The Effects of Oleic Acid and Palmitic Acid on Porcine Muscle Satellite Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Preparation of PA and OA Samples
2.3. PMSC Culture and Treatments
2.4. Cell Viability Assay
2.5. Giemsa Staining
2.6. Flow Cytometry
2.7. Apoptosis Assay Using Acridine Orange/Ethidium Bromide Staining Methods
2.8. DAPI Staining
2.9. Wound-Healing Assay
2.10. Oil Red O Staining
2.11. Analysis of Myotube Formation
2.12. Measurement of Triacylglycerol Content
2.13. RNA Extraction and qRT-PCR
2.14. Protein Extraction and Western Blot Analysis
2.15. Statistical Analysis
3. Results
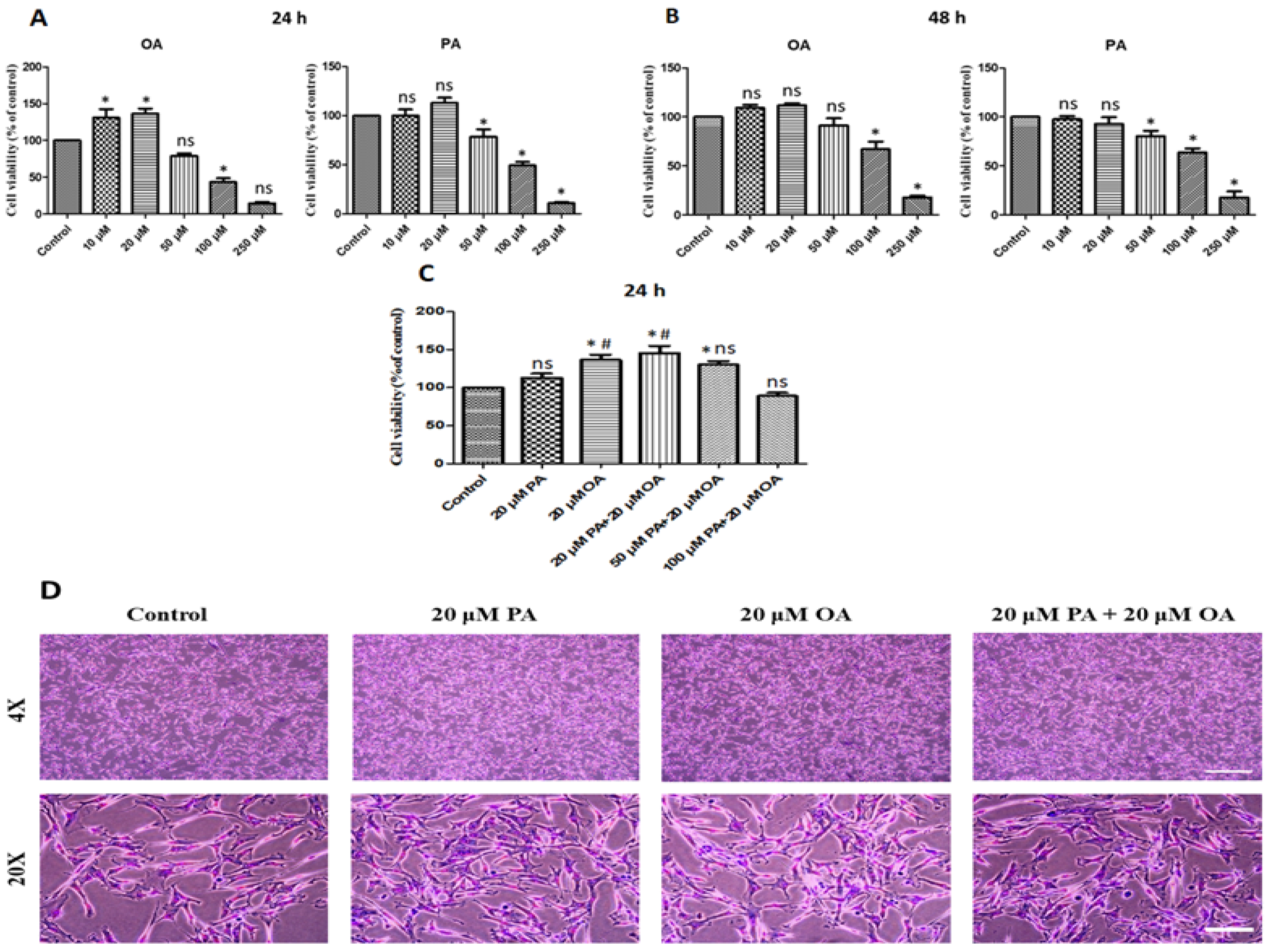
3.1. Effects of PA and OA on Cell Proliferation
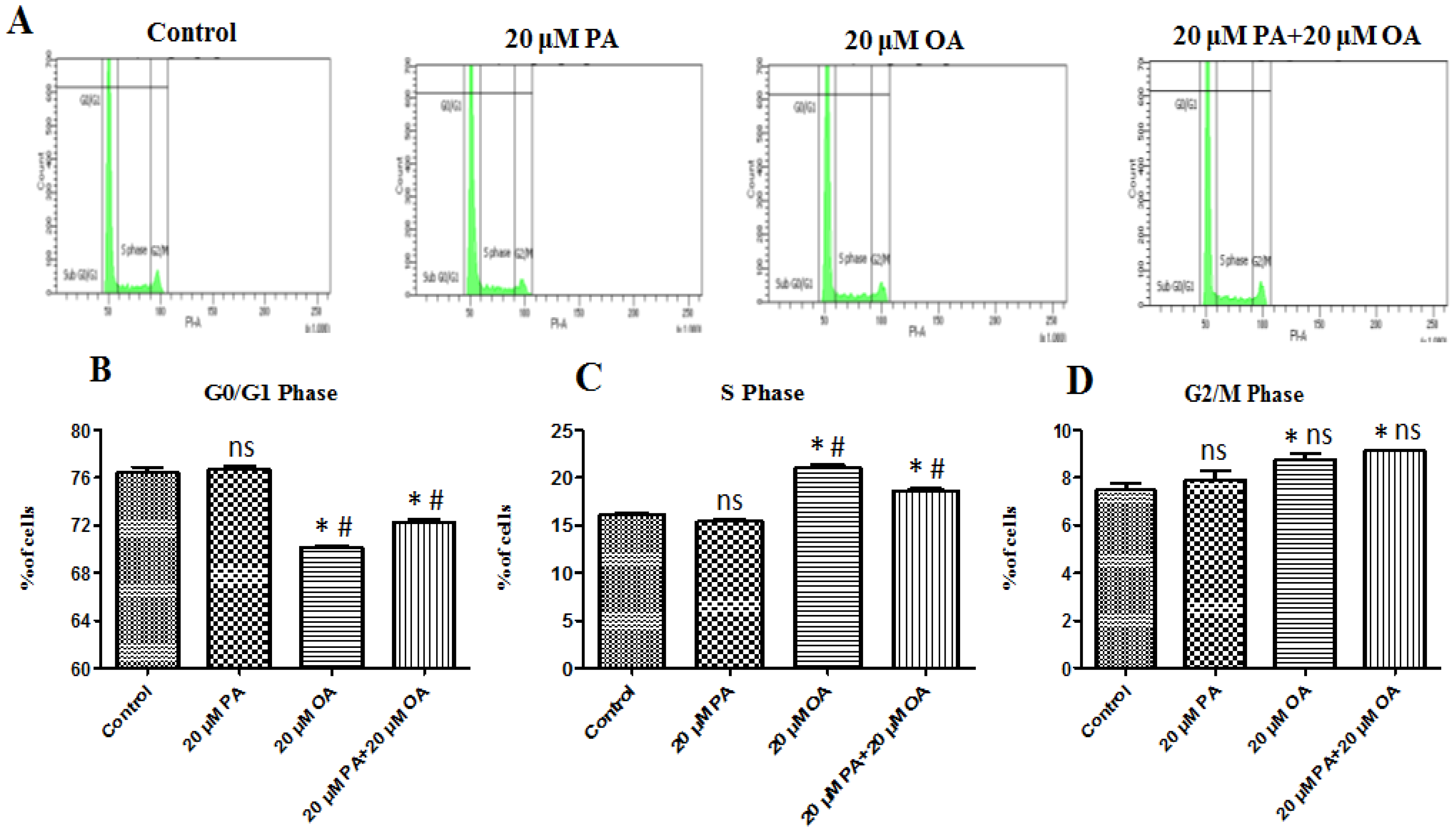
3.2. Effects of PA and OA on the Cell Cycle
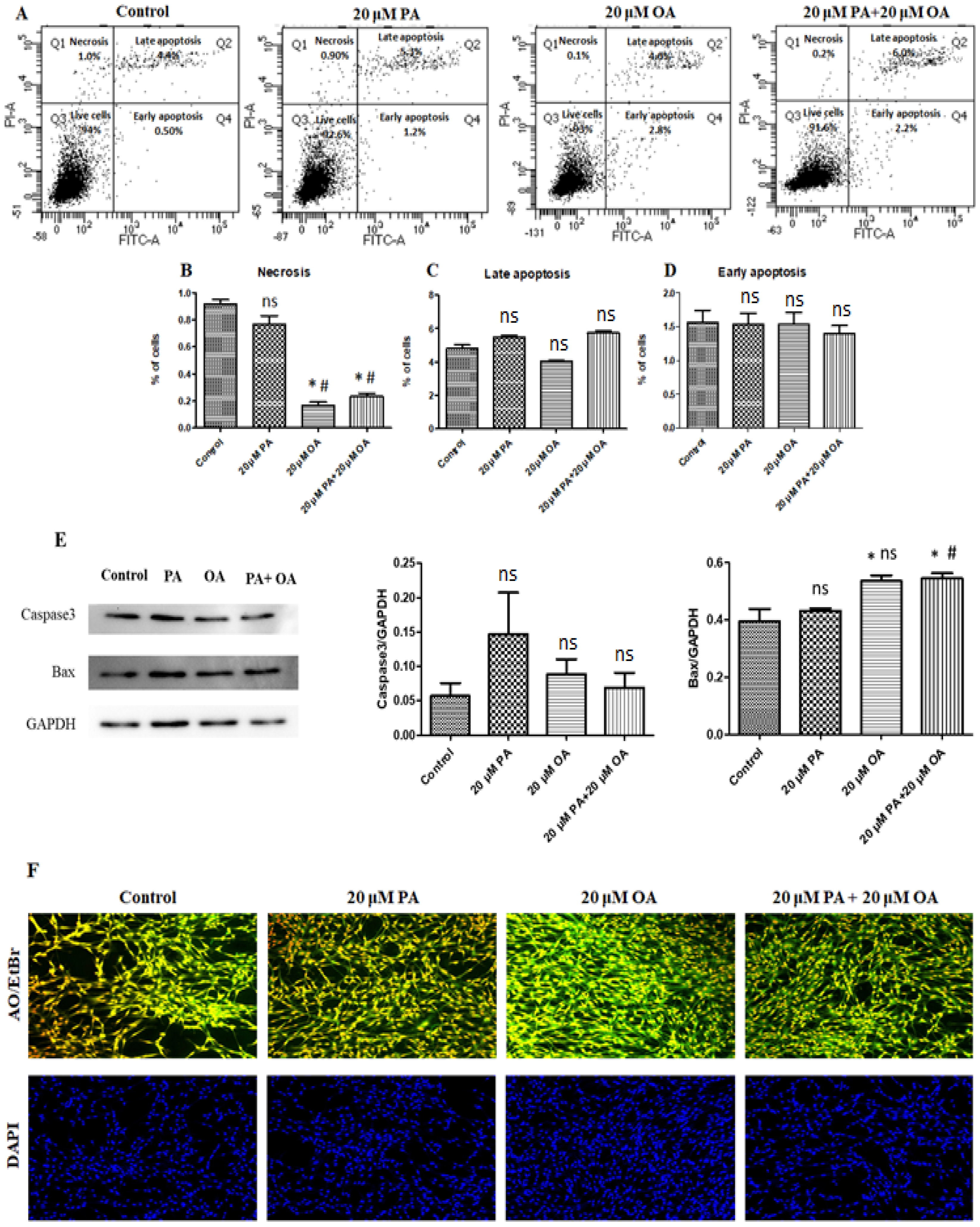
3.3. Effects of PA and OA on Apoptosis and Necrosis
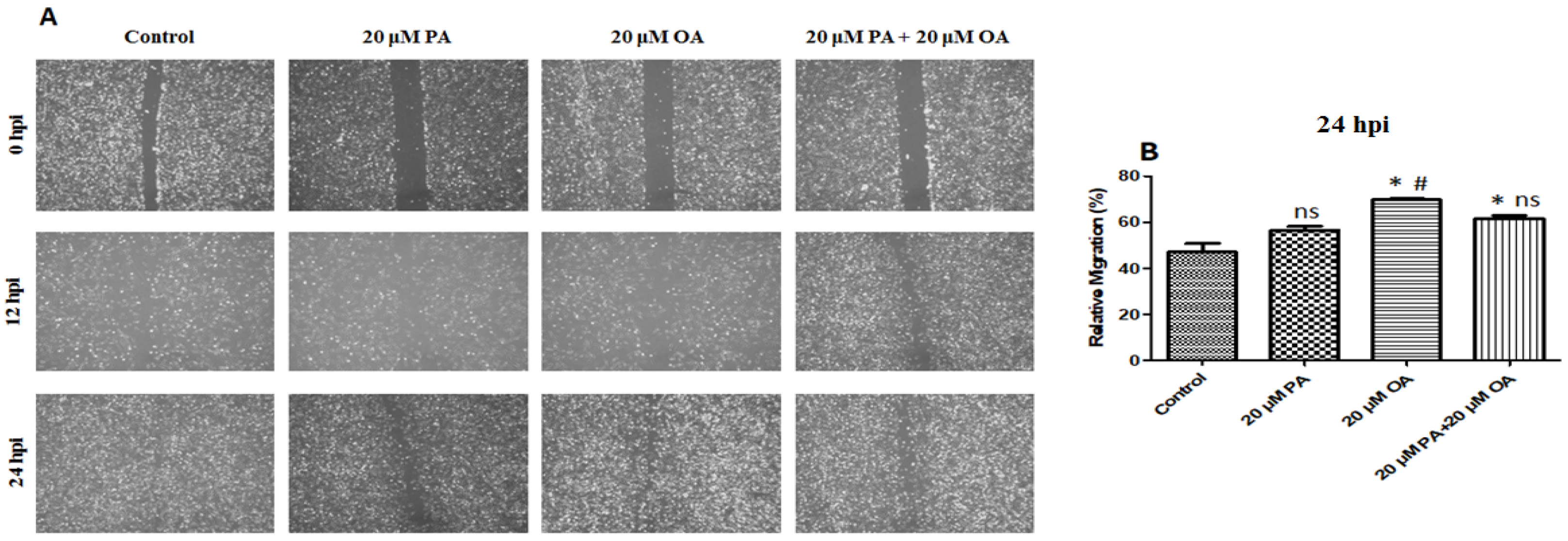
3.4. Effects of PA and OA on Migration of PMSCs
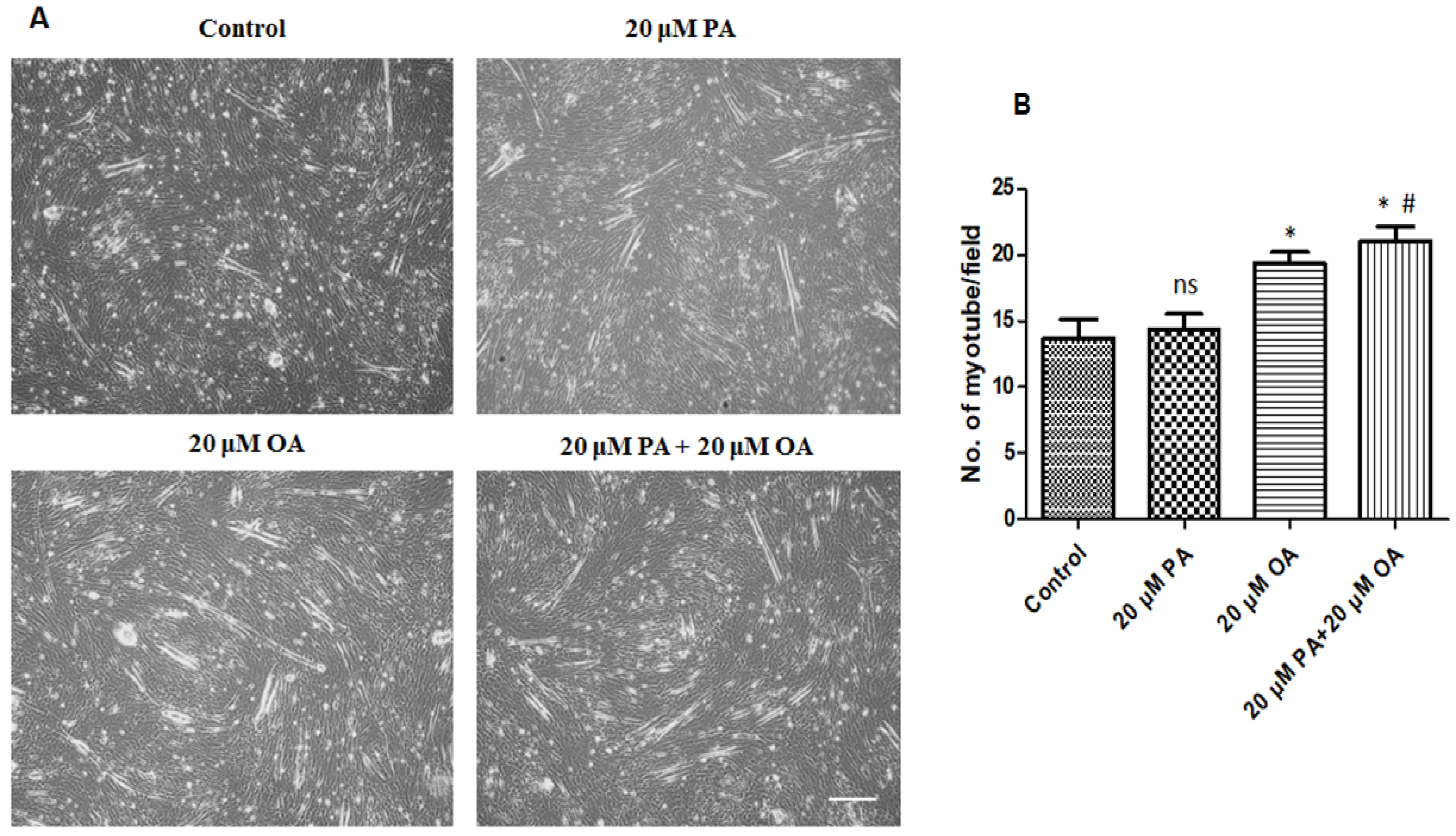
3.5. Effects of PA and OA on Myotube Formation
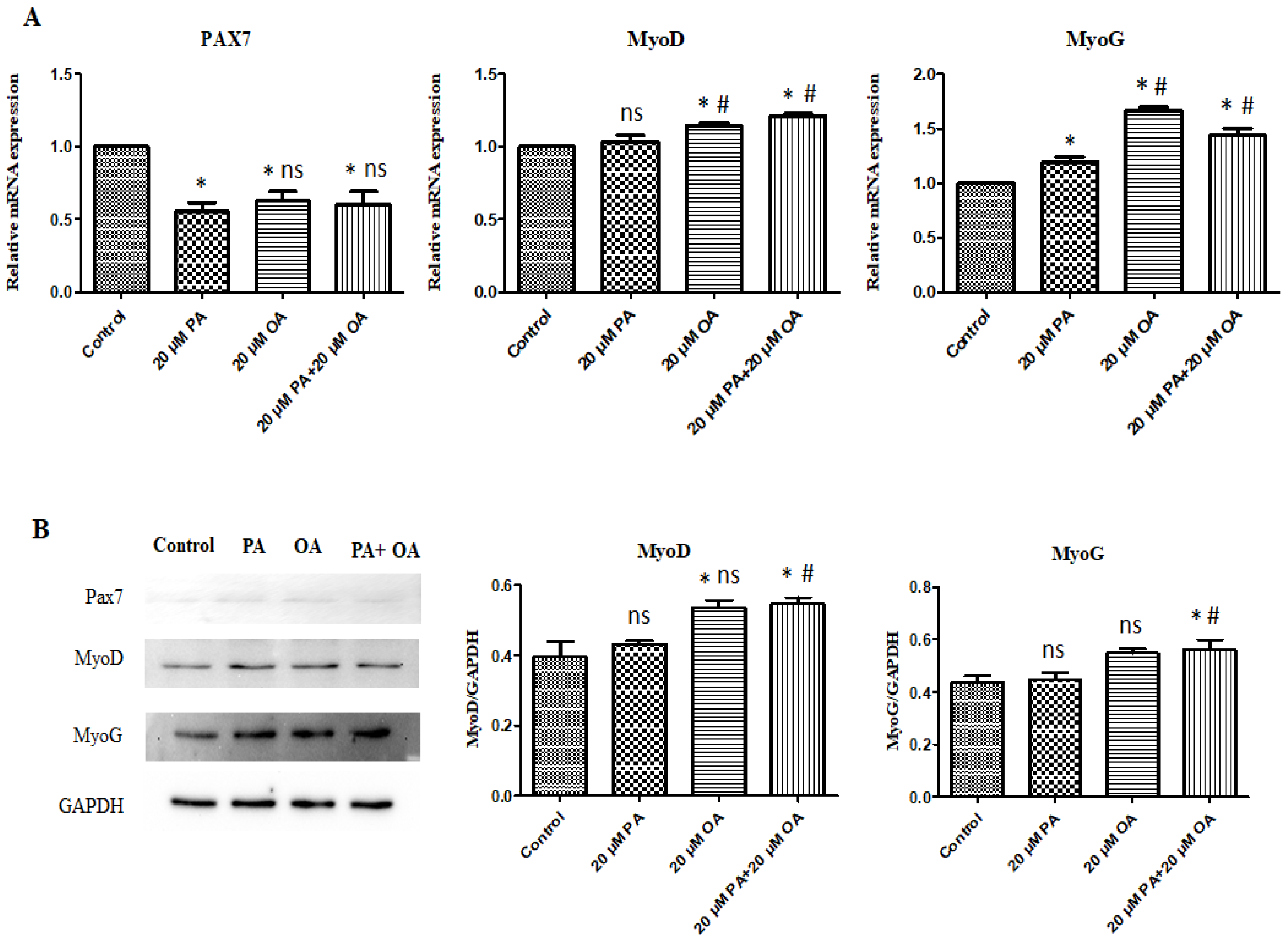
3.6. Effects of PA and OA on the mRNA and Protein Expression of Myogenic Differentiation-Related Genes
3.7. Effects of PA and OA on TAG Accumulation in PMSCs
3.8. Effects of PA and OA on the mRNA Expression of Adipogenic Differentiation-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, N.S.; Shu, G.; Xie, Q.P.; Zhu, X.T.; Gao, P.; Zhou, G.X.; Wang, S.; Wang, L.N.; Xi, Q.Y.; Zhang, Y.L.; et al. Myristic Acid (MA) Promotes Adipogenic Gene Expression and the Differentiation of Porcine Intramuscular Adipocyte Precursor Cells. J. Integr. Agric. 2014, 13, 2488–2499. [Google Scholar] [CrossRef][Green Version]
- Schonfeld, P.; Wojtczak, L. Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.S.; Flux, C.; Salter, A.M.; Brameld, J.M. Effects of fatty acids on skeletal muscle cell differentiation in vitro. Br. J. Nutr. 2006, 95, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Huard, J.; Li, Y.; Fu, F.H. Muscle injuries and repair: Current trends in research. J. Bone Jt. Surg. 2002, 84, 822–832. [Google Scholar] [CrossRef]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, K.; Zeng, X.; Zhou, X.; Yan, F.; Yang, S.; Zhao, A. Isolation and identification of goose skeletal muscle satellite cells and preliminary study on the function of C1q and tumor necrosis factor-related protein 3 gene. Anim. Biosci. 2021, 34, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, R.P.; Fernyhough, M.E.; Liu, X.; McFarland, D.C.; Velleman, S.G.; Hausman, G.J.; Dodson, M.V. Extrinsic regulation of domestic animal-derived myogenic satellite cells II. Domest. Anim. Endocrinol. 2009, 36, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Tachibana, H.; Morinaga, Y.; Fujimura, Y.; Yamada, K. Modulation of proliferation and differentiation of C2C12 skeletal muscle cells by fatty acids. Life Sci. 2009, 84, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Imperlini, E.; Nigro, E.; Montagnese, C.; Daniele, A.; Orrù, S.; Buono, P. Biological and nutritional properties of palm oil and palmitic acid: Effects on health. Molecules 2015, 20, 17339–17361. [Google Scholar] [CrossRef]
- Librán-Pérez, M.; Pereiro, P.; Figueras, A.; Novoa, B. Antiviral activity of palmitic acid via autophagic flux inhibition in zebrafish (Danio rerio). Fish. Shellfish Immunol. 2019, 95, 595–605. [Google Scholar] [CrossRef]
- Mayneris-Perxachs, J.; Guerendiain, M.; Castellote, A.I.; Estruch, R.; Covas, M.I.; Fitó, M.; Salas-Salvadó, J.; Martínez-González, M.A.; Aros, F.; Lamuela-Raventós, R.M. Plasma fatty acid composition, estimated desaturase activities, and their relation with the metabolic syndrome in a population at high risk of cardiovascular disease. Clin. Nutr. 2014, 33, 90–97. [Google Scholar] [CrossRef]
- Sawada, K.; Kawabata, K.; Yamashita, T.; Kawasaki, K.; Yamamoto, N.; Ashida, H. Ameliorative effects of polyunsaturated fatty acids against palmitic acid-induced insulin resistance in L6 skeletal muscle cells. Lipids Health Dis. 2012, 11, 36. [Google Scholar] [CrossRef]
- Moon, M.L.; Joesting, J.J.; Lawson, M.A.; Chiu, G.S.; Blevins, N.A.; Kwakwa, K.A.; Freund, G.G. The saturated fatty acid, palmitic acid, induces anxiety-like behavior in mice. Metabolism 2014, 63, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhang, J.; Zhang, Q.; Permatasari, F.; Xu, Y.; Wu, D.; Yin, Z.; Luo, D. Palmitic acid induces production of proinflammatory cytokines interleukin-6, interleukin-1β, and tumor necrosis factor-α via a NF-κB-dependent mechanism in HaCaT keratinocytes. Mediat. Inflamm. 2013, 2013, 530429. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.H.; Lin, C.I.; Liao, H.; Chen, Y.H.; Lin, S.H. Palmitic acid-induced neuron cell cycle G2/M arrest and endoplasmic reticular stress through protein palmitoylation in SH SY5Y human neuroblastoma cells. Int. J. Mol. Sci. 2014, 15, 20876–20899. [Google Scholar] [CrossRef]
- Pereira, D.M.; Correia-da-Silva, G.; Valentão, P.; Teixeira, N.; Andrade, P.B. Palmitic acid and ergosta-7, 22-dien-3-ol contribute to the apoptotic effect and cell cycle arrest of an extract from Marthasterias glacialis L. in neuroblastoma cells. Mar. Drugs 2014, 12, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Hardy, S.; Langelier, Y.; Prentki, M. Oleate activates phosphatidylinositol 3-kinase and promotes proliferation and reduces apoptosis of MDA-MB-231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res. 2000, 60, 6353–6358. [Google Scholar]
- Lin, L.; Ding, Y.; Wang, Y.; Wang, Z.; Yin, X.; Yan, G.; Zhang, L.; Yang, P.; Shen, H. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology 2017, 66, 432–448. [Google Scholar] [CrossRef]
- Takahashi, H.K.; Cambiaghi, T.D.; Luchessi, A.D.; Hirabara, S.M.; Vinolo, M.A.; Newsholme, P.; Curi, R. Activation of survival and apoptotic signaling pathways in lymphocytes exposed to palmitic acid. J. Cell Physiol. 2012, 227, 339–350. [Google Scholar] [CrossRef]
- Gorjao, R.; Cury-Boaventura, M.F.; de Lima, T.M.; Curi, R. Regulation of human lymphocyte proliferation by fatty acids. Cell Biochem. Funct. 2007, 25, 305–315. [Google Scholar] [CrossRef]
- Lima, T.M.; Kanunfre, C.C.; Pompeia, C.; Verlengia, R.; Curi, R. Ranking the toxicity of fatty acids on Jurkat and Raji cells by flow cytometric analysis. Toxicol. In Vitro 2002, 16, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Meier, K.E.; Jaffa, A.A.; Rosenzweing, S.A.; Egan, B.M. Oleic acid and angiotensin II induce a synergistic mitogenic response in vascular smooth muscle cells. Hypertension 1998, 31, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.N.; Alexander, R.W.; Runge, M.S. Linoleic acid and its metabolites, hydroperoxyoctadecadienoic acids, stimulate c-fos, c-jun and c-myc mRNA expression, MAP kinase activation and growth in rat aortic smooth muscle cells. J. Clin. Investig. 1995, 96, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.M.; Yanase, T.; Nishi, Y.; Tanaka, A.; Saito, M.; Jin, C.H.; Mukasa, C.; Okabe, T.; Nomura, M.; Goto, K.; et al. Saturated FFAs, palmitic acid and stearic acid, induce apoptosis in human granulosa cells. Endocrinology 2001, 142, 3590–3597. [Google Scholar] [CrossRef] [PubMed]
- Eitel, K.; Staiger, H.; Brendel, M.D.; Brandhorst, D.; Bretzel, R.G.; Haring, H.U.; Kellerer, M. Different role of saturated and unsaturated fatty acids in beta-cell apoptosis. Biochem. Biophys. Res. Commun. 2002, 299, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Akazawa, Y.; Cazanave, S.; Mott, J.L.; Elmi, N.; Bronk, S.F.; Kohno, S.; Charlton, M.R.; Gores, G.J. Palmitoleate attenuates palmitate induced Bim and PUMA up-regulation and hepatocyte lipoapoptosis. J. Hepatol. 2010, 52, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Akahane, K.; Nakamura, J.; Naruse, K.; Kamiya, H.; Himeno, T.; Nakamura, N.; Shibata, T.; Kondo, M.; Nagasaki, H.; et al. Palmitate induces apoptosis in Schwann cells via both ceramide-dependent and independent pathways. Neuroscience 2011, 176, 188–198. [Google Scholar] [CrossRef]
- Pennisi, E.M.; Garibaldi, M.; Antonini, G. Lipid myopathies. J. Clin. Med. 2018, 7, 472. [Google Scholar] [CrossRef] [PubMed]
- Frago, L.M.; Canelles, S.; Freire-Regatillo, A.; Argente-Arizón, P.; Barrios, V.; Argente, J.; Garcia-Segura, L.M.; Chowen, J.A. Estradiol uses different mechanisms in astrocytes from the hippocampus of male and female rats to protect against damage induced by palmitic acid. Front. Mol. Neurosci. 2017, 10, 330–346. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Song, K.D.; Kim, S.J.; Kim, D.C.; Lee, S.C.; Son, Y.J.; Choi, H.W.; Shim, K.S. Growth factors improve the proliferation of Jeju black pig muscle cells by regulating myogenic differentiation and growth-related genes. Anim. Biosci. 2021, 34, 1392–1402. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Choe, H.; Shim, K.S. Insect peptide CopA3 promotes proliferation and PAX7 and MYOD expression in porcine muscle satellite cells. J. Anim. Sci. Technol. 2022, 64, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, W.; Ao, L.; Wang, Z.; Hao, X.; Zhang, H. Benzo[a]pyrene-7,8-diol-9,10-epoxide suppresses the migration and invasion of human extravillous trophoblast HTR-8/SVneo cells by downregulating MMP2 through inhibition of FAK/SRC/PI3K/AKT pathway. Toxicology 2017, 386, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Liu, P.; Li, H.; Ding, S. Large-Scale Expansion of Porcine Adipose-Derived Stem Cells Based on Microcarriers System for Cultured Meat Production. Foods 2022, 11, 3364. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.; Shim, K.S. Effects of heat stress exposure on porcine muscle satellite cells. J. Therm. Biol. 2023, 114, 103569. [Google Scholar] [CrossRef] [PubMed]
- Baik, M.; Nguyen, T.H.; Jeong, J.Y.; Piao, M.Y.; Kang, H.J. Effects of Castration on Expression of Lipid Metabolism Genes in the Liver of Korean Cattle. Asian-Australas. J. Anim. Sci. 2015, 28, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Repa, J.J.; Evans, R.M.; Mangelsdorf, D.J. Nuclear receptors and lipid physiology: Opening the X-files. Science 2001, 294, 1866–1870. [Google Scholar] [CrossRef] [PubMed]
- Maurin, A.C.; Chavassieux, P.M.; Vericel, E.; Meunier, P.J. Role of polyunsaturated fatty acids in the inhibitory effect of human adipocytes on osteoblastic proliferation. Bone 2002, 31, 260–266. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, S. Cell cycle regulation. Wormbook 2005, 1–16. [Google Scholar] [CrossRef]
- Yonezawa, T.; Haga, S.; Kobayashi, Y.; Katoh, K.; Obara, Y. Unsaturated fatty acids promote proliferation via ERK1/2 and Akt pathway in bovine mammary epithelial cells. Biochem. Biophys. Res. Commun. 2008, 367, 729–735. [Google Scholar] [CrossRef]
- Lu, G.; Morinelli, T.A.; Meier, K.E.; Rosenzweing, S.A.; Egan, B.M. Oleic acid-induced mitogenic signaling in vascular smooth muscle cells. A role for protein kinase C. Circ. Res. 1996, 79, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Mattern, H.M.; Hardin, C.D. Vascular metabolic dysfunction and lipotoxicity. Physiol. Res. 2007, 56, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zhu, M.; Liu, X.; Chen, X.; Yuan, Y.; Li, L.; Liu, J.; Lu, Y.; Cheng, J.; Chen, Y. Oleic acid ameliorates palmitic acid induced hepatocellular lipotoxicity by inhibition of ER stress and pyroptosis. Nutr. Metab. 2020, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yang, M.; Li, G.; Zhang, X.; Huang, Y.; Tang, Y. Effects of palmitic acid and eicosapentaenoic acid on angiogenesis of porcine vascular endothelial cells. Vet. Med. Sci. 2021, 7, 2260–2267. [Google Scholar] [CrossRef] [PubMed]
- Grabiec, K.; Gajewska, M.; Milewska, M.; Błaszczyk, M.; Grzelkowska-Kowalczyk, K. The influence of high glucose and high insulin on mechanisms controlling cell cycle progression and arrest in mouse C2C12 myoblasts: The comparison with IGF-I effect. J. Endocrinol. Investig. 2014, 37, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, X.; Lin, L.; Wen, C.; Rao, S. Effects of fatty acids on proliferation and differentiation of myoblast. Wei Sheng Yan Jiu = J. Hyg. Res. 2012, 41, 883–888. [Google Scholar]
- Cossu, G.; Borello, U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999, 18, 6867–6872. [Google Scholar] [CrossRef]
- Weintraub, H.; Davis, R.; Tapscott, S.; Thayer, M.; Krause, M.; Benezra, R.; Blackwell, T.K.; Turner, D.; Rupp, R.; Hollenberg, S.; et al. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science 1991, 251, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Sun, J.; Li, Q.; Wang, Y.; Wang, E.; Jin, H.; Hua, H.; Jin, Q.; Li, X. Study on the effect of oleic acid-induced lipogenic differentiation of skeletal muscle satellite cells in Yanbian cattle and related mechanisms. Animals 2023, 13, 3618. [Google Scholar] [CrossRef]
- Zhao, D.D.; Liu, C.C.; Jia, M.Y.; Yang, Y.; Ye, F.; Yan, Y.Q. Progress in the study of upstream transcriptional regulatory elements of muscle-specific gene promoters. Chin. J. Cell Biol. 2012, 34, 500–505. (In Chinese) [Google Scholar]
- Hasty, K.A.; Wu, H.; Byrne, M.; Goldring, M.B.; Seyer, J.M.; Jaenisch, R.; Krane, S.M.; Mainardi, C.L. Susceptibility of type I collagen containing mutated alpha 1(1) chains to cleavage by human neutrophil collagenase. Matrix 1993, 13, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Apponi, L.H.; Corbett, A.H.; Pavlath, G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011, 32, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Yan, Y.; Zhang, J.F.; Sun, J.F.; Sun, B.; Yan, C.G.; Choi, S.H.; Johnson, B.J.; Kim, J.K.; Smith, S.B. Oleic acid in the absence of a PPARγ agonist increases adipogenic gene expression in bovine muscle satellite cells. J. Anim. Sci. 2019, 97, 4114–4123. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.E.; Rittweger, J. Adaptive processes in skeletal muscle: Molecular regulators and genetic influences. J. Musculoskelet. Neuronal Interact. 2006, 6, 73–86. [Google Scholar] [PubMed]
- Fujimoto, Y.; Onoduka, J.; Homma, K.J.; Yamaguchi, S.; Mori, M.; Higashi, Y.; Makita, M.; Kinoshita, T.; Noda, J.I.; Itabe, H.; et al. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of Acyl-CoA synthetase. Biol. Pharm. Bull. 2006, 29, 2174–2180. [Google Scholar] [CrossRef]
- Kohjima, M.; Enjoji, M.; Higuchi, N.; Kato, M.; Kotoh, K.; Nakashima, M.; Nakamuta, M. The effects of unsaturated fatty acids on lipid metabolism in HepG2 cells. In Vitro Cell. Dev. Biol. -Anim. 2009, 45, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Belal, S.A.; Kang, D.R.; Sivakumar, A.S.; Choe, H.S.; Shim, K.S. Effect of long chain fatty acids on triacylglycerol accumulation, fatty acid composition and related gene expression in primary cultured bovine satellite cells. Anim. Biotechnol. 2019, 30, 323–331. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Q.; Nogoy, K.M.; Sun, J.; Sun, B.; Wang, Y.; Tang, L.; Yu, J.; Jin, X.; Li, X.; et al. Effect of palmitoleic acid on the differentiation of bovine skeletal muscle satellite cells. J. Anim. Sci. Technol. 2021, 63, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Mersmann, H.J.; Ding, S.T. Fatty acids modulate porcine adipocyte differentiation and transcripts for transcription factors and adipocyte-characteristic proteins. J. Nutr. Biochem. 2001, 12, 101–108. [Google Scholar] [CrossRef]
- Sanosaka, M.; Minashima, T.; Suzuki, K.; Watanabe, K.; Ohwada, S.; Hagino, A.; Rose, M.T.; Yamaguchi, T.; Aso, H. A combination of octanoate and oleate promotes in vitro differentiation of porcine intramuscular adipocytes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 149, 285–292. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belal, S.A.; Lee, J.; Park, J.; Kang, D.; Shim, K. The Effects of Oleic Acid and Palmitic Acid on Porcine Muscle Satellite Cells. Foods 2024, 13, 2200. https://doi.org/10.3390/foods13142200
Belal SA, Lee J, Park J, Kang D, Shim K. The Effects of Oleic Acid and Palmitic Acid on Porcine Muscle Satellite Cells. Foods. 2024; 13(14):2200. https://doi.org/10.3390/foods13142200
Chicago/Turabian StyleBelal, Shah Ahmed, Jeongeun Lee, Jinryong Park, Darae Kang, and Kwanseob Shim. 2024. "The Effects of Oleic Acid and Palmitic Acid on Porcine Muscle Satellite Cells" Foods 13, no. 14: 2200. https://doi.org/10.3390/foods13142200
APA StyleBelal, S. A., Lee, J., Park, J., Kang, D., & Shim, K. (2024). The Effects of Oleic Acid and Palmitic Acid on Porcine Muscle Satellite Cells. Foods, 13(14), 2200. https://doi.org/10.3390/foods13142200




