Susceptibility of Foodborne Pathogens to Milk-Origin Lactic Acid Bacteria Supernatants: A Comprehensive Meta-Regression Study
Abstract
1. Introduction
2. Materials and Methods
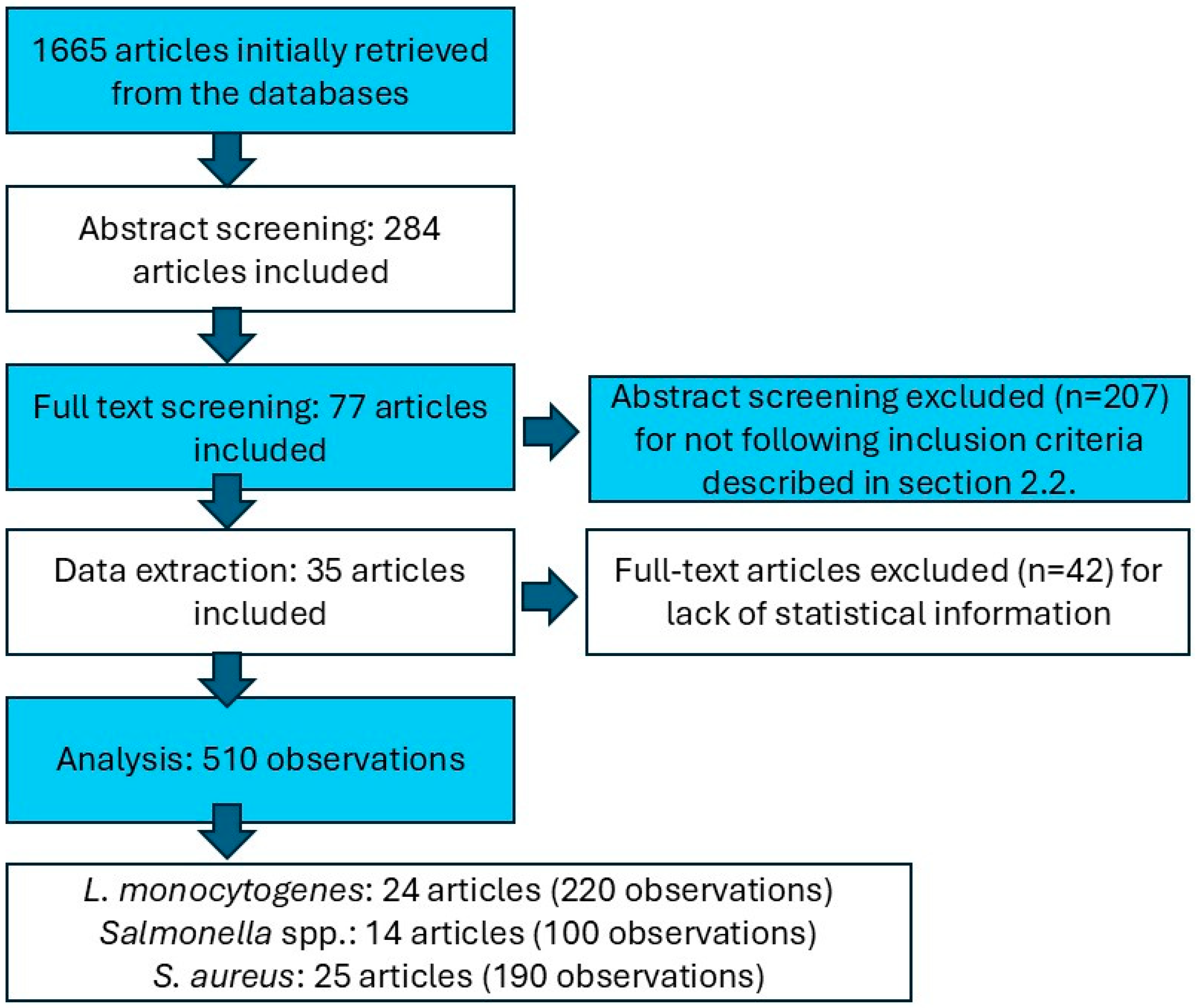
2.1. Bibliographic Search
2.2. Data Extraction
2.3. Meta-Regression Modeling
2.3.1. Overarching Meta-Regression Models
2.3.2. Pathogen-Specific Meta-Regression Models
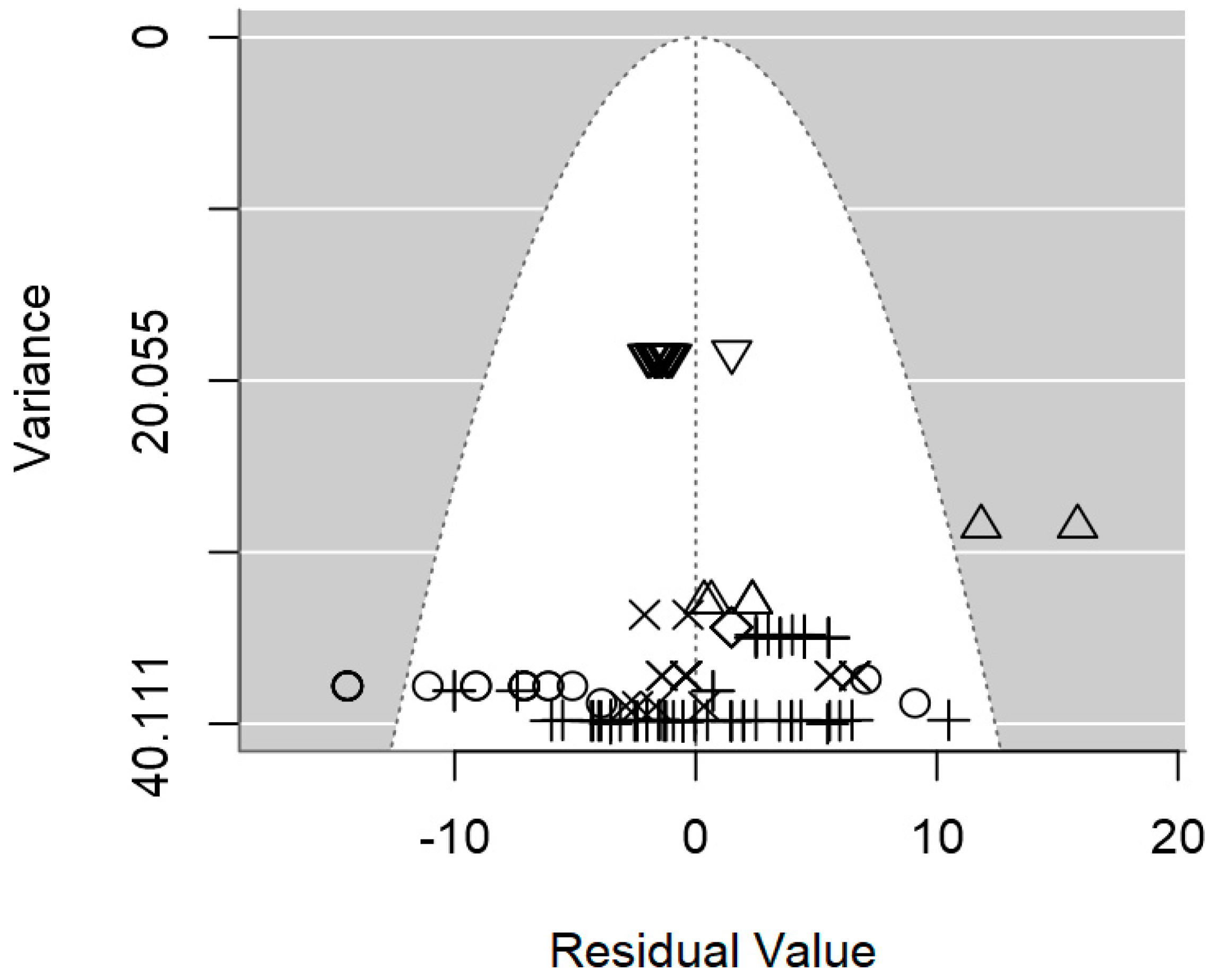
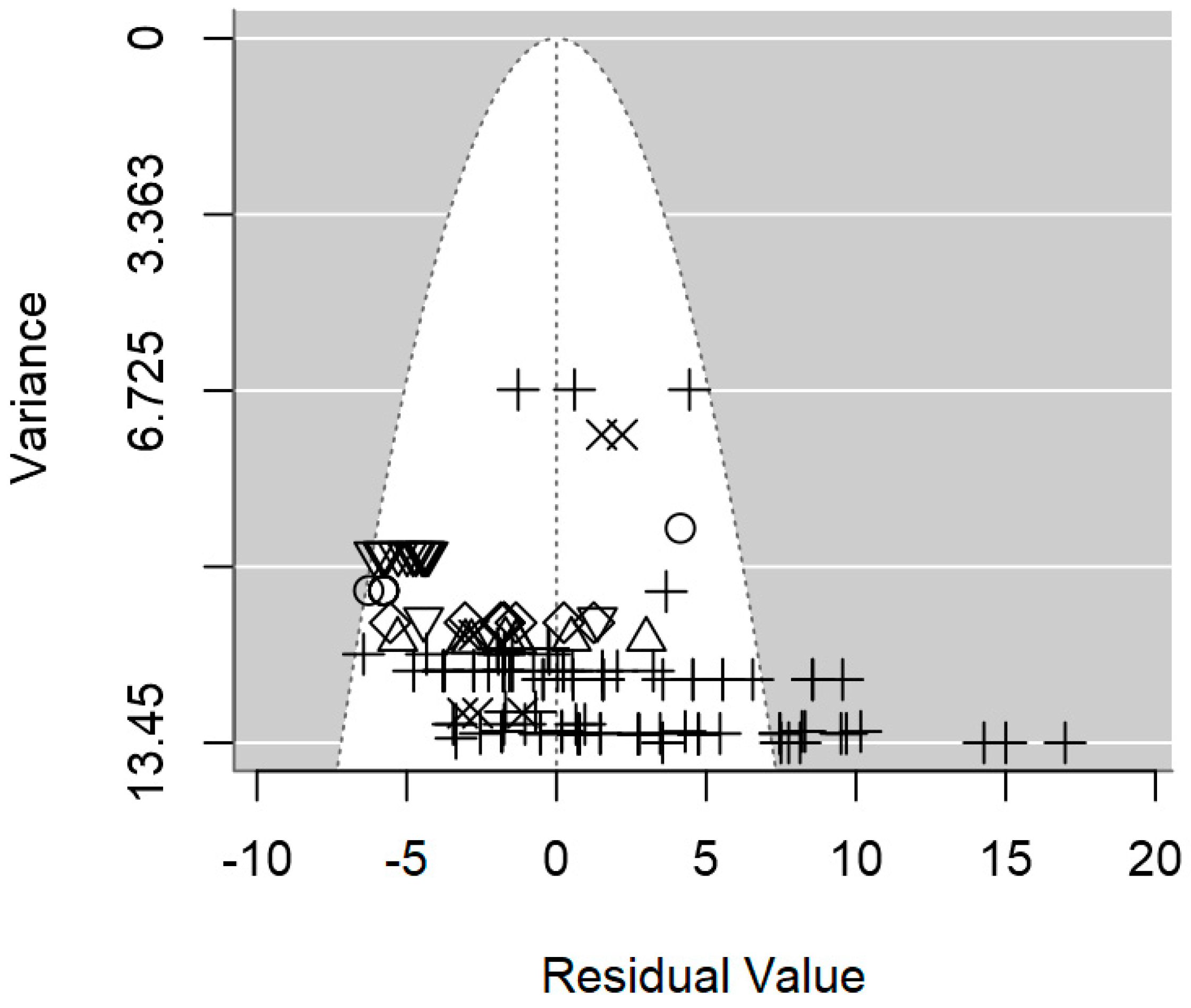
2.4. Assessment of Heterogeneity and Publication Bias
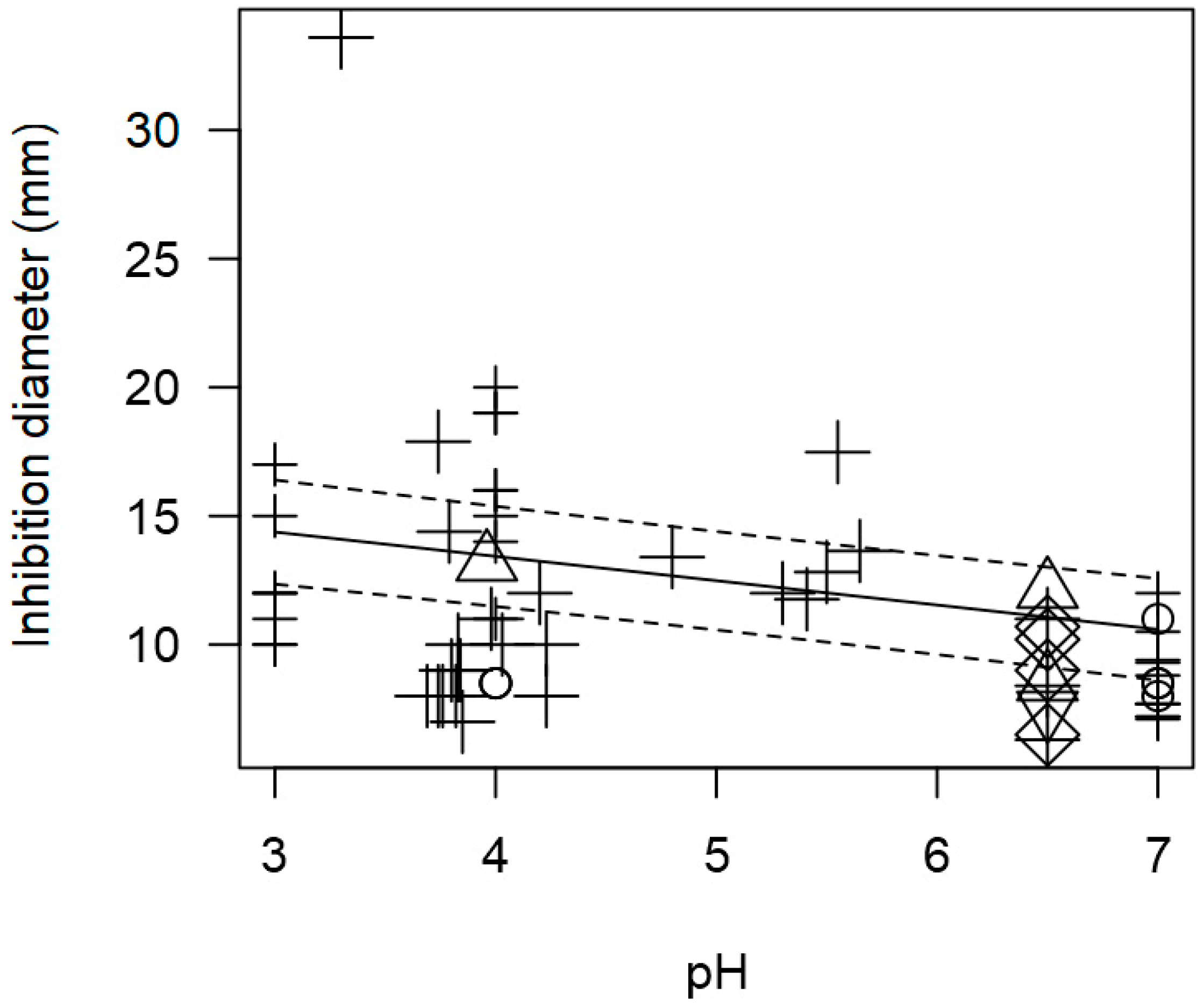
3. Results
3.1. Description of the Meta-Analytical Data
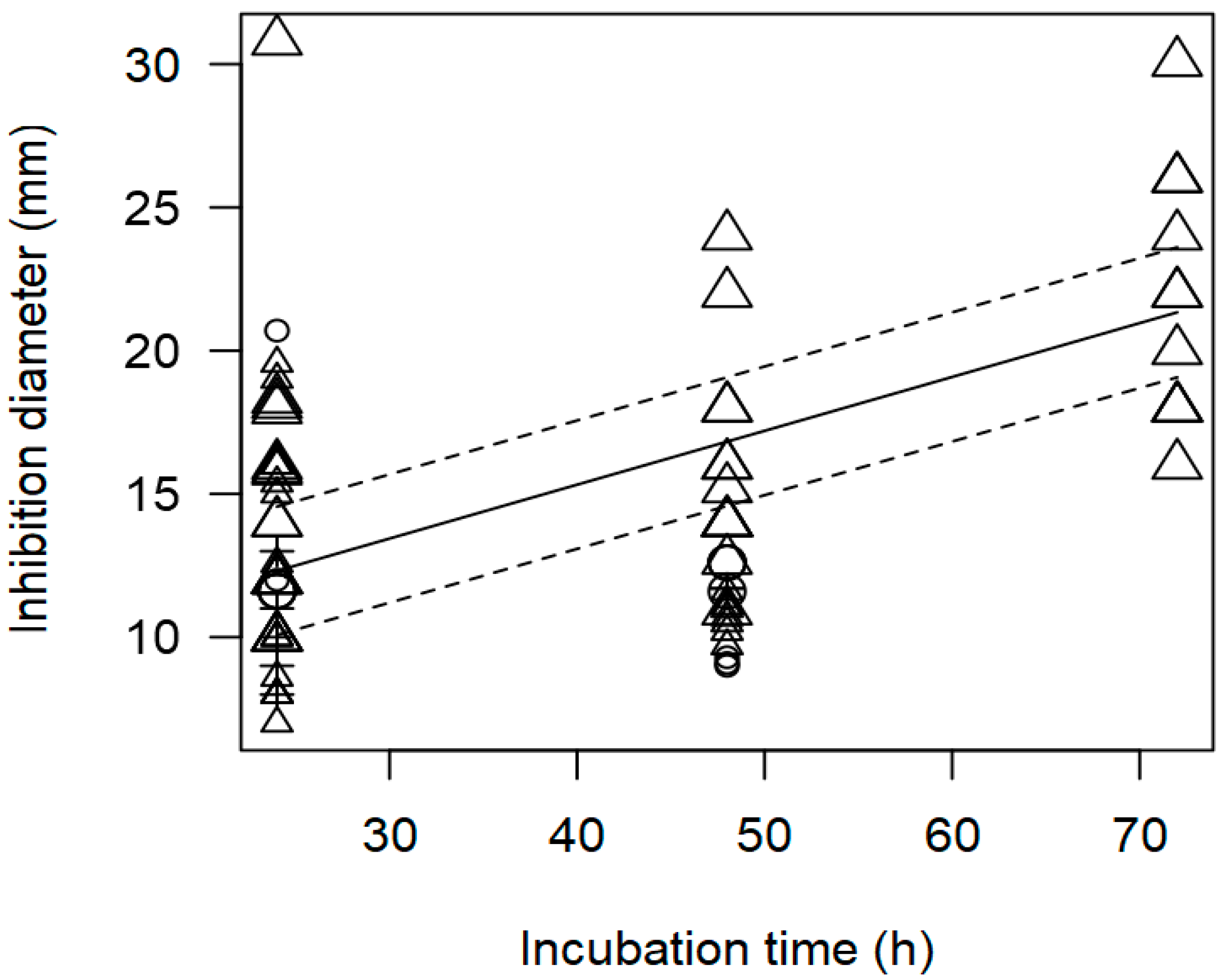
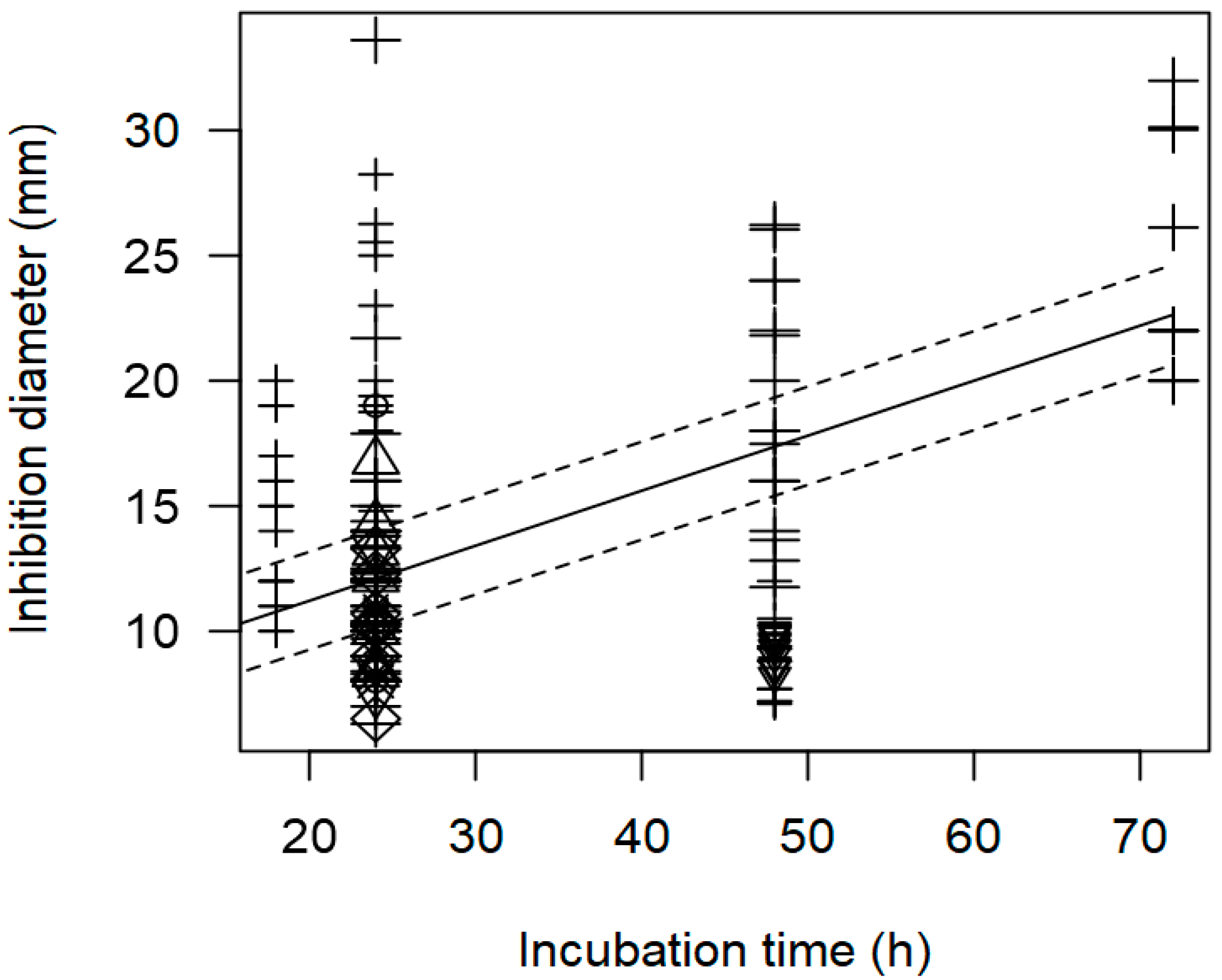
3.2. Overarching Meta-Regression Models
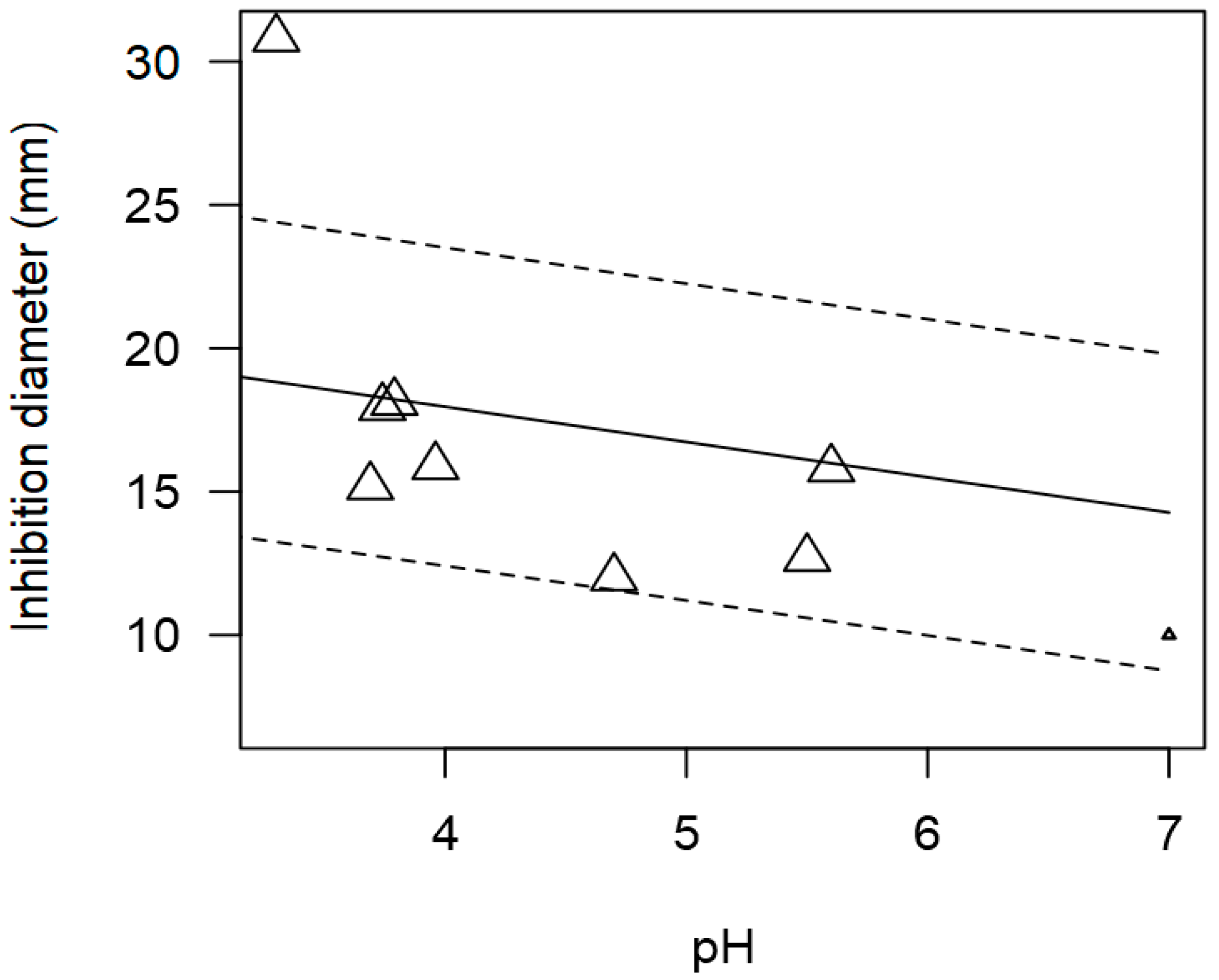
3.3. Pathogen-Specific Meta-Regression Models
3.3.1. Listeria monocytogenes
3.3.2. Salmonella spp.
3.3.3. Staphylococcus aureus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EUROSTAT. Milk and Milk Product Statistics, Eurostat Statistics Explained; EUROSTAT: Luxembourg, 2022; Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Milk_and_milk_product_statistics (accessed on 25 March 2024).
- Cardinali, F.; Osimani, A.; Taccari, M.; Milanović, V.; Garofalo, C.; Clementi, F.; Polverigiani, S.; Zitti, S.; Raffaelli, N.; Mozzon, M.; et al. Impact of Thistle Rennet from Carlina Acanthifolia All. subsp. Acanthifolia on Bacterial Diversity and Dynamics of a Specialty Italian Raw Ewes’ Milk Cheese. Int. J. Food Microbiol. 2017, 255, 7–16. [Google Scholar] [CrossRef]
- Cardinali, F.; Ferrocino, I.; Milanović, V.; Belleggia, L.; Corvaglia, M.R.; Garofalo, C.; Foligni, R.; Mannozzi, C.; Mozzon, M.; Cocolin, L.; et al. Microbial Communities and Volatile Profile of Queijo de Azeitão PDO Cheese, a Traditional Mediterranean Thistle-Curdled Cheese from Portugal. Food Res. Int. 2021, 147, 110537. [Google Scholar] [CrossRef]
- Silva, B.N.; Fernandes, N.; Carvalho, L.; Faria, A.S.; Teixeira, J.A.; Rodrigues, C.; Gonzales-Barron, U.; Cadavez, V. Lactic Acid Bacteria from Artisanal Raw Goat Milk Cheeses: Technological Properties and Antimicrobial Potential. Ital. J. Food Saf. 2023, 12, 11559. [Google Scholar] [CrossRef]
- Nero, L.A.; Andretta, M.; Almeida, T.T.; Ferreira, L.R.; Camargo, A.C.; Yamatogi, R.S.; Carvalho, A.F.; Call, D.R. Lactic Microbiota of the Minas Artisanal Cheese Produced in the Serro Region, Minas Gerais, Brazil. LWT 2021, 148, 111698. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Pineda, A.P.A.; Campos, G.Z.; Pimentel-Filho, N.J.; Franco, B.D.G.d.M.; Pinto, U.M. Brazilian Artisanal Cheeses: Diversity, Microbiological Safety, and Challenges for the Sector. Front. Microbiol. 2021, 12, 666922. [Google Scholar] [CrossRef]
- Favaro, L.; Basaglia, M.; Casella, S.; Hue, I.; Dousset, X.; Dora Gombossy de Melo Franco, B.; Todorov, S.D. Bacteriocinogenic Potential and Safety Evaluation of Non-Starter Enterococcus Faecium Strains Isolated from Home Made White Brine Cheese. Food Microbiol. 2014, 38, 228–239. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef]
- Fung, F.; Wang, H.-S.; Menon, S. Food Safety in the 21st Century. Biomed. J. 2018, 41, 88–95. [Google Scholar] [CrossRef]
- European Commission Rapid Alert System for Food and Feed (RASFF). Available online: https://webgate.ec.europa.eu/rasff-window/screen/search (accessed on 25 July 2024).
- Delgado, J.; Álvarez, M.; Cebrián, E.; Martín, I.; Roncero, E.; Rodríguez, M. Biocontrol of Pathogen Microorganisms in Ripened Foods of Animal Origin. Microorganisms 2023, 11, 1578. [Google Scholar] [CrossRef]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Vandera, E.; Kakouri, A.; Koukkou, A.-I.; Samelis, J. Major Ecological Shifts within the Dominant Nonstarter Lactic Acid Bacteria in Mature Greek Graviera Cheese as Affected by the Starter Culture Type. Int. J. Food Microbiol. 2019, 290, 15–26. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Sarkar, T.; Pati, S.; Basu, D.; Abdul Kari, Z.; Wei, L.S.; Smaoui, S.; Wen Goh, K.; et al. Bacteriocin: A Natural Approach for Food Safety and Food Security. Front. Bioeng. Biotechnol. 2022, 10, 1005918. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Godolphin, P.J.; Bath, P.M.; Montgomery, A.A. Short Email with Attachment versus Long Email without Attachment When Contacting Authors to Request Unpublished Data for a Systematic Review: A Nested Randomised Trial. BMJ Open 2019, 9, e025273. [Google Scholar] [CrossRef]
- Microsoft Corporation. Microsoft Excel Online, Spreadsheet Software. Available online: https://office.microsoft.com/excel (accessed on 15 January 2024).
- Priya, B.K.; Reddy, D.A.; Rani, A.D.; Kalahasthi, N.; Soliman, W.G.; Reddy, D.V.R.K. Automatic Inhibition Zone Diameter Measurement for Disc Diffusion Test Using Image Segmentation. IETE J. Res. 2023, 69, 5708–5725. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 20 January 2024).
- Viechtbauer, W. Conducting Meta-Analyses in R with the Metafor Package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Doleman, B.; Freeman, S.C.; Lund, J.N.; Williams, J.P.; Sutton, A.J. Funnel Plots May Show Asymmetry in the Absence of Publication Bias with Continuous Outcomes Dependent on Baseline Risk: Presentation of a New Publication Bias Test. Res. Synth. Methods 2020, 11, 522–534. [Google Scholar] [CrossRef]
- Alvarado, C.; García-Almendárez, B.E.; Martin, S.E.; Regalado, C. Anti-Listeria Monocytogenes Bacteriocin-like Inhibitory Substances from Enterococcus Faecium UQ31 Isolated from Artisan Mexican-Style Cheese. Curr. Microbiol. 2005, 51, 110–115. [Google Scholar] [CrossRef]
- Azizi, F.; Habibi Najafi, M.B.; Edalatian Dovom, M.R. The Biodiversity of Lactobacillus spp. from Iranian Raw Milk Motal Cheese and Antibacterial Evaluation Based on Bacteriocin-Encoding Genes. AMB Express 2017, 7, 176. [Google Scholar] [CrossRef]
- Bian, X.; Evivie, S.E.; Muhammad, Z.; Luo, G.-W.; Liang, H.-Z.; Wang, N.-N.; Huo, G.-C. In Vitro Assessment of the Antimicrobial Potentials of Lactobacillus Helveticus Strains Isolated from Traditional Cheese in Sinkiang China against Food-Borne Pathogens. Food Funct. 2016, 7, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Câmara, S.P.; Dapkevicius, A.; Riquelme, C.; Elias, R.B.; Silva, C.C.G.; Malcata, F.X.; Dapkevicius, M.L.N.E. Potential of Lactic Acid Bacteria from Pico Cheese for Starter Culture Development. Food Sci. Technol. Int. 2019, 25, 303–317. [Google Scholar] [CrossRef]
- Ferrari, I.D.S.; de Souza, J.V.; Ramos, C.L.; da Costa, M.M.; Schwan, R.F.; Dias, F.S. Selection of Autochthonous Lactic Acid Bacteria from Goat Dairies and Their Addition to Evaluate the Inhibition of Salmonella Typhi in Artisanal Cheese. Food Microbiol. 2016, 60, 29–38. [Google Scholar] [CrossRef]
- Hejazi, M.A.; Ghafouri-Fard, S.; Eslami, S.; Afshar, D.; Barzegari, A.; Khorshidian, N. Polyphasic Characterization of Enterococcus Strains Isolated from Traditional Moghan Cheese in Iran. J. Food Saf. 2019, 39, e12631. [Google Scholar] [CrossRef]
- Heredia-Castro, P.Y.; Méndez-Romero, J.I.; Hernández-Mendoza, A.; Acedo-Félix, E.; González-Córdova, A.F.; Vallejo-Cordoba, B. Antimicrobial Activity and Partial Characterization of Bacteriocin-like Inhibitory Substances Produced by Lactobacillus spp. Isolated from Artisanal Mexican Cheese. J. Dairy Sci. 2015, 98, 8285–8293. [Google Scholar] [CrossRef]
- Jutinico-Shubach, A.; Gutiérrez-Cortés, C.; Suarez, H. Antilisterial Activity of Chitosan-Based Edible Coating Incorporating Cell-Free Supernatant from Pediococcus Pentosaceus 147 on the Preservation of Fresh Cheese. J. Food Process. Preserv. 2020, 44, e14715. [Google Scholar] [CrossRef]
- Kačániová, M.; Borotová, P.; Terentjeva, M.; Kunová, S.; Felšöciová, S.; Haščík, P.; Lopašovský, L.; Štefániková, J. Bryndza Cheese of Slovak Origin as Potential Resources of Probiotic Bacteria. Potravin. Slovak J. Food Sci. 2020, 14, 641–646. [Google Scholar] [CrossRef]
- Kanak, E.K.; Yilmaz, S.Ö. Maldi-Tof Mass Spectrometry for the Identification and Detection of Antimicrobial Activity of Lactic Acid Bacteria Isolated from Local Cheeses. Food Sci. Technol. 2019, 39, 462–469. [Google Scholar] [CrossRef]
- Kanak, E.K.; Yilmaz, S.Ö. Identification, Antibacterial and Antifungal Effects, Antibiotic Resistance of Some Lactic Acid Bacteria. Food Sci. Technol. 2021, 41, 174–182. [Google Scholar] [CrossRef]
- Leboš Pavunc, A.; Kos, B.; Beganović, J.; Uroić, K.; Bučan, D.; Šušković, J. Antibiotic Susceptibility and Antimicrobial Activity of Autochthonous Starter Cultures as Safety Parameters for Fresh Cheese Production. Mljekarstvo 2013, 63, 185–194. [Google Scholar]
- Loessner, M.; Guenther, S.; Steffan, S.; Scherer, S. A Pediocin-Producing Lactobacillus Plantarum Strain Inhibits Listeria Monocytogenes in a Multispecies Cheese Surface Microbial Ripening Consortium. Appl. Environ. Microbiol. 2003, 69, 1854–1857. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, Z.; Ramzan, R.; Abdelazez, A.; Amjad, A.; Afzaal, M.; Zhang, S.; Pan, S. Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus Plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese Against Foodborne Pathogens. Pathogens 2019, 8, 71. [Google Scholar] [CrossRef]
- Nespolo, C.R.; Brandelli, A. Production of Bacteriocin-like Substances by Lactic Acid Bacteria Isolated from Regional Ovine Cheese. Braz. J. Microbiol. 2010, 41, 1009–1018. [Google Scholar] [CrossRef]
- Ołdak, A.; Zielińska, D.; Kołożyn-Krajewska, D. Comparison of antagonistic activity of lactic acid bacteria isolated from various types of traditional food. Zywnosc. Nauka Technol. Jakosc/Food Sci. Technol. Qual. 2019, 26, 60–72. [Google Scholar] [CrossRef]
- Partovi, R.; Gandomi, H.; Akhondzadeh Basti, A. Safety Aspects of Lactobacillus Plantarum Strains Isolated from Siahmazgi Cheese. Ank. Univ. Vet. Fak. Derg. 2019, 66, 337–342. [Google Scholar] [CrossRef]
- Settanni, L.; Franciosi, E.; Cavazza, A.; Cocconcelli, P.S.; Poznanski, E. Extension of Tosèla Cheese Shelf-Life Using Non-Starter Lactic Acid Bacteria. Food Microbiol. 2011, 28, 883–890. [Google Scholar] [CrossRef]
- Sip, A.; Więckowicz, M.; Olejnik-Schmidt, A.; Grajek, W. Anti-Listeria Activity of Lactic Acid Bacteria Isolated from Golka, a Regional Cheese Produced in Poland. Food Control 2012, 26, 117–124. [Google Scholar] [CrossRef]
- Toğay, S.Ö.; Ay, M.; Güneşer, O.; Yüceer, Y.K. Investigation of Antimicrobial Activity and entA and entB Genes in Enterococcus Faecium and Enterococcus Faecalis Strains Isolated from Naturally Fermented Turkish White Cheeses. Food Sci. Biotechnol. 2016, 25, 1633–1637. [Google Scholar] [CrossRef]
- Tulini, F.L.; Winkelströter, L.K.; De Martinis, E.C.P. Identification and Evaluation of the Probiotic Potential of Lactobacillus Paraplantarum FT259, a Bacteriocinogenic Strain Isolated from Brazilian Semi-Hard Artisanal Cheese. Anaerobe 2013, 22, 57–63. [Google Scholar] [CrossRef]
- Vandera, E.; Tsirka, G.; Kakouri, A.; Koukkou, A.-I.; Samelis, J. Approaches for Enhancing in Situ Detection of Enterocin Genes in Thermized Milk, and Selective Isolation of Enterocin-Producing Enterococcus Faecium from Baird-Parker Agar. Int. J. Food Microbiol. 2018, 281, 23–31. [Google Scholar] [CrossRef]
- Vataščinová, T.; Pipová, M.; Fraqueza, M.J.R.; Maľa, P.; Dudriková, E.; Drážovská, M.; Lauková, A. Short Communication: Antimicrobial Potential of Lactobacillus Plantarum Strains Isolated from Slovak Raw Sheep Milk Cheeses. J. Dairy Sci. 2020, 103, 6900–6903. [Google Scholar] [CrossRef] [PubMed]
- Vera Pingitore, E.; Todorov, S.D.; Sesma, F.; Gombossy de Melo Franco, B.D. Application of Bacteriocinogenic Enterococcus Mundtii CRL35 and Enterococcus Faecium ST88Ch in the Control of Listeria Monocytogenes in Fresh Minas Cheese. Food Microbiol. 2012, 32, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Champiri, I.D.; Bamzadeh, Z.; Rahimi, E.; Rouhi, L. Isolation and Identification of Lactobacillus Brevis from Local Cheese of Bazoft and Evaluation of Antimicrobial Activity against Some Pathogenic Microorganisms. Iran. J. Med. Microbiol. 2022, 16, 17–34. [Google Scholar] [CrossRef]
- Erdoǧrul, Ö.; Erbilir, F. Isolation and Characterization of Lactobacillus Bulgaricus and Lactobacillus Casei from Various Foods. Turk. J. Biol. 2006, 30, 39–44. [Google Scholar]
- Heredia-Castro, P.Y.; Reyes-Díaz, R.; Rendón-Rosales, M.Á.; Beltrán-Barrientos, L.M.; Torres-Llanez, M.J.; Estrada-Montoya, M.C.; Hernández-Mendoza, A.; González-Córdova, A.F.; Vallejo-Cordoba, B. Novel Bacteriocins Produced by Lactobacillus Fermentum Strains with Bacteriostatic Effects in Milk against Selected Indicator Microorganisms. J. Dairy Sci. 2021, 104, 4033–4043. [Google Scholar] [CrossRef]
- Hojjati, M.; Behabahani, B.A.; Falah, F. Aggregation, Adherence, Anti-Adhesion and Antagonistic Activity Properties Relating to Surface Charge of Probiotic Lactobacillus Brevis Gp104 against Staphylococcus Aureus. Microb. Pathog. 2020, 147, 104420. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Goda, H.A.; Abdel-Hamid, M.; Renye, J.A., Jr.; Yang, P.; Huang, Z.; Zeng, Q.-K.; Li, L. Partial Characterization of Probiotic Lactic Acid Bacteria Isolated from Chinese Dairy Products. Int. J. Food Prop. 2021, 24, 446–456. [Google Scholar] [CrossRef]
- Potočnjak, M.; Pušić, P.; Frece, J.; Abram, M.; Jankovic, T.; Gobin, I. Three New Lactobacillus Plantarum Strains in the Probiotic Toolbox against Gut Pathogen Salmonella Enterica Serotype Typhimurium. Food Technol. Biotechnol. 2017, 55, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Slyvka, I.; Tsisaryk, O.; Musii, L.; Kushnir, I.; Koziorowski, M.; Koziorowska, A. Identification and Investigation of Properties of Strains Enterococcus spp. Isolated from Artisanal Carpathian Cheese. Biocatal. Agric. Biotechnol. 2022, 39, 102259. [Google Scholar] [CrossRef]
- Ait Chait, Y.; Gunenc, A.; Hosseinian, F.; Bendali, F. Antipathogenic and Probiotic Potential of Lactobacillus Brevis Strains Newly Isolated from Algerian Artisanal Cheeses. Folia Microbiol. 2021, 66, 429–440. [Google Scholar] [CrossRef]
- Jabbari, V.; Khiabani, M.S.; Mokarram, R.R.; Hassanzadeh, A.M.; Ahmadi, E.; Gharenaghadeh, S.; Karimi, N.; Kafil, H.S. Lactobacillus Plantarum as a Probiotic Potential from Kouzeh Cheese (Traditional Iranian Cheese) and Its Antimicrobial Activity. Probiotics Antimicrob. Proteins 2017, 9, 189–193. [Google Scholar] [CrossRef]
- Karami, S.; Roayaei, M.; Hamzavi, H.; Bahmani, M.; Hassanzad-Azar, H.; Leila, M.; Rafieian-Kopaei, M. Isolation and Identification of Probiotic Lactobacillus from Local Dairy and Evaluating Their Antagonistic Effect on Pathogens. Int. J. Pharm. Investig. 2017, 7, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.; Kheadr, E.; El-Ziney, M.; Dabour, N. Lactobacillus plantarum Protective Cultures to Improve Safety and Quality of Wheyless Domiati-like Cheese. J. Food Process. Preserv. 2022, 46, e16416. [Google Scholar] [CrossRef]
- Madi, N.; Boushaba, R. Identification of Potential Biopreservative Lactic Acid Bacteria Strains Isolated from Algerian Cow’s Milk and Demonstration of Antagonism against S. aureus in Cheese. Food Sci. Technol. Res. 2017, 23, 679–688. [Google Scholar] [CrossRef]
- Ngamsomchat, A.; Kaewkod, T.; Konkit, M.; Tragoolpua, Y.; Bovonsombut, S.; Chitov, T. Characterisation of Lactobacillus Plantarum of Dairy-Product Origin for Probiotic Chèvre Cheese Production. Foods 2022, 11, 934. [Google Scholar] [CrossRef] [PubMed]
- Vahabzadeh, S.; Özpinar, H. Investigation of Some Biochemical Properties, Antimicrobial Activity and Antibiotic Resistances of Kefir Supernatants and Lactococcus Lactis Ssp. Lactis Strains Isolated from Raw Cow Milk and Cheese Samples. Kafkas Univ. Vet. Fak. Derg. 2018, 24, 443–450. [Google Scholar] [CrossRef]
- Parente, E.; Zotta, T.; Ricciardi, A. Microbial Association Networks in Cheese: A Meta-Analysis. bioRxiv 2021. bioRxiv:2021.07.21.453196. [Google Scholar] [CrossRef]
- García-Cano, I.; Rocha-Mendoza, D.; Kosmerl, E.; Zhang, L.; Jiménez-Flores, R. Technically Relevant Enzymes and Proteins Produced by LAB Suitable for Industrial and Biological Activity. Appl. Microbiol. Biotechnol. 2020, 104, 1401–1422. [Google Scholar] [CrossRef]
- Walsh, A.M.; Macori, G.; Kilcawley, K.N.; Cotter, P.D. Meta-Analysis of Cheese Microbiomes Highlights Contributions to Multiple Aspects of Quality. Nat. Food 2020, 1, 500–510. [Google Scholar] [CrossRef]
- Fu, Y.; Mu, D.; Qiao, W.; Zhu, D.; Wang, X.; Liu, F.; Xu, H.; Saris, P.; Kuipers, O.P.; Qiao, M. Co-Expression of Nisin Z and Leucocin C as a Basis for Effective Protection against Listeria Monocytogenes in Pasteurized Milk. Front. Microbiol. 2018, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.O.; Erazo Flores, B.J.; Skeens, J.; Kent, D.; Murphy, S.I.; Wiedmann, M.; Guariglia-Oropeza, V. Nevertheless, She Resisted-Role of the Environment on Listeria Monocytogenes Sensitivity to Nisin Treatment in a Laboratory Cheese Model. Front. Microbiol. 2020, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Taye, Y.; Degu, T.; Fesseha, H.; Mathewos, M. Isolation and Identification of Lactic Acid Bacteria from Cow Milk and Milk Products. ScientificWorldJournal 2021, 2021, 4697445. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.N.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Meta-Regression Models Describing the Effects of Essential Oils and Added Lactic Acid Bacteria on Pathogen Inactivation in Cheese. Microb. Risk Anal. 2021, 18, 100131. [Google Scholar] [CrossRef]
- Lam, L.H.; Monack, D.M. Intraspecies Competition for Niches in the Distal Gut Dictate Transmission during Persistent Salmonella Infection. PLoS Pathog. 2014, 10, e1004527. [Google Scholar] [CrossRef]
- Shafique, B.; Ranjha, M.M.A.N.; Murtaza, M.A.; Walayat, N.; Nawaz, A.; Khalid, W.; Mahmood, S.; Nadeem, M.; Manzoor, M.F.; Ameer, K.; et al. Recent Trends and Applications of Nanoencapsulated Bacteriocins against Microbes in Food Quality and Safety. Microorganisms 2022, 11, 85. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
| Pathogen 1 | LAB Genus | Method | Pooled Inhibition Diameter 2,3 (SE) [mm] | n | N | |
|---|---|---|---|---|---|---|
| Listeria monocytogenes A | Enterococcus | Overall | 15.90 A (2.138) | 38 | 24 | |
| Well | 19.47 a (2.409) | 28 | ||||
| Disk | 10.98 c (1.962) | 10 | ||||
| Lacticaseibacillus | Overall | 13.95 B (2.313) | 8 | |||
| Well | 28.57 a (5.058) | 4 | ||||
| Disk | 11.73 c (1.835) | 4 | ||||
| Lactobacillus | Overall | 11.96 C (1.886) | 92 | |||
| Spot | 14.87 b (2.883) | 14 | ||||
| Well | 12.02 c (2.541) | 18 | ||||
| Disk | 10.88 c (1.598) | 60 | ||||
| Lactococcus | Overall | 13.12 B (2.129) | 20 | |||
| Spot | 17.75 a (2.076) | 14 | ||||
| Disk | 10.23 c (1.792) | 6 | ||||
| Leuconostoc | Overall | 10.25 C (2.712) | 5 | |||
| Spot | 14.04 b (3.045) | 4 | ||||
| Pediococcus | Overall | 8.735 C (4.663) | 14 | |||
| Well | 8.800 c (3.025) | 14 | ||||
| Salmonella spp B | Enterococcus | Overall | 12.00 A (1.195) | 13 | 14 | |
| Well | 13.39 a (1.408) | 12 | ||||
| Lactobacillus | Overall | 12.36 A (1.148) | 69 | |||
| Well | 13.58 a (1.362) | 18 | ||||
| Disk | 11.13 b (1.361) | 51 | ||||
| Lactococcus | Overall | 12.76 A (1.194) | 7 | |||
| Disk | 10.82 b (1.433) | 5 | ||||
| Staphylococcus aureus B | Enterococcus | Overall | 11.01 A (2.105) | 10 | 25 | |
| Well | 14.16 a (0.716) | 4 | ||||
| Disk | 5.28 d (0.366) | 6 | ||||
| Lacticaseibacillus | Overall | 11.89 A (0.573) | 9 | |||
| Well | 12.36 b (1.501) | 5 | ||||
| Disk | 12.00 b (1.197) | 4 | ||||
| Lactobacillus | Overall | 11.35 A (1.096) | 133 | |||
| Well | 10.00 c (1.772) | 67 | ||||
| Disk | 12.03 b (1.394) | 66 | ||||
| Lactococcus | Overall | 11.33 A (9.578) | 7 | |||
| Disk | 11.24 b (0.529) | 5 | ||||
| Leuconostoc | Overall | 7.173 B (0.287) | 8 | |||
| Well | 7.198 (0.304) | 8 | ||||
| Pediococcus | Overall | 7.173 B (0.225) | 15 | |||
| Well | 7.161 (0.232) | 15 | ||||
| Parameter | Estimate | Standard Error | p-Value | n | Heterogeneity Analysis 1 |
|---|---|---|---|---|---|
| Intercept | 45.67 | 12.41 | 0.002 | s2 = 53.0 | |
| LAB genus | τ2 = 65.2 | ||||
| Lacticaseibacillus | −1.217 | 2.112 | 0.565 | 8 | τ2res = 43.1 |
| Lactobacillus | −3.375 | 1.580 | 0.033 | 92 | R2 = 33.8% |
| Lactococcus | −2.248 | 1.787 | 0.208 | 20 | I2 = 55.1% |
| Leuconostoc | −5.140 | 2.528 | 0.042 | 5 | |
| Enterococcus | - | - | - | 81 | |
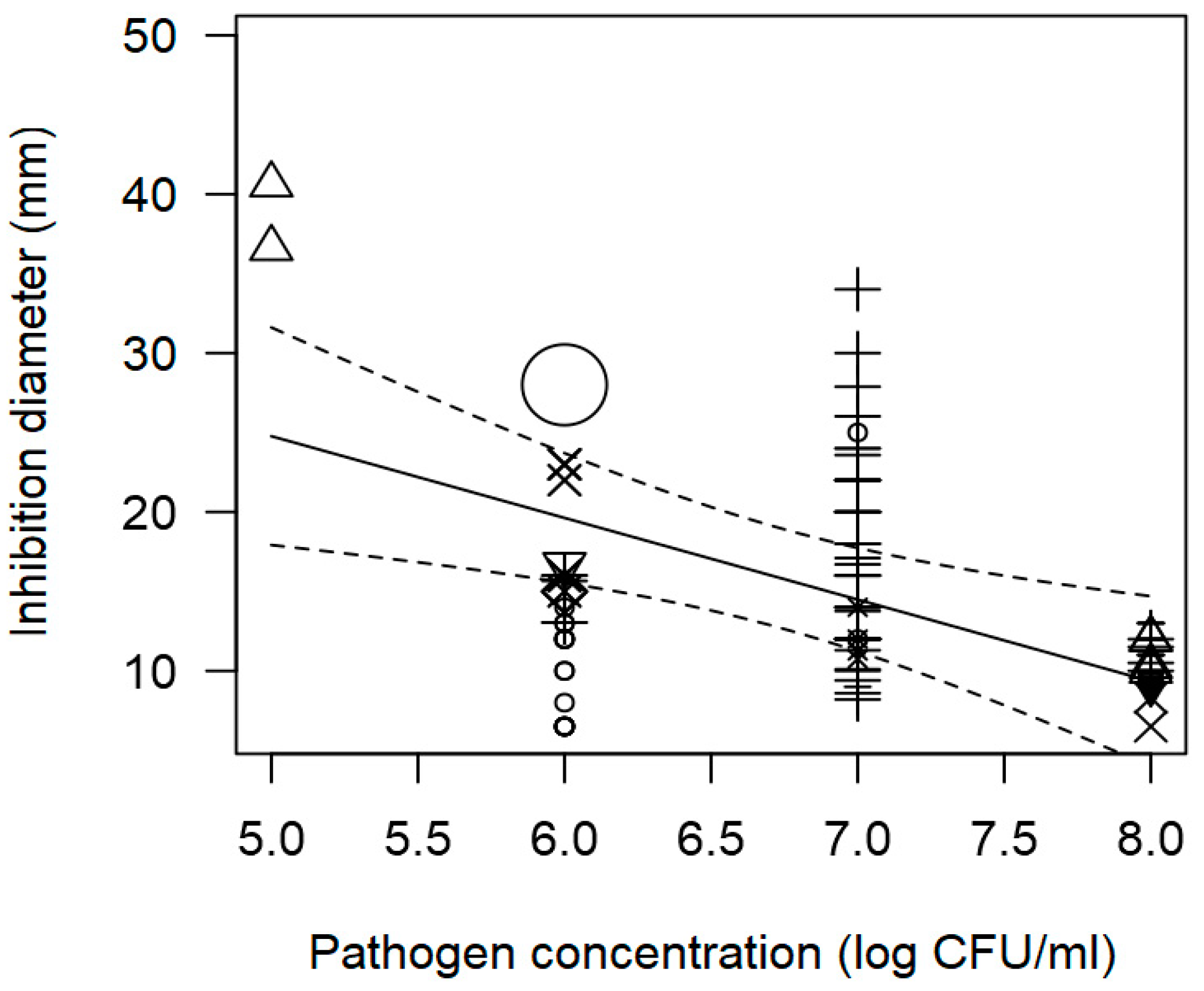
| Pathogen concentration | −5.037 | 1.840 | 0.006 | Publication bias | |
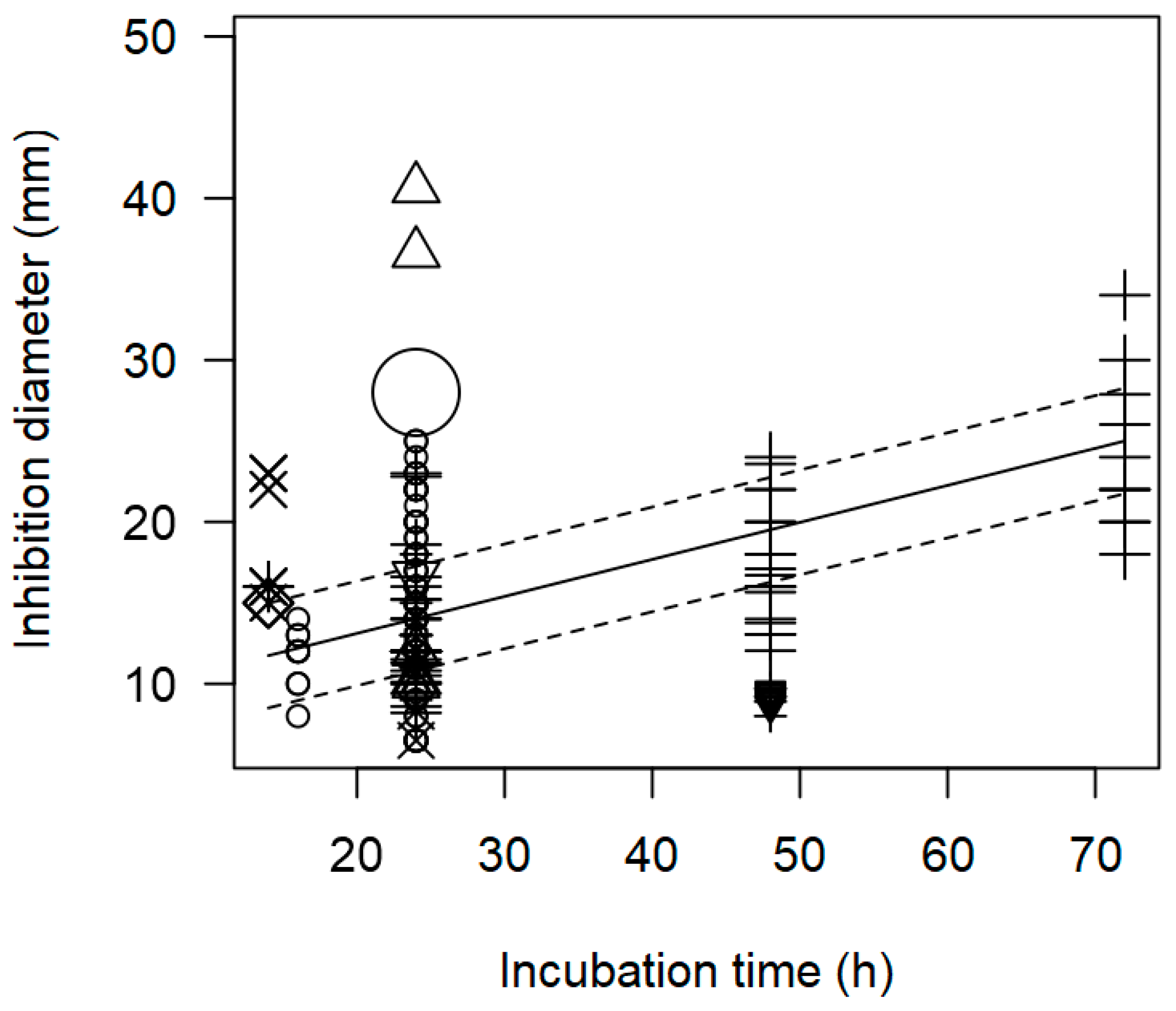
| Incubation time | 0.229 | 0.229 | <0.0001 | p = 0.977 |
| Parameter | Estimate | Standard Error | p-Value | n | Heterogeneity Analysis 1 |
|---|---|---|---|---|---|
| Intercept | 10.41 | 2.250 | <0.0001 | ||
| Susceptibility method | |||||
| Disk diffusion | −2.529 | 0.979 | 0.009 | s2 = 25.1 | |
| Agar | τ2 = 12.8 | ||||
| MH | −1.540 | 2.920 | 0.589 | 11 | I2 = 33.6% |
| MRS | −8.114 | 2.273 | 0.001 | 20 | τ2res = 6.1 |
| Nutrient | 3.575 | 2.553 | 0.161 | 5 | R2 = 52.3% |
| TSA | −2.111 | 2.037 | 0.300 | 10 | |
| BHI | - | - | - | 43 | Publication bias |
| Incubation time | 0.188 | 0.005 | <0.0001 | p = 0.217 |
| Parameter | Estimate | Standard Error | p-Value | n | Heterogeneity Analysis 1 |
|---|---|---|---|---|---|
| Intercept | 22.04 | 2.545 | <0.0001 | ||
| LAB genus | s2 = 31.8 | ||||
| Lacticaseibacillus | −0.923 | 1.481 | 0.533 | 9 | τ2 = 20.2 |
| Lactobacillus | −1.295 | 1.208 | 0.283 | 133 | I2 = 38.8% |
| Lactococcus | −1.758 | 1.842 | 0.340 | 7 | τ2res = 16.2 |
| Leuconostoc | −5.666 | 1.835 | 0.002 | 8 | R2 = 19.8% |
| Pediococcus | −7.335 | 2.154 | 0.001 | 15 | |
| Enterococcus | - | - | - | 10 | |
| Pathogen concentration | −1.888 | 0.297 | <0.0001 | Publication bias | |
| Incubation time | 0.220 | 0.005 | <0.0001 | p = 0.492 | |
| Susceptibility method | |||||
| Disk diffusion | −2.373 | 0.483 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, N.; Loforte, Y.; Cadavez, V.; Gonzales-Barron, U. Susceptibility of Foodborne Pathogens to Milk-Origin Lactic Acid Bacteria Supernatants: A Comprehensive Meta-Regression Study. Foods 2024, 13, 2635. https://doi.org/10.3390/foods13162635
Fernandes N, Loforte Y, Cadavez V, Gonzales-Barron U. Susceptibility of Foodborne Pathogens to Milk-Origin Lactic Acid Bacteria Supernatants: A Comprehensive Meta-Regression Study. Foods. 2024; 13(16):2635. https://doi.org/10.3390/foods13162635
Chicago/Turabian StyleFernandes, Nathália, Yara Loforte, Vasco Cadavez, and Ursula Gonzales-Barron. 2024. "Susceptibility of Foodborne Pathogens to Milk-Origin Lactic Acid Bacteria Supernatants: A Comprehensive Meta-Regression Study" Foods 13, no. 16: 2635. https://doi.org/10.3390/foods13162635
APA StyleFernandes, N., Loforte, Y., Cadavez, V., & Gonzales-Barron, U. (2024). Susceptibility of Foodborne Pathogens to Milk-Origin Lactic Acid Bacteria Supernatants: A Comprehensive Meta-Regression Study. Foods, 13(16), 2635. https://doi.org/10.3390/foods13162635








