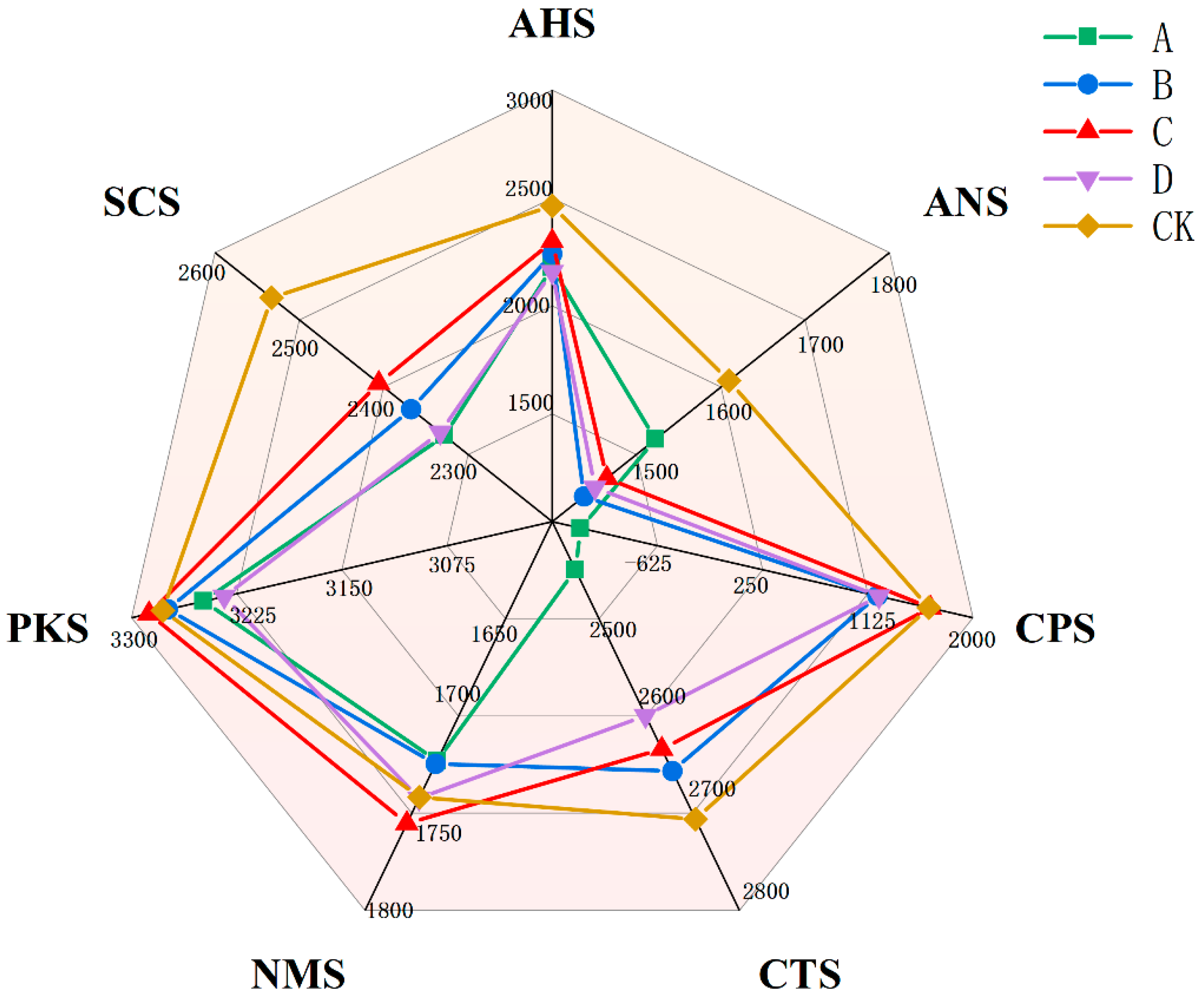
3.3. Electronic Tongue Detection
3.4. Volatile Compounds
3.5. Amino Acid Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Yuan, C.J.; Gao, X.L.; Kang, Y.L.; Huang, M.Q.; Wu, J.H.; Liu, Y.P.; Zhang, J.L.; Li, H.H.; Zhang, Y.Y. Characterization of key aroma compounds in Huangjiu from northern China by sensory-directed flavor analysis. Food Res. Int. 2020, 134, 109238. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Zheng, H.J.; Meng, K.; Yu, H.F.; Xie, G.F.; Zhang, Y.H.; Yang, X.Y.; Chen, J.L.; Xu, Z.Q.; Lin, Z.C.; et al. Quantitative study on core bacteria producing flavor substances in Huangjiu (Chinese yellow rice wine). LWT-Food Sci. Technol. 2022, 168, 113900. [Google Scholar] [CrossRef]
- Liang, Z.C.; Lin, X.Z.; He, Z.G.; Su, H.; Li, W.X.; Ren, X.Y. Amino acid and microbial community dynamics during the fermentation of Hong Qu glutinous rice wine. Food Microbiol. 2020, 90, 103467. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhang, J.; Ni, T.J.; Lin, N.; Meng, L.P.; Gao, F.D.; Luo, H.Q.; Liu, X.T.; Chi, J.F.; Guo, H.Y. Yellow Wine Polyphenolic Compounds prevents Doxorubicin-induced cardiotoxicity through activation of the Nrf2 signalling pathway. J. Cell. Mol. Med. 2019, 23, 6034–6047. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.D.; Tang, J.; Chen, Q.; Wu, P.G.; Han, J.L. Evaluation of direct sampling method for trace elements analysis in Chinese rice wine by ICP-OES. Eur. Food Res. Technol. 2013, 236, 531–535. [Google Scholar] [CrossRef]
- Evers, M.S.; Roullier-Gall, C.; Morge, C.; Sparrow, C.; Gobert, A.; Alexandre, H. Vitamins in wine: Which, what for, and how much? Compr. Rev. Food Sci. Food Saf. 2021, 20, 2991–3035. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Mao, J.; Chen, Y.Q.; Meng, X.Y.; Ji, Z.W. Extraction optimization of polysaccharides from Chinese rice wine from the Shaoxing region and evaluation of its immunity activities. J. Sci. Food Agric. 2015, 95, 1991–1996. [Google Scholar] [CrossRef]
- Han, F.L.; Xu, Y. Identification of Low Molecular Weight Peptides in Chinese Rice Wine (Huang Jiu) by UPLC-ESI-MS/MS. J. Inst. Brew. 2011, 117, 238–250. [Google Scholar] [CrossRef]
- Cai, H.Y.; Zhang, Q.; Shen, L.Z.; Luo, J.; Zhu, R.Y.; Mao, J.W.; Zhao, M.J.; Cai, C.G. Phenolic profile and antioxidant activity of Chinese rice wine fermented with different rice materials and starters. LWT-Food Sci. Technol. 2019, 111, 226–234. [Google Scholar] [CrossRef]
- Jiang, L.; Su, W.; Mu, Y.; Mu, Y. Major Metabolites and Microbial Community of Fermented Black Glutinous Rice Wine with Different Starters. Front. Microbiol. 2020, 11, 593. [Google Scholar] [CrossRef]
- Ji, Z.W.; Jin, J.S.; Yu, G.S.; Mou, R.; Mao, J.; Liu, S.P.; Zhou, Z.L.; Peng, L. Characteristic of filamentous fungal diversity and dynamics associated with wheat Qu and the traditional fermentation of Chinese rice wine. Int. J. Food Sci. Technol. 2018, 53, 1611–1621. [Google Scholar] [CrossRef]
- Zhang, B.; Kong, L.Q.; Cao, Y.; Xie, G.F.; Guan, Z.B.; Lu, J. Metaproteomic characterisation of a Shaoxing rice wine “wheat Qu” extract. Food Chem. 2012, 134, 387–391. [Google Scholar] [CrossRef]
- Yang, Y.J.; Xia, Y.J.; Wang, G.Q.; Zhang, H.; Xiong, Z.Q.; Yu, J.S.; Yu, H.Y.; Ai, L.Z. Comparison of oenological property, volatile profile, and sensory characteristic of Chinese rice wine fermented by different starters during brewing. Int. J. Food Prop. 2018, 20, S3195–S3211. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y. Effect of ‘wheat Qu’ on the fermentation processes and volatile flavour-active compounds of Chinese rice wine (Huangjiu). J. Inst. Brew. 2013, 119, 71–77. [Google Scholar] [CrossRef]
- Chai, C.; Lim, G.S.; Kim, Y.J.; Oh, S.W. Microbial community changes in Makgeolli during brewing. J. Inst. Brew. 2015, 121, 304–308. [Google Scholar] [CrossRef]
- Luangkhlaypho, A.; Pattaragulwanit, K.; Leepipatpiboon, N.; Yompakdee, C. Development of a defined starter culture mixture for the fermentation of sato, a Thai rice-based alcoholic beverage. Scienceasia 2014, 40, 125–134. [Google Scholar] [CrossRef]
- Lei, Y.; Cai, W.; Wang, Y.; Guo, Z.; Shan, C. Fungal communities and their correlation with the sensory quality of rice wine from the Xiaogan and Dazhou regions in China. LWT 2024, 191, 115575. [Google Scholar] [CrossRef]
- Chen, L.; Wang, S.; Liu, B.; Feng, S. Flavor characteristics of hulless barley wine fermented with mixed starters by molds and yeasts isolated from Jiuqu. Food Biosci. 2023, 52, 102349. [Google Scholar] [CrossRef]
- Guo, L.; Luo, Y.; Zhou, Y.; Bianba, C.; Guo, H.; Zhao, Y.; Fu, H. Exploring microbial dynamics associated with flavours production during highland barley wine fermentation. Food Res. Int. 2020, 130, 108971. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Zeng, W.; Zhou, J.; Du, G. Correlation between the microbial community and ethyl carbamate generated during Huzhou rice wine fermentation. Food Res. Int. 2022, 154, 111001. [Google Scholar] [CrossRef] [PubMed]
- Londoño-Hernández, L.; Ramírez-Toro, C.; Ruiz, H.A.; Ascacio-Valdés, J.A.; Aguilar-Gonzalez, M.A.; Rodríguez-Herrera, R.; Aguilar, C.N. Rhizopus oryzae—Ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 2017, 257, 110–127. [Google Scholar] [CrossRef] [PubMed]
- Loira, I.; Morata, A.; Comuzzo, P.; Callejo, M.J.; González, C.; Calderón, F.; Suárez-Lepe, J.A. Use of Schizosaccharomyces pombe and Torulaspora delbrueckii strains in mixed and sequential fermentations to improve red wine sensory quality. Food Res. Int. 2015, 76, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Larroque, M.N.; Carrau, F.; Fariña, L.; Boido, E.; Dellacassa, E.; Medina, K. Effect of Saccharomyces and non-Saccharomyces native yeasts on beer aroma compounds. Int. J. Food Microbiol. 2021, 337, 108953. [Google Scholar] [CrossRef] [PubMed]
- Ly, S.; Mith, H.; Tarayre, C.; Taminiau, B.; Daube, G.; Fauconnier, M.L.; Delvigne, F. Impact of Microbial Composition of Cambodian Traditional Dried Starters (Dombea) on Flavor Compounds of Rice Wine: Combining Amplicon Sequencing with HP-SPME-GCMS. Front. Microbiol. 2018, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Nguyen, T.T.H.; Jin, J.H.; Lim, J.; Lee, J.; Piao, M.Z.; Mok, I.; Kim, D. Brewing of glucuronic acid-enriched apple cider with enhanced antioxidant activities through the co-fermentation of yeast (Saccharomyces cerevisiae and Pichia kudriavzevii) and bacteria (Lactobacillus plantarum). Food Sci. Biotechnol. 2021, 30, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Ciosek, A.; Fulara, K.; Hrabia, O.; Satora, P.; Poreda, A. Chemical Composition of Sour Beer Resulting from Supplementation the Fermentation Medium with Magnesium and Zinc Ions. Biomolecules 2020, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.B.; Du, J.; Cao, C.L.; Cai, G.L.; Sun, J.Y.; Wu, D.H.; Lu, J. Development of a novel multi-strain wheat Qu with high enzyme activities for Huangjiu fermentation. J. Sci. Food Agric. 2021, 101, 4808–4817. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, L.; Zhou, Y.; He, S.D.; Li, J.L.; Sun, H.J.; Yao, S.F.; Xu, S.Y. Effect of mixed moulds starters on volatile flavor compounds in rice wine. LWT-Food Sci. Technol. 2019, 112, 108215. [Google Scholar] [CrossRef]
- Ivey, M.; Massel, M.; Phister, T.G. Microbial Interactions in Food Fermentations. Annu. Rev. Food Sci. Technol. 2013, 4, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Li, D.N.; Ren, L.X.; Song, S.Q.; Ma, X.; Rong, Y.Z. Effects of simultaneous and sequential cofermentation of Wickerhamomyces anomalus and Saccharomyces cerevisiae on physicochemical and flavor properties of rice wine. Food Sci. Nutr. 2021, 9, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xu, Y.; Wang, D. Effects of simultaneous and sequential mixed fermentation of non-Saccharomyces strains and a Saccharomyces cerevisiae strain on the fermentation process and volatile compounds of Mijiu. Int. J. Food Sci. Technol. 2023, 58, 6576–6587. [Google Scholar] [CrossRef]
- Li, P.P.; Su, R.; Wang, Q.; Liu, K.Y.; Yang, H.; Du, W.; Li, Z.A.; Chen, S.; Xu, B.; Yang, W. Comparison of fungal communities and nonvolatile flavor components in black Huangjiu formed using different inoculation fermentation methods. Front. Microbiol. 2022, 13, 955825. [Google Scholar] [CrossRef]
- Yoo, Y.J.; Hong, J.; Hatch, R.T. Comparison of alpha-amylase activities from different assay methods. Biotechnol. Bioeng. 1987, 30, 147–151. [Google Scholar] [CrossRef]
- Li, Z.M.; Bai, Z.H.; Wang, D.L.; Zhang, W.J.; Zhang, M.; Lin, F.; Gao, L.P.; Hui, B.D.; Zhang, H.X. Cultivable bacterial diversity and amylase production in three typical Daqus of Chinese spirits. Int. J. Food Sci. Technol. 2014, 49, 776–786. [Google Scholar] [CrossRef]
- Wei, J.P.; Zhang, Y.X.; Yuan, Y.H.; Dai, L.; Yue, T.L. Characteristic fruit wine production via reciprocal selection of juice and non-Saccharomyces species. Food Microbiol. 2019, 79, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.P.; Zhang, Y.X.; Qiu, Y.; Guo, H.; Ju, H.M.; Wang, Y.W.; Yuan, Y.H.; Yue, T.L. Chemical composition, sensorial properties, and aroma-active compounds of ciders fermented with Hanseniaspora osmophila and Torulaspora quercuum in co- and sequential fermentations. Food Chem. 2020, 306, 125623. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Toko, K. Electronic Tongues—A Review. IEEE Sens. J. 2013, 13, 3001–3011. [Google Scholar] [CrossRef]
- Qian, M.; Ruan, F.; Zhao, W.; Dong, H.; Bai, W.; Li, X.; Huang, X.; Li, Y. The dynamics of physicochemical properties, microbial community, and flavor metabolites during the fermentation of semi-dry Hakka rice wine and traditional sweet rice wine. Food Chem. 2023, 416, 135844. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Siddique, F.; Mahmood, M.S.; Ahmed, S.I. A Review of the Microbiological Aspect of α-amylase Production. Int. J. Agric. Biol. 2013, 15, 1029–1034. [Google Scholar]
- Kim, M.S.; Park, J.T.; Kim, Y.W.; Lee, H.S.; Nyawira, R.; Shin, H.S.; Park, C.S.; Yoo, S.H.; Kim, Y.R.; Moon, T.W.; et al. Properties of a novel thermostable glucoamylase from the hyperthermophilic archaeon Sulfolobus solfataricus in relation to starch processing. Appl. Environ. Microbiol. 2004, 70, 3933–3940. [Google Scholar] [CrossRef]
- Pan, S.K.; Wu, S.J.; Kim, J.M. Preparation of glucosamine by hydrolysis of chitosan with commercial α-amylase and glucoamylase. J. Zhejiang Univ. Sci. B 2011, 12, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Shiraga, S.; Ueda, M.; Takahashi, S.; Tanaka, A. Construction of the combinatorial library of Rhizopus oryzae lipase mutated in the lid domain by displaying on yeast cell surface. J. Mol. Catal. B Enzym. 2002, 17, 167–173. [Google Scholar] [CrossRef]
- Nishise, H.; Fuji, A.; Ueno, M.; Vongsuvanlert, V.; Tani, Y. Production of raw cassava starch-digestive glucoamylase by Rhizopus sp. in liquid culture. J. Ferment. Technol. 1988, 66, 397–402. [Google Scholar] [CrossRef]
- Huang, Z.R.; Hong, J.L.; Xu, J.X.; Li, L.; Guo, W.L.; Pan, Y.Y.; Chen, S.J.; Bai, W.D.; Rao, P.F.; Ni, L.; et al. Exploring core functional microbiota responsible for the production of volatile flavour during the traditional brewing of Wuyi Hong Qu glutinous rice wine. Food Microbiol. 2018, 76, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.Y.; Zhang, T.; Zhang, Q.; Luo, J.; Cai, C.G.; Mao, J.W. Microbial diversity and chemical analysis of the starters used in traditional Chinese sweet rice wine. Food Microbiol. 2018, 73, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadi, S.; de Hoog, G.S.; Meis, J.F.; Walther, G. Species boundaries and nomenclature of Rhizopus arrhizus (syn. R. oryzae). Mycoses 2014, 57, 108–127. [Google Scholar] [CrossRef] [PubMed]
- Turgut, T.; Diler, A. The effect of addition Eriobotrya japonica L. marmalade on physicochemical, microbiological, and sensory properties of probiotic yogurts. Front. Nutr. 2023, 10, 1151037. [Google Scholar] [CrossRef] [PubMed]
- Kamel, D.G.; Othman, A.A.; Osman, D.M.; Hammam, A.R.A. Probiotic yogurt supplemented with nanopowdered eggshell: Shelf-life stability, physicochemical, and sensory characteristics. Food Sci. Nutr. 2021, 9, 1736–1742. [Google Scholar] [CrossRef] [PubMed]
- Montanuci, F.D.; Pimentel, T.C.; Garcia, S.; Prudencio, S.H. Effect of starter culture and inulin addition on microbial viability, texture, and chemical characteristics of whole or skim milk Kefir. Food Sci Technol. 2012, 32, 850–861. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y.; Qian, M.C. Comparison of the aromatic profile of traditional and modern types of Huang Jiu (Chinese rice wine) by aroma extract dilution analysis and chemical analysis. Flavour Fragr. J. 2018, 33, 263–271. [Google Scholar] [CrossRef]
- Yang, Y.J.; Xia, Y.J.; Wang, G.Q.; Yu, J.S.; Ai, L.Z. Effect of mixed yeast starter on volatile flavor compounds in Chinese rice wine during different brewing stages. LWT-Food Sci. Technol. 2017, 78, 373–381. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; González-Arenzana, L.; Portu, J.; Garijo, P.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R. Wine aroma evolution throughout alcoholic fermentation sequentially inoculated with non-Saccharomyces/Saccharomyces yeasts. Food Res. Int. 2018, 112, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Zdaniewicz, M.; Satora, P.; Pater, A.; Bogacz, S. Low Lactic Acid-Producing Strain of Lachancea thermotolerans as a New Starter for Beer Production. Biomolecules 2020, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Li, Y.; Lou, Y.; Zhao, Y.; Feng, X.; Li, P.; Laaksonen, O.; Yang, B.; Capozzi, V.; Liu, S. Selecting autochthonous lactic acid bacteria for co-inoculation in Chinese bayberry wine production: Stress response, starter cultures application and volatilomic study. Food Res. Int. 2024, 178, 113976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.L.; Ji, Z.W.; Liu, S.P.; Han, X.; Zheng, F.P.; Mao, J. Characterization of the volatile compounds of huangjiu using comprehensive two-dimensional gas chromatography coupled to time of flight mass spectrometry (GC × GC-TOFMS). J. Food Process. Preserv. 2019, 43, e14159. [Google Scholar] [CrossRef]
- Fujii, T.; Yoshimoto, H.; Nagasawa, N.; Bogaki, T.; Tamai, Y.; Hamachi, M. Nucleotide sequences of alcohol acetyltransferase genes from lager brewing yeast, Saccharomyces carlsbergensis. Yeast 1996, 12, 593–598. [Google Scholar] [CrossRef]
- Inoue, Y.; Trevanichi, S.; Fukuda, K.; Izawa, S.; Wakai, Y.; Kimura, A. Roles of Esterase and Alcohol Acetyltransferase on Production of Isoamyl Acetate in Hansenula mrakii. J. Agric. Food Chem. 1997, 45, 644–649. [Google Scholar] [CrossRef]
- Mason, A.B.; Dufour, J.P. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast 2000, 16, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Avila, O.; Sánchez, A.; Font, X.; Barrena, R. Bioprocesses for 2-phenylethanol and 2-phenylethyl acetate production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 9991–10004. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Zhang, H.; Lu, X.Y.; Hong, Z.; Bin, Z.G. Advances in 2-phenylethanol production from engineered microorganisms. Biotechnol. Adv. 2019, 37, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-H.; Chai, L.-J.; Wang, H.-M.; Lu, Z.-M.; Zhang, X.-J.; Xiao, C.; Wang, S.-T.; Shen, C.-H.; Shi, J.-S.; Xu, Z.-H. Community-level bioaugmentation results in enzymatic activity- and aroma-enhanced Daqu through altering microbial community structure and metabolic function. Food Biosci. 2024, 57, 103630. [Google Scholar] [CrossRef]
- Kim, B.; Cho, B.R.; Hahn, J.S. Metabolic Engineering of Saccharomyces cerevisiae for the Production of 2-Phenylethanol via Ehrlich Pathway. Biotechnol. Bioeng. 2014, 111, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.F.; Gu, B.T.; Xiong, D.W.; Huang, G.C.; Huang, X.P.; Liu, L.; Xiao, J. A Transcriptomic Analysis of Saccharomyces cerevisiae Under the Stress of 2-Phenylethanol. Curr. Microbiol. 2018, 75, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, M.; Ren, T.; Wang, J.; Niu, C.; Zheng, F.; Li, Q. Effect of Saccharomyces cerevisiae and non-Saccharomyces strains on alcoholic fermentation behavior and aroma profile of yellow-fleshed peach wine. LWT 2022, 155, 112993. [Google Scholar] [CrossRef]
- Chen, L.H.; Ren, L.X.; Li, D.N.; Ma, X. Analysis of microbiomes in three traditional starters and volatile components of the Chinese rice wines. Food Sci. Biotechnol. 2021, 30, 87–96. [Google Scholar] [CrossRef]
- Tian, Y.T.; Huang, J.M.; Xie, T.T.; Huang, L.Q.; Zhuang, W.J.; Zheng, Y.F.; Zheng, B.D. Oenological characteristics, amino acids and volatile profiles of Hongqu rice wines during pottery storage: Effects of high hydrostatic pressure processing. Food Chem. 2016, 203, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Ying, Y.B.; Li, B.B.; Zheng, Y.F.; Qing, Z.G. Multivariate classification of rice wines according to ageing time and brand based on amino acid profiles. Food Chem. 2011, 129, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.X.; Mao, J.; Meng, X.Y.; Li, X.Z.; Liu, Y.Y.; Feng, H. Changes in flavour characteristics and bacterial diversity during traditional fermentation of Chinese rice wines from Shaoxing region. Food Control 2014, 44, 58–63. [Google Scholar] [CrossRef]
- Zeng, X.A.; Yu, S.J.; Zhang, L.; Chen, X.D. The effects of AC electric field on wine maturation. Innov. Food Sci. Emerg. Technol. 2008, 9, 463–468. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Rotzoll, N.; Dunkel, A.; Hofmann, T. Quantitative studies, taste reconstitution, and omission experiments on the key taste compounds in morel mushrooms (Morchella deliciosa Fr.). J. Agric. Food Chem. 2006, 54, 2705–2711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Z.; Pan, Y.F.; Zou, W.; Zhou, L.H.; Wu, Z.Y.; Zhang, W.X. Nutritive assessment of amino acids for three Chinese Zajius produced from hull-less barley. J. Inst. Brew. 2017, 123, 587–593. [Google Scholar] [CrossRef]
- Santos, M.C.; Nunes, C.; Rocha, M.A.M.; Rodrigues, A.; Rocha, S.M.; Saraiva, J.A.; Coimbra, M.A. Impact of high pressure treatments on the physicochemical properties of a sulphur dioxide-free white wine during bottle storage: Evidence for Mail lard reaction acceleration. Innov. Food Sci. Emerg. Technol. 2013, 20, 51–58. [Google Scholar] [CrossRef]
- Chen, X.R.; Wang, Z.Y.; Guo, X.N.; Liu, S.; He, X.P. Regulation of general amino acid permeases Gap1p, GATA transcription factors Gln3p and Gat1p on 2-phenylethanol biosynthesis via Ehrlich pathway. J. Biotechnol. 2017, 242, 83–91. [Google Scholar] [CrossRef]
- Celinska, E.; Borkowska, M.; Bialas, W.; Kubiak, M.; Korpys, P.; Archacka, M.; Ledesma-Amaro, R.; Nicaud, J.M. Genetic engineering of Ehrlich pathway modulates production of higher alcohols in engineered Yarrowia lipolytica. FEMS Yeast Res. 2019, 19, foy122. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, G.; Knijnenburg, T.A.; Liti, G.; Louis, E.J.; Pronk, J.T.; Daran, J.M. Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast 2015, 32, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.X.; Diao, R.Q.; Zhang, J.H.; Cao, M.Y.; Gao, H.L.; Tang, B.B. Tetramethyl pyrazine exerts anti-apoptotic and antioxidant effects in a mouse model of MPTP-induced Parkinson’s disease via regulation of the expressions of Bax, Bcl-2, Nrf2 and GCLC. Trop. J. Pharm. Res. 2021, 20, 893–898. [Google Scholar] [CrossRef]
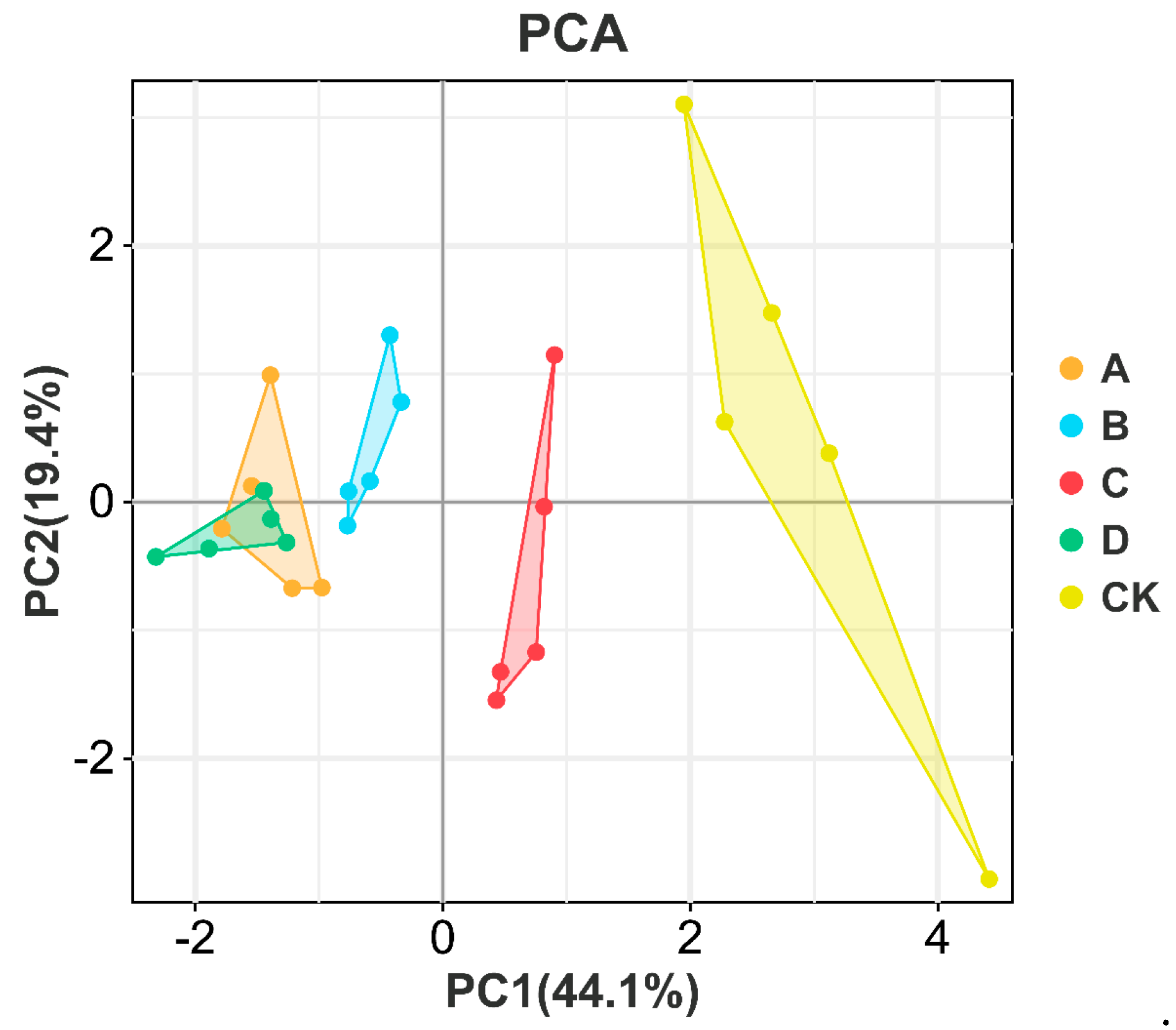

| Clusters | Mixed Bacteria Ratio (R. arrhizus:S. cerevisiae:P. kudriavzevii:L. rhamnosus) | Reduced Sugar (g/100 mL) | Total Acid (g/L) | Amino Acid Nitrogen (g/L) | Alcohol Content (%vol) | Sense Value | |||
|---|---|---|---|---|---|---|---|---|---|
| CK | Commercially available koji | 6.503 ± 0.6590 a | 7.175 ± 0.137 a | 0.658 ± 0.014 a | 10.5 ± 0.500 d | 78.5 | |||
| A | 4 | 2 | 1 | 1 | 3.592 ± 0.2776 bc | 6.023 ± 0.668 bc | 0.294 ± 0.024 cd | 12.0 ± 0.252 c | 79.6 |
| B | 5 | 1 | 1 | 1 | 3.023 ± 0.2002 c | 5.375 ± 0.187 c | 0.266 ± 0.014 d | 15.1 ± 0.173 a | 80.1 |
| C | 4 | 1 | 1 | 2 | 3.977 ± 0.0544 b | 6.065 ± 0.156 bc | 0.313 ± 0.021 c | 13.1 ± 0.153 b | 83.1 |
| D | 4.5 | 2.5 | 1 | 0.5 | 3.889 ± 0.3446 b | 6.155 ± 0.476 b | 0.331 ± 0.035 b | 13.4 ± 0.416 b | 80.3 |
| Volatile Compound | CAS | Chemical Formula | Relative Content/% | |||||
|---|---|---|---|---|---|---|---|---|
| CK | A | B | C | D | ||||
| Alcohol compounds (6) | 1-Propanol, 2,2-dimethyl- | 75-84-3 | C5H12O | 0.34 | / | / | / | / |
| Phenylethyl Alcohol | 60-12-8 | C8H10O | 29.71 | 14.76 | 14.2 | 13.12 | 16.29 | |
| 2-Naphthalenemethanol, 2,3,4,4a,5,6,7,8-octahydro-.alpha.,.alpha.,4a,8-tetramethyl-, [2R-(2.alpha.,4a.beta.,8.beta.)]- | 63891-61-2 | C15H26O | / | 0.08 | / | / | / | |
| 1,1,3,3,5,5,7,7,9,9-Decamethyl-9-(2-methylpropoxy)pentasiloxan-1-ol | ND | C14H40O6Si5 | / | / | / | 0.03 | / | |
| 1,1,3,3,5,5,7,7-Octamethyl-7-(2-methylpropoxy)tetrasiloxan-1-ol | C12H34O5Si4 | / | 0.02 | / | / | / | ||
| 1-Heptanol, 2,4-dimethyl-, | 98982-97-9 | C9H20O | 2.77 | 1.7 | / | 1.11 | / | |
| Ester compounds (37) | 1-Butanol, 3-methyl-, propanoate | 105-68-0 | C8H16O2 | / | / | / | 0.27 | / |
| 1-Butanol, 2-methyl-, propanoate | 2438-20-2 | C8H16O2 | / | / | / | 0.1 | / | |
| Acetic acid, pentyl ester | 628-63-7 | C7H14O2 | 0.57 | 1.19 | 0.67 | 0.92 | 0.49 | |
| Hexanoic acid, ethyl ester | 123-66-0 | C8H16O2 | / | 0.3 | / | 0.13 | 0.08 | |
| 2-Propenoic acid, 2-methyl-, pentyl ester | 2849-98-1 | C9H16O2 | 0.1 | / | / | / | / | |
| Formic acid, octyl ester | 112-32-3 | C9H18O2 | 0.07 | / | / | / | / | |
| Butanedioic acid, diethyl ester | 123-25-1 | C8H14O4 | 0.2 | 1.02 | 0.8 | 0.61 | 0.81 | |
| Octanoic acid, ethyl ester | 106-32-1 | C10H20O2 | 0.41 | 0.82 | 0.47 | 0.57 | 0.45 | |
| Acetic acid, 2-phenylethyl ester | 103-45-7 | C10H12O2 | 1.72 | 5.08 | 5.84 | 4.33 | 6.35 | |
| Nonanoic acid, ethyl ester | 123-29-5 | C11H22O2 | 0.23 | 0.15 | / | 0.09 | / | |
| Decanoic acid, ethyl ester | 110-38-3 | C12H24O2 | 0.23 | 1.33 | 1.03 | 1.76 | 0.97 | |
| 3,4-Dihydroxymandelic acid, ethyl ester, tri-TMS | ND | C19H36O5Si3 | 0.27 | 0.47 | 2.11 | 0.27 | 1.47 | |
| Methyl 2-methyl-2-(methoxy-3-hydroxypropoxy)amino-propanoate | 76664-32-9 | C9H19NO5 | 0.05 | / | / | / | / | |
| Dodecanoic acid, ethyl ester | 106-33-2 | C14H28O2 | 0.44 | / | 0.91 | 1.51 | 1.07 | |
| Tetradecanoic acid, ethyl ester | 124-06-1 | C16H32O2 | 2.62 | 2.38 | 2.67 | 3.51 | 3.23 | |
| Formic acid, undecyl ester | C12H24O2 | 0.11 | / | / | / | / | ||
| Pentadecanoic acid, ethyl ester | 41114-00-5 | C17H34O2 | 0.15 | 0.12 | / | / | / | |
| Ethyl 9-hexadecenoate | 54546-22-4 | C18H34O2 | / | 0.46 | 0.42 | 0.56 | 0.59 | |
| Hexadecanoic acid, ethyl ester | 628-97-7 | C18H36O2 | 31.65 | 43.22 | 41.86 | 42.51 | 34.06 | |
| i-Propyl 14-methyl-pentadecanoate | ND | C19H38O2 | 0.37 | 0.13 | 0.09 | / | / | |
| Hexadecanoic acid, 2-methylpropyl ester | 110-34-9 | C20H40O2 | 0.08 | 0.11 | 0.12 | 0.2 | / | |
| Heptadecanoic acid, ethyl ester | 14010-23-2 | C19H38O2 | / | 0.09 | 0.75 | 0.12 | 0.05 | |
| Butyl 9,12-octadecadienoate | ND | C22H40O2 | 3.8 | / | / | / | / | |
| Ethyl Oleate | 111-62-6 | C20H38O2 | 3.8 | 12.67 | 12.26 | 9.19 | 9.99 | |
| 9,12-Octadecadienoic acid, ethyl ester | 7619-08-1 | C20H36O2 | / | / | 0.5 | / | 0.58 | |
| (Z)-Ethyl pentadec-9-enoate | 56219-09-1 | C17H32O2 | / | / | 0.1 | 0.2 | 0.14 | |
| Ethyl 13-methyl-tetradecanoate | 64317-63-1 | C17H34O2 | / | / | 0.13 | / | 0.14 | |
| trans,trans-9,12-Octadecadienoic acid, propyl ester | ND | C21H38O2 | / | / | 9.61 | / | / | |
| O-Butylisourea | 57536-14-8 | C5H12N2O | / | / | / | 1.91 | / | |
| Arsenous acid, tris(trimethylsilyl) ester | 55429-29-3 | C9H27AsO3Si3 | / | / | / | 0.02 | / | |
| Octadecanoic acid, ethyl ester | 111-61-5 | C20H40O2 | / | / | / | 0.12 | / | |
| Pentadecanoic acid, 3-methylbutyl ester | 2306-91-4 | C15H30O2 | / | / | / | 0.1 | / | |
| 1-Undecanol, acetate | 1731-81-3 | C13H26O2 | / | / | / | 0.06 | / | |
| l-(+)-Ascorbic acid 2,6-dihexadecanoate | 28474-90-0 | C38H68O8 | 2.73 | / | / | 1.97 | / | |
| Heptadecanoic acid, 15-methyl-, ethyl ester | 57274-46-1 | C20H40O2 | / | / | / | / | 1.74 | |
| Decanoic acid, pentyl ester | 5933-87-9 | C15H30O2 | / | / | / | / | 0.13 | |
| cis-10-Pentadecenoic acid, propyl ester | ND | C18H34O2 | / | / | / | / | 0.08 | |
| Acid compounds (5) | DL-Allothreonine | 144-98-9 | C4H9NO3 | 3.8 | / | / | / | / |
| Butanoic acid, 4-butoxy- | 55724-73-7 | C8H16O3 | 0.02 | / | / | / | / | |
| Tetradecanoic acid | 544-63-8 | C14H28O2 | 0.24 | / | / | 0.05 | 0.09 | |
| 11-Bromoundecanoic acid | 2834-05-1 | C11H21BrO2 | / | 0.07 | / | / | / | |
| 2,5-Dihydroxybenzoic acid, 3TMS derivative | 3618-20-0 | C16H30O4Si3 | / | / | 0.8 | 0.07 | 1.16 | |
| Alkane compounds (10) | Dodecane, 2,6,11-trimethyl- | 31295-56-4 | C15H32 | / | / | / | / | 0.12 |
| Heptane, 3-ethyl-5-methylene- | 52896-90-9 | C10H20 | 0.07 | / | / | / | / | |
| Decane, 2,3,6-trimethyl- | 62238-12-4 | C13H28 | 0.16 | / | / | / | / | |
| Eicosane | 112-95-8 | C20H42 | 0.32 | 0.18 | / | / | / | |
| Cyclopentane, (4-octyldodecyl)- | 5638-09-5 | C25H50 | 0.1 | / | / | / | / | |
| 2,6,10-Trimethyltridecane | 3891-99-4 | C16H34 | 0.2 | / | / | / | / | |
| Decane, 2,3,7-trimethyl- | 62238-13-5 | C13H28 | / | 0.13 | 0.05 | / | / | |
| 3,6-Dioxa-2,4,5,7-tetrasilaoctane, 2,2,4,4,5,5,7,7-octamethyl- | ND | C10H30O2Si4 | / | 0.16 | 0.07 | / | 0.14 | |
| Cyclohexane, 1,2-dimethyl-3-pentyl-4-propyl- | 62376-17-4 | C16H32 | / | 0.03 | / | / | / | |
| 2-Methyltetracosane | 1560-78-7 | C25H52 | / | / | 0.38 | 0.42 | / | |
| Ketones (3) | 3-Octanone | 106-68-3 | C8H16O | 0.1 | / | / | / | / |
| 2H-Benzocyclohepten-2-one, decahydro-9a-methyl-, trans- | 55103-67-8 | C12H20O | 0.61 | / | / | / | / | |
| Neronine, 4.beta.,5-dihydro- | 19483-30-8 | C18H21NO6 | / | 0.08 | / | / | / | |
| Other compounds (16) | Oxime-, methoxy-phenyl-_ | ND | C8H9NO2 | 0.97 | / | / | 0.1 | 0.14 |
| 2′,6′-Dihydroxyacetophenone, acetate | ND | C10H10O4 | 0.14 | / | / | / | / | |
| i-Propyl tricosanoate | ND | C26H52O2 | 0.22 | / | / | / | / | |
| Benzeneethanamine, N-[(pentafluorophenyl)methylene]-.beta.,3,4-tris[(trimethylsilyl)oxy]- | 55429-13-5 | C24H34F5NO3Si3 | 0.2 | 0.06 | / | 0.04 | 0.04 | |
| 2H-3,9a-Methano-1-benzoxepin, octahydro-2,2,5a,9-tetramethyl-, [3R-(3.alpha.,5a.alpha.,9.alpha.,9a.alpha.)]- | 5956-09-2 | C15H26O | / | / | 0.03 | / | 0.05 | |
| 1,2-Ethanediamine, N-(phenylmethyl)- | 4152-09-4 | C9H14N2 | / | / | / | 0.09 | / | |
| (Z,Z)-.alpha.-Farnesene | ND | C15H24 | / | / | / | 0.22 | / | |
| 6-epi-shyobunol | 69350-61-4 | C15H26O | / | / | 0.43 | 0.78 | / | |
| Ethyl 9.cis.,11.trans.-octadecadienoate | ND | C20H36O2 | / | / | / | 0.87 | 1.32 | |
| 3-N-Nitroso-solanocapsine | ND | C27H45N3O3 | / | / | / | 0.18 | / | |
| Citronellol epoxide (R or S) | ND | C10H20O2 | / | / | / | 0.11 | / | |
| n-Propyl 9,12-octadecadienoate | ND | C21H38O2 | / | / | / | 8.86 | 9.93 | |
| Thiophene, tetrahydro-2-methyl- | 1795-09-1 | C5H10S | / | / | / | / | 0.13 | |
| .tau.-Muurolol | 19912-62-0 | C15H26O | / | / | / | / | 0.2 | |
| 2(1H)-Naphthalenone, octahydro-4a,5-dimethyl-, (4a.alpha.,5.alpha.,8a.beta.)- | 51557-64-3 | C12H20O | / | / | / | / | 0.91 | |
| Gamolenic acid | 506-26-3 | C18H30O2 | / | / | / | / | 0.14 | |
| Amino Acids | CK | A | B | C | D |
|---|---|---|---|---|---|
| (mg/mL) | |||||
| Asp | 1.383 ± 0.033 a | 1.18 ± 0.057 bc | 1.103 ± 0.062 c | 1.215 ± 0.050 b | 1.201 ± 0.041 bc |
| Thr | 0.686 ± 0.014 a | 0.628 ± 0.027 bc | 0.593 ± 0.012 c | 0.675 ± 0.023 a | 0.657 ± 0.018 ab |
| Ser | 0.845 ± 0.025 a | 0.737 ± 0.037 bc | 0.692 ± 0.023 c | 0.776 ± 0.035 b | 0.772 ± 0.028 b |
| Glu | 3.061 ± 0.079 a | 2.647 ± 0.117 bc | 2.48 ± 0.068 c | 2.734 ± 0.097 b | 2.711 ± 0.068 b |
| Gly | 0.787 ± 0.026 a | 0.699 ± 0.043 c | 0.681 ± 0.028 c | 0.733 ± 0.031 bc | 0.764 ± 0.022 a |
| Ala | 0.872 ± 0.027 a | 0.709 ± 0.046 b | 0.61 ± 0.025 c | 0.717 ± 0.043 b | 0.75 ± 0.024 b |
| Cys | 0.453 ± 0.019 a | 0.407 ± 0.013 b | 0.435 ± 0.014 ab | 0.436 ± 0.025 ab | 0.466 ± 0.016 a |
| Val | 0.721 ± 0.031 a | 0.627 ± 0.023 b | 0.578 ± 0.012 b | 0.634 ± 0.034 b | 0.63 ± 0.035 b |
| Met | 0.083 ± 0.016 a | 0.072 ± 0.015 a | 0.091 ± 0.009 a | 0.087 ± 0.008 a | 0.081 ± 0.007 a |
| Ile | 0.558 ± 0.015 a | 0.452 ± 0.023 b | 0.44 ± 0.012 b | 0.457 ± 0.035 b | 0.456 ± 0.011 b |
| Leu | 1.132 ± 0.044 a | 0.948 ± 0.037 b | 0.85 ± 0.022 c | 0.923 ± 0.048 bc | 0.937 ± 0.030 b |
| Tyr | 0.754 ± 0.013 a | 0.673 ± 0.018 bc | 0.631 ± 0.012 c | 0.687 ± 0.036 b | 0.67 ± 0.025 bc |
| Phe | 0.85 ± 0.024 a | 0.694 ± 0.022 bc | 0.654 ± 0.036 c | 0.705 ± 0.021 b | 0.712 ± 0.019 b |
| His | 0.493 ± 0.017 a | 0.436 ± 0.013 bc | 0.405 ± 0.012 c | 0.456 ± 0.026 b | 0.452 ± 0.018 b |
| Lys | 0.927 ± 0.012 a | 0.66 ± 0.023 b | 0.611 ± 0.026 c | 0.696 ± 0.035 b | 0.697 ± 0.021 b |
| Arg | 1.271 ± 0.033 ab | 1.26 ± 0.053 ab | 1.174 ± 0.069 b | 1.329 ± 0.054 a | 1.356 ± 0.022 a |
| Pro | 1.034 ± 0.048 a | 0.888 ± 0.037 b | 0.724 ± 0.024 c | 0.826 ± 0.043 b | 0.828 ± 0.035 b |
| total | 15.91 ± 0.641 a | 13.717 ± 0.364 bc | 12.752 ± 0.298 c | 14.086 ± 0.357 b | 14.14 ± 0.338 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Song, C.; Zhao, J.; Xiong, Z.; Peng, L.; Zou, L.; Liu, B.; Li, Q. Effect of a New Fermentation Strain Combination on the Fermentation Process and Quality of Highland Barley Yellow Wine. Foods 2024, 13, 2193. https://doi.org/10.3390/foods13142193
Chen X, Song C, Zhao J, Xiong Z, Peng L, Zou L, Liu B, Li Q. Effect of a New Fermentation Strain Combination on the Fermentation Process and Quality of Highland Barley Yellow Wine. Foods. 2024; 13(14):2193. https://doi.org/10.3390/foods13142193
Chicago/Turabian StyleChen, Xiaodie, Chuan Song, Jian Zhao, Zhuang Xiong, Lianxin Peng, Liang Zou, Bingliang Liu, and Qiang Li. 2024. "Effect of a New Fermentation Strain Combination on the Fermentation Process and Quality of Highland Barley Yellow Wine" Foods 13, no. 14: 2193. https://doi.org/10.3390/foods13142193
APA StyleChen, X., Song, C., Zhao, J., Xiong, Z., Peng, L., Zou, L., Liu, B., & Li, Q. (2024). Effect of a New Fermentation Strain Combination on the Fermentation Process and Quality of Highland Barley Yellow Wine. Foods, 13(14), 2193. https://doi.org/10.3390/foods13142193









