Pueraria lobata–Prunus mume Complex Alleviates Alcoholic Liver Disease by Regulating Lipid Metabolism and Inhibiting Inflammation: A Transcriptome and Gut Microbiota Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Liquid Chromatography–Mass Spectrometry (LC-MS)
2.2. Animals and Model Establishment
2.3. Transcriptome and 16S rRNA Analysis
2.4. Serum Biochemical Analysis
2.5. Tissue Staining and Immunohistochemistry
2.6. Western Blotting
2.7. Statistical Analysis
3. Results and Discussion
3.1. Component Analysis of PPC and Liver Transcriptomics Results in Mice with ALD
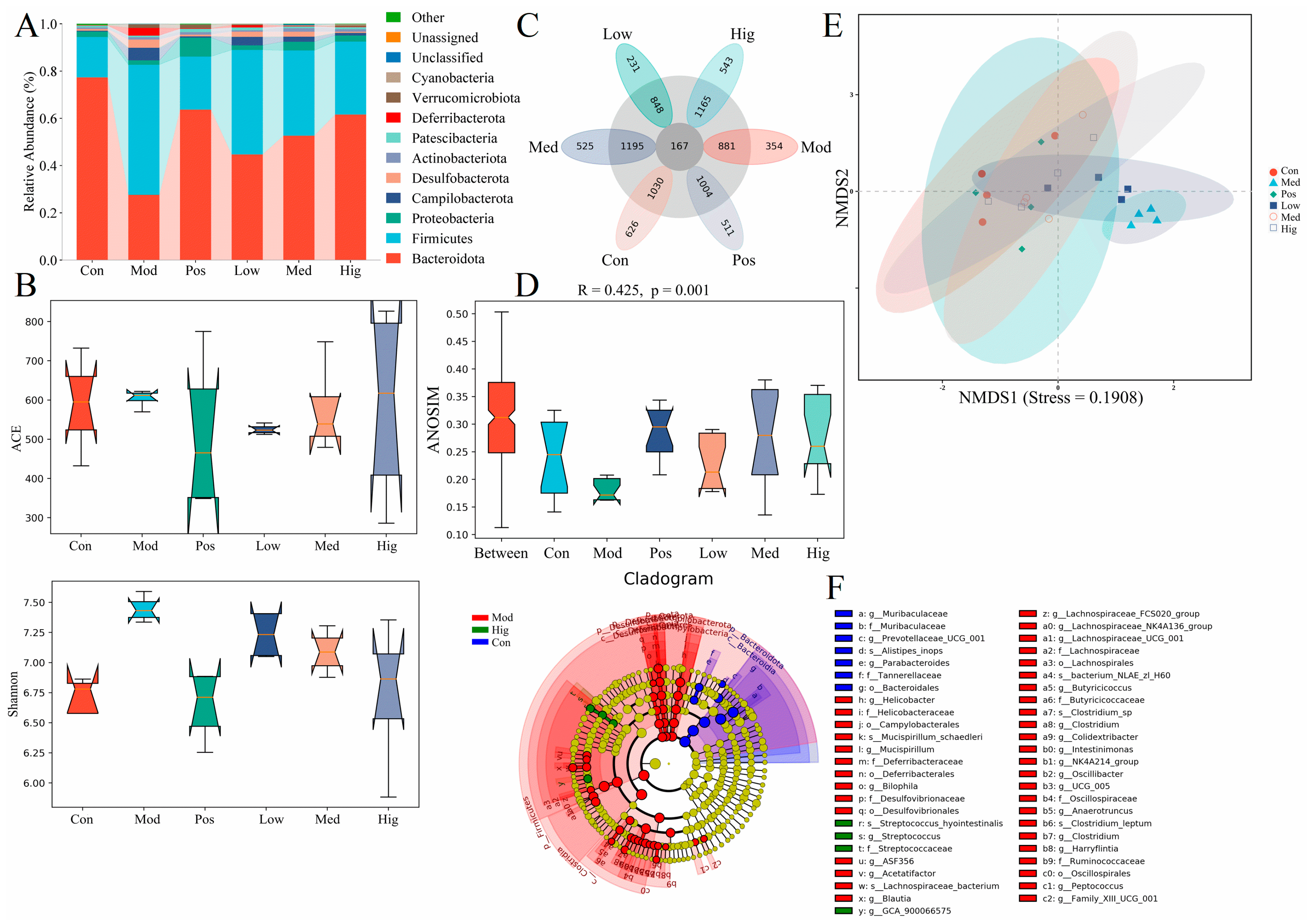
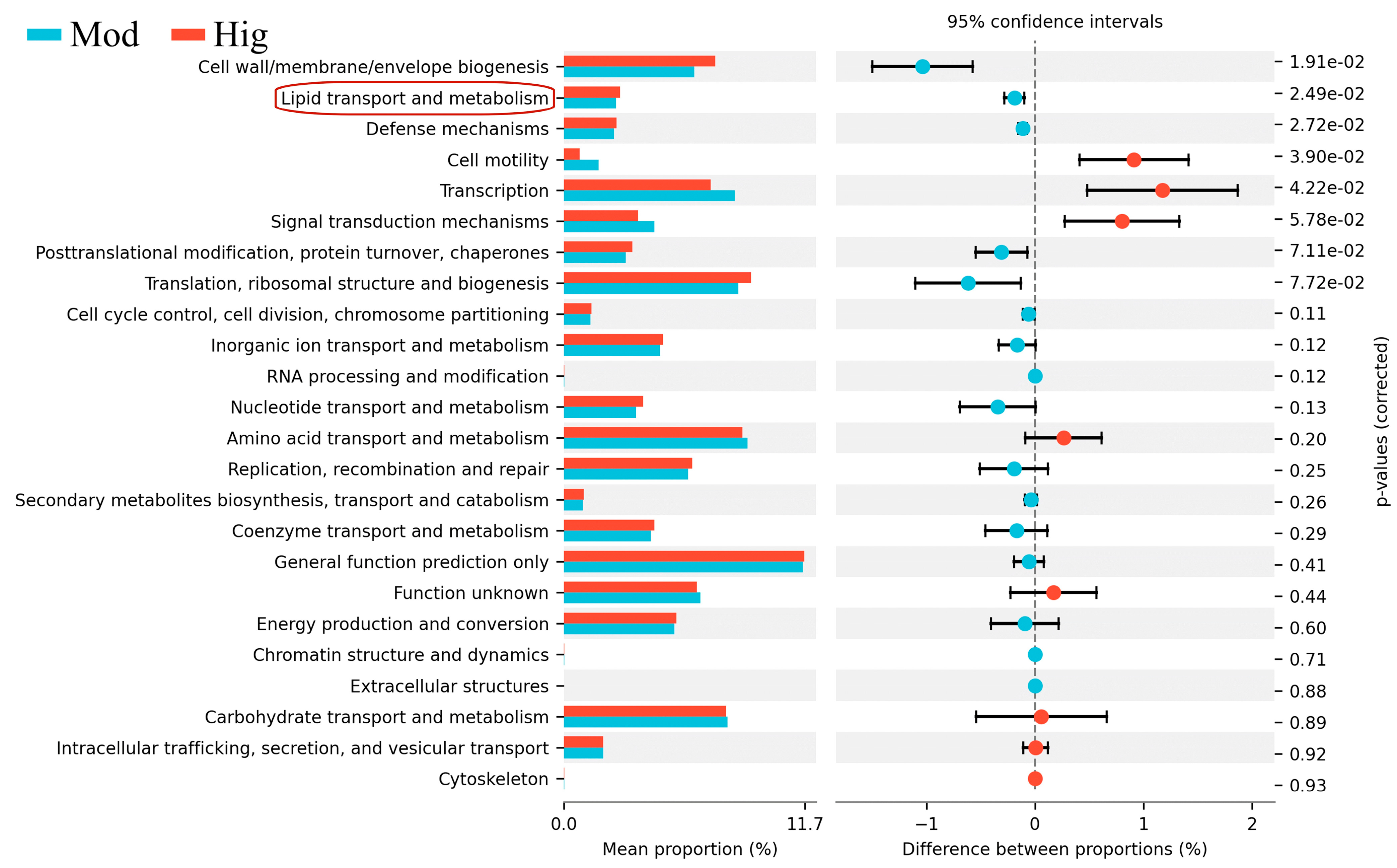
3.2. 16S rRNA Results of Mice with ALD with/without PPC Administration
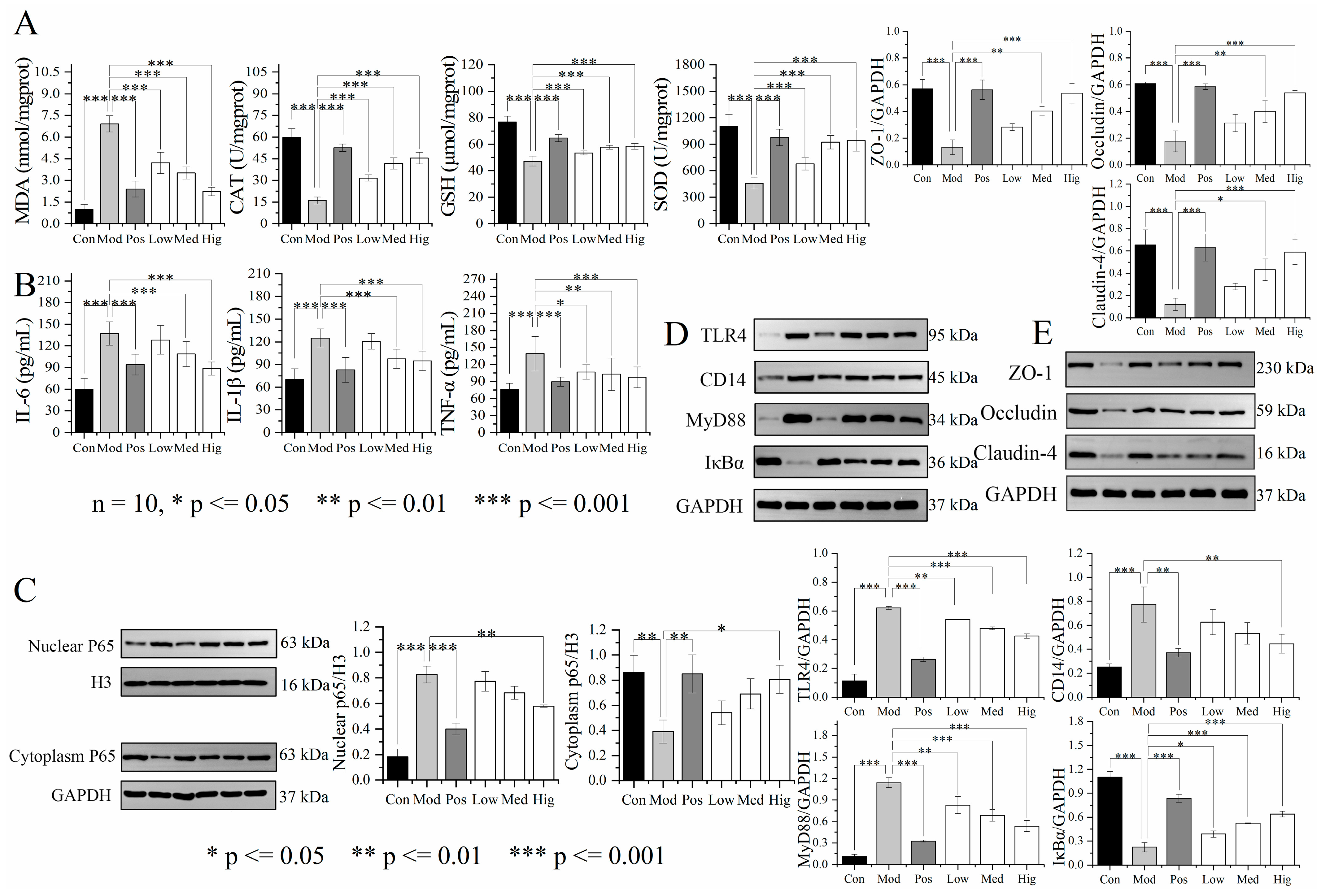
3.3. Validation of Liver Transcriptomics and Gut Microbiota Results
3.4. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zou, J.; Yang, R.; Feng, R.; Liu, J.; Wan, J. Ginsenoside Rk2, a dehydroprotopanaxadiol saponin, alleviates alcoholic liver disease via regulating NLRP3 and NLRP6 inflammasome signaling pathways in mice. J. Pharm. Anal. 2023, 13, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Dogra, A.; Li, F. Small-molecule chemical probes for the potential therapeutic targets in alcoholic liver diseases. Liver Res. 2023, 7, 177–188. [Google Scholar] [CrossRef]
- Qiu, L.; Feng, R.; Wu, Q.; Wan, J.; Zhang, Q. Total saponins from Panax japonicus attenuate acute alcoholic liver oxidative stress and hepatosteatosis by p62-related Nrf2 pathway and AMPK-ACC/PPARα axis in vivo and in vitro. J. Ethnopharmacol. 2023, 317, 116785. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Liu, C.; Liu, T.; Tian, M.; Wu, N.; Yu, Z.; Zhao, F.; Qi, J.; Zhu, Q. FNDC3B protects steatosis and ferroptosis via the AMPK pathway in alcoholic fatty liver disease. Free Radic. Biol. Med. 2022, 193, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Chen, J.; Liu, C.; Xia, F.; Xiao, Z.; Zhang, X.; Wan, J. A combination of Pueraria lobata and Silybum marianum protects against alcoholic liver disease in mice. Phytomedicine 2019, 58, 152824. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhao, X.; Bo, J.; Lv, Z.; Li, G.; Chen, Y.; Liang, J.; Zhang, C.; Jin, X.; Liu, C.; et al. A polysaccharide from Alhagi honey protects the intestinal barrier and regulates the Nrf2/HO-1-TLR4/MAPK signaling pathway to treat alcoholic liver disease in mice. J. Ethnopharmacol. 2024, 321, 117552. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Li, S.; Yan, Q.; Hu, X.; Li, L.; Zhou, H.; Pan, L.; Li, J.; Shen, C.; Xu, T. Natural products, extracts and formulations comprehensive therapy for the improvement of motor function in alcoholic liver disease. Pharmacol. Res. 2019, 150, 104501. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yuan, M.; Zhang, C.; Liu, H.; Liu, J.; Wei, A.; Ye, Q.; Zeng, B.; Li, M.; Guo, Y.; et al. Puerariae Lobatae radix flavonoids and puerarin alleviate alcoholic liver injury in zebrafish by regulating alcohol and lipid metabolism. Biomed. Pharmacother. 2021, 134, 111121. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, S.; Wu, L.; Yang, K.; Yang, F.; Yang, J.; Hu, S.; Yao, Y.; Xia, X.; Liu, Y.; et al. Puerarin inhibits inflammation and lipid accumulation in alcoholic liver disease through regulating MMP8. Chin. J. Nat. Med. 2023, 21, 670–681. [Google Scholar] [CrossRef]
- Pan, J.H.; Lee, K.Y.; Kim, J.H.; Shin, H.; Lee, J.H.; Kim, Y.J. Prunus mume Sieb et Zucc. fruit ameliorates alcoholic liver injury in mice by inhibiting apoptosis and inflammation through oxidative stress. J. Funct. Foods 2016, 25, 135–148. [Google Scholar] [CrossRef]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef]
- Yan, X.; Lee, S.; Li, W.; Jang, H.; Kim, Y. Terpenes and sterols from the fruits of Prunus mume and their inhibitory effects on osteoclast differentiation by suppressing tartrate-resistant acid phosphatase activity. Arch. Pharm. Res. 2015, 38, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wei, S.; Cao, X.; Wang, S.; Chang, Y.; Ouyang, H.; He, J. Multi-component pharmacokinetic study of prunus mume fructus extract after oral administration in rats using UPLC-MS/MS. Front. Pharmacol. 2022, 13, 954692. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Ko, B.; Kim, D.J.; Tak, J.; Han, C.Y.; Cho, J.; Kim, W.; Kim, S.G. Induction of the hepatic aryl hydrocarbon receptor by alcohol dysregulates autophagy and phospholipid metabolism via PPP2R2D. Nat. Commun. 2022, 13, 6080. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, M.; Wang, Q.; Huang, H.; Zhao, Y.; Du, F.; Chen, Y.; Shen, J.; Luo, H.; Zhao, Q.; et al. Pueraria lobata starch regulates gut microbiota and alleviates high-fat high-cholesterol diet induced non-alcoholic fatty liver disease in mice. Food. Res. Int. 2022, 157, 111401. [Google Scholar] [CrossRef] [PubMed]
- Nicola, S.; Jacques, I.; Levi, W.; Dirk, G.; Larisa, M.; Wendy, S.G.; Curtis, H. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar]
- Sebastian, M.; Teresa, P.; Qin, H.; Katharina, G.; Rüdiger, W.; Christa, F.; Beate, K.S.; Gunda, M.; Felix, S.; Thomas, B.; et al. Carcinogenic etheno DNA adducts in alcoholic liver disease: Correlation with cytochrome P-4502E1 and fibrosis. Alcohol. Clin. Exp. Res. 2018, 42, 252–259. [Google Scholar]
- Elise, S.; Leonardo, B.; Nan, W.; Burcin, E.; Keisaku, S.; Emily, L.; Ludovica, C.; Chen, L.; Sugeily, R.L.; Xu, W.; et al. Kupffer cells: Inflammation pathways and cell-cell interactions in alcohol-associated liver disease. Am. J. Pathol. 2020, 190, 2185–2193. [Google Scholar]
- Daniel, Z.; Abel, W.Z.; Abdeen, E.E.; Mahmoud, S.; Tao, M.; Li, R.; Wu, T.; Xu, X. Red raspberry supplementation mitigates alcohol-induced liver injury associated with gut microbiota alteration and intestinal barrier dysfunction in mice. Food. Funct. 2023, 14, 1209–1226. [Google Scholar]
- Zhao, W.; Peng, D.; Li, W.; Chen, S.; Liu, B.; Huang, P.; Wu, J.; Du, B.; Li, P. Probiotic-fermented Pueraria lobata (Willd) Ohwi alleviates alcoholic liver injury by enhancing antioxidant defense and modulating gut microbiota. J. Sci. Food Agric. 2022, 102, 6877–6888. [Google Scholar] [CrossRef]
- Wang, T.; Wang, Z.; Yang, Z.; Cui, X.; Yan, L.; Xu, Z.; Liu, X. Effect of the fermentation broth of the mixture of Pueraria lobata, Lonicera japonica, and Crataegus pinnatifida by Lactobacillus rhamnosus 217-1 on liver health and intestinal flora in mice with alcoholic liver disease induced by liquor. Front. Microbiol. 2021, 12, 722171. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, M.; Jeon, W.K. 16S rRNA gene-based microbiome taxonomic profiling for evaluating the effect of mumefural on fecal microbiota in C57BL/6 mice. Nat. Prod. Commun. 2024, 19, 1934578X241241960. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, W.; Liu, C.; Wang, C.; Hu, B.; Guo, Y.; Cheng, Y.; Qian, H. Ameliorative effects of chlorogenic acid on alcoholic liver injury in mice via gut microbiota informatics. Eur. J. Pharmacol. 2022, 928, 175096. [Google Scholar] [CrossRef]
- Zhong, X.; Cui, P.; Jiang, J.; Ning, C.; Liang, B.; Zhou, J.; Tian, L.; Zhang, Y.; Lei, T.; Zuo, T.; et al. Streptococcus, the predominant bacterium to predict the severity of liver injury in alcoholic liver disease. Front. Cell. Infect. Microbiol. 2021, 11, 649060. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.N.; Vetoshkina, D.V.; Marenkova, T.V.; Borisova-Mubarakshina, M.M. Antioxidants of non-enzymatic nature: Their function in higher plant cells and the ways of boosting their biosynthesis. Antioxidants 2023, 12, 2014. [Google Scholar] [CrossRef]
- Chen, X.; David, D.K. Flavonoid composition of orange peel extract ameliorates alcohol-induced tight junction dysfunction in Caco-2 monolayer. Food. Chem. Toxicol. 2017, 105, 398–406. [Google Scholar] [CrossRef]
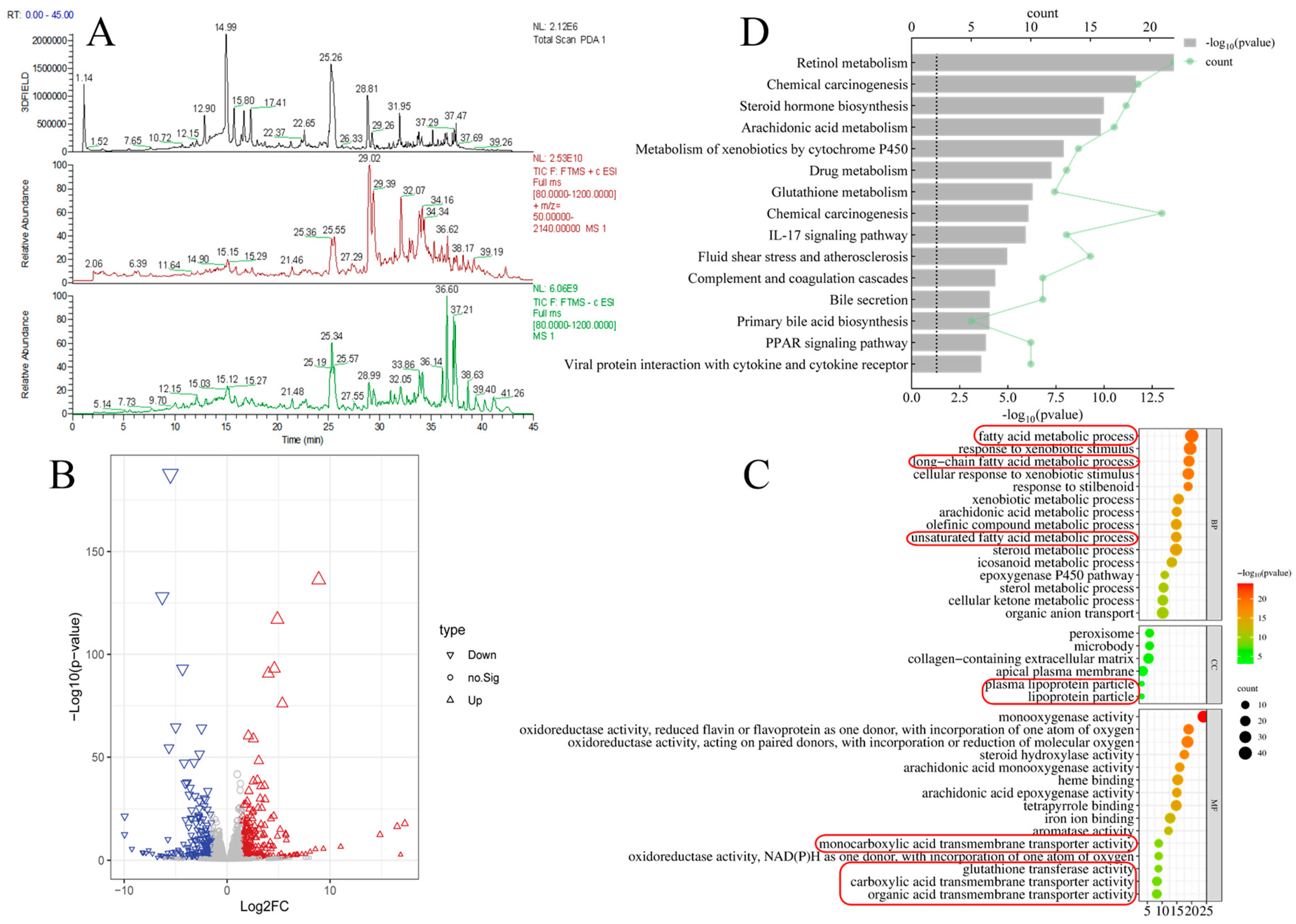


| No | Retention Time (min) | Compound Name | Molecular Weight | Adduct Ion | Molecular Formula | MS/MS Fragments |
|---|---|---|---|---|---|---|
| 1 | 12.90 | 3′-Hydroxypuerarin | 431.0995 | [M−H]− | C21H20O10 | 431.0991, 311.0566, 283.0616 |
| 2 | 14.99 | Puerarin | 415.1047 | [M−H]− | C21H20O9 | 415.1043, 295.0615, 267.0677 |
| 3 | 15.80 | 3′-Methoxypuerarin | 445.1153 | [M−H]− | C22H22O10 | 445.1148, 325.0723, 282.0539 |
| 4 | 16.77 | Puerarin apioside | 547.1473 | [M−H]− | C26H28O13 | 547.1467, 295.0616, 267.0667 |
| 5 | 17.41 | Daidzin | 417.1183 | [M+H]+ | C21H20O9 | 297.0748, 255.0656, 199.0754 |
| 6 | 25.07 | Daidzein | 253.0512 | [M−H]− | C15H10O4 | 253.0509, 209.0609, 135.0078 |
| 7 | 25.57 | Glycitein | 283.0621 | [M−H]+ | C16H12O5 | 283.0616, 268.0381, 211.0394 |
| 8 | 29.02 | Luteolin | 285.0414 | [M−H]− | C15H10O6 | 285.0409, 255.0300, 151.0031 |
| 9 | 36.51 | Ursolic acid | 455.3546 | [M−H]− | C30H48O3 | 455.3540, 407.3370, 238.0641 |
| 10 | 37.43 | Palmitic acid | 255.2334 | [M−H]− | C16H32O2 | 255.2332, 233.1788, 193.8156 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Huang, Q.; Zeng, Y.; Chen, D.; Jia, Z.; Han, B.; Huang, X.; Wang, Q.; Hu, X.; Liao, M.; et al. Pueraria lobata–Prunus mume Complex Alleviates Alcoholic Liver Disease by Regulating Lipid Metabolism and Inhibiting Inflammation: A Transcriptome and Gut Microbiota Analysis. Foods 2024, 13, 2431. https://doi.org/10.3390/foods13152431
Gao R, Huang Q, Zeng Y, Chen D, Jia Z, Han B, Huang X, Wang Q, Hu X, Liao M, et al. Pueraria lobata–Prunus mume Complex Alleviates Alcoholic Liver Disease by Regulating Lipid Metabolism and Inhibiting Inflammation: A Transcriptome and Gut Microbiota Analysis. Foods. 2024; 13(15):2431. https://doi.org/10.3390/foods13152431
Chicago/Turabian StyleGao, Ruixi, Qi Huang, Yanfeng Zeng, Dandan Chen, Ziming Jia, Bingchen Han, Xianju Huang, Qiang Wang, Xin Hu, Maochuan Liao, and et al. 2024. "Pueraria lobata–Prunus mume Complex Alleviates Alcoholic Liver Disease by Regulating Lipid Metabolism and Inhibiting Inflammation: A Transcriptome and Gut Microbiota Analysis" Foods 13, no. 15: 2431. https://doi.org/10.3390/foods13152431
APA StyleGao, R., Huang, Q., Zeng, Y., Chen, D., Jia, Z., Han, B., Huang, X., Wang, Q., Hu, X., Liao, M., & Li, J. (2024). Pueraria lobata–Prunus mume Complex Alleviates Alcoholic Liver Disease by Regulating Lipid Metabolism and Inhibiting Inflammation: A Transcriptome and Gut Microbiota Analysis. Foods, 13(15), 2431. https://doi.org/10.3390/foods13152431







