Antioxidant Activity of Protein Hydrolysates from Redlip Mullet (Chelon haematocheilus) Muscle and Byproducts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Fish Sample
2.3. Preparation of Protein Hydrolysates
2.4. Determination of Protein Content
2.5. Determination of α-Amino Acid Content and the Degree of Hydrolysis
2.6. Determination of Antioxidant Activity
2.6.1. DPPH• Scavenging Activity
2.6.2. ABTS• Scavenging Activity
2.6.3. SOD-like Activity
2.7. Statistical Analysis
3. Results
3.1. Degree of Hydrolysis
3.1.1. White Meat
3.1.2. Red Meat
3.1.3. Frames
3.1.4. Head
3.1.5. Viscera
3.1.6. Fins
3.1.7. Skin
3.1.8. Scales
3.2. Protein Content
3.2.1. White Meat
3.2.2. Red Meat
3.2.3. Frames
3.2.4. Head
3.2.5. Viscera
3.2.6. Fins
3.2.7. Skin
3.2.8. Scales
3.3. DPPH• Scavenging Activity
3.3.1. White Meat
3.3.2. Red Meat
3.3.3. Frames
3.3.4. Head
3.3.5. Viscera
3.3.6. Fins
3.3.7. Skin
3.3.8. Scales
3.4. ABTS• Scavenging Activity
3.5. SOD-like Activity
3.5.1. White Meat
3.5.2. Red Meat
3.5.3. Frames
3.5.4. Head
3.5.5. Viscera
3.5.6. Fins
3.5.7. Skin
3.5.8. Scales
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barrow, C.; Shahidi, F. Marine Nutraceuticals and Functional Foods, 1st ed.; CRC Press: Boca Raton, FL, USA, 2007; pp. 1–512. [Google Scholar]
- Gao, M.-T.; Hirata, M.; Toorisaka, E.; Hano, T. Acid-hydrolysis of fish wastes for lactic acid fermentation. Bioresour. Technol. 2006, 97, 2414–2420. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.M.I.; Park, Y.-J.; An, J.H.; Choi, S.-J.; Kim, J.-H.; Baek, M.-K.; Kim, A.; Sohn, J.H.; Choi, J.S. Antioxidant properties of Scomber japonicus hydrolysates prepared by enzymatic hydrolysis. J. Aquat. Food Prod. Technol. 2018, 27, 107–121. [Google Scholar] [CrossRef]
- Choi, J.-I.; Kim, J.-H.; Lee, J.-W. Physiological properties of tuna cooking drip hydrolysate prepared with gamma irradiation. Process Biochem. 2011, 46, 1875–1878. [Google Scholar] [CrossRef]
- Ahmed, R.; Chun, B.-S. Subcritical water hydrolysis for the production of bioactive peptides from tuna skin collagen. J. Supercrit. Fluids 2018, 141, 88–96. [Google Scholar] [CrossRef]
- Wang, W.; Li, Z.; Liu, J.; Wang, Y.; Liu, S.; Sun, M. Comparison between thermal hydrolysis and enzymatic proteolysis processes for the preparation of tilapia skin. Czech J. Food Sci. 2013, 31, 1–4. [Google Scholar] [CrossRef]
- Prabha, J.; Narikimelli, A.; Sajini, M.I.; Vincent, S. Optimization for autolysis assisted production of fish protein hydrolysate from underutilized fish Pellona ditchela. IJSER 2014, 4, 1863. [Google Scholar]
- Jemil, I.; Jridi, M.; Nasri, R.; Ktari, N.; Slama-Ben Salem, R.B.; Mehiri, M.; Hajji, M.; Nasri, M. Functional, antioxidant and antibacterial properties of protein hydrolysates prepared from fish meat fermented by Bacillus subtilis A26. Process Biochem. 2014, 49, 963–972. [Google Scholar] [CrossRef]
- Bhat, Z.F.; Kumar, S.; Bhat, H.F. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef]
- Chakniramol, S.; Wierschem, A.; Cho, M.-G.; Bashir, K.M.I. Physiological and clinical aspects of bioactive peptides from marine animals. Antioxidants 2022, 11, 1021. [Google Scholar] [CrossRef]
- Lee, J.K.; Byun, H.-G. Characterization of antioxidative peptide purified from black eelpout (Lycodes diapterus) hydrolysate. Fish. Aquat. Sci. 2019, 22, 22. [Google Scholar] [CrossRef]
- Bashir, K.M.I.; Sohn, J.H.; Kim, J.-S.; Choi, J.-S. Identification and characterization of novel antioxidant peptides from mackerel (Scomber japonicus) muscle protein hydrolysates. Food Chem. 2020, 323, 126809. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Mendis, E.; Byun, H.-G.; Kim, S.-K. Purification and in vitro antioxidative effects of giant squid muscle peptides on free radical-mediated oxidative systems. J. Nutr. Biochem. 2005, 16, 562–569. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Brogan, S.E.; Olivera, B.M. Pain therapeutics from cone snail venoms: From Ziconotide to novel non-opioid pathways. J. Proteom. 2019, 190, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Najafian, L.; Babji, A.S. A review of fish-derived antioxidant and antimicrobial peptides: Their production, assessment, and applications. Peptides 2012, 33, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Kundam, D.; Acham, I.O.; Girgih, A. Bioactive compounds in fish and their health benefits. Asian Food Sci. J. 2018, 4, 1–14. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020; In brief; FAO: Rome, Italy, 2020. [Google Scholar]
- Ahn, C.B.; Kim, J.G.; Je, J.Y. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014, 147, 78–83. [Google Scholar] [CrossRef]
- Jai ganesh, R.; Nazeer, R.A.; Sampath Kumar, N.S. Purification and identification of antioxidant peptide from black pomfret, Parastromateus niger (Bloch, 1975) viscera protein hydrolysate. Food Sci. Biotechnol. 2011, 20, 1087. [Google Scholar] [CrossRef]
- Barkia, A.; Bougatef, A.; Khaled, H.B.; Nasri, M. Antioxidant activities of sardinelle heads and/or viscera protein hydrolysates prepared by enzymatic treatment. J. Food Biochem. 2010, 34, 303–320. [Google Scholar] [CrossRef]
- Han, H.-J.; Lee, N.S.; Kim, M.S.; Jung, S.H. An outbreak of Lactococcus garvieae infection in cage-cultured red lip mullet Chelon haematocheilus with green liver syndrome. Fish. Aquat. Sci. 2015, 8, 333–339. [Google Scholar] [CrossRef]
- Ministry of Maritime Affairs and Fisheries. Statistical Yearbook of Maritime Affairs and Fisheries; Ministry of Maritime Affairs and Fisheries: Sejong-si, Republic of Korea, 2012. [Google Scholar]
- Ko, E.Y.; Park, J.O.; Lee, K.S. Review of fish name on the fishes of the family Mugilidae in Korea and resource utilization. J. Mar. Life Sci. 2019, 4, 96–105. [Google Scholar]
- Benjakul, S.; Morrissey, M.T. Protein hydrolysates from pacific whiting solid wastes. J. Agric. Food Chem. 1997, 45, 3423–3430. [Google Scholar] [CrossRef]
- Baek, H.H.; Cadwallader, K. Enzymatic hydrolysis of crayfish processing by-products. J. Food Sci. 2006, 60, 929–935. [Google Scholar] [CrossRef]
- Dudonné, S.; Vitrac, X.; Coutière, P.; Woillez, M.; Mérillon, J.M. Comparative study of antioxidant properties and total phenolic content of 30 plant extracts of industrial interest using DPPH, ABTS, FRAP, SOD, and ORAC assays. J. Agric. Food Chem. 2009, 57, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ou, B.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef] [PubMed]
- Buenger, J.; Ackermann, H.; Jentzsch, A.; Mehling, A.; Pfitzner, I.; Reiffen, K.A.; Schroeder, K.R.; Wollenweber, U. An interlaboratory comparison of methods used to assess antioxidant potentials. Int. J. Cosmet. Sci. 2006, 28, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Wong, C.C.; Cheng, K.W.; Chen, F. Antioxidant properties in vitro and total phenolic contents in methanol extracts from medicinal plants. LWT 2008, 41, 385–390. [Google Scholar] [CrossRef]
- Kuda, T.; Tsunekawa, M.; Goto, H.; Araki, Y. Antioxidant properties of four edible algae harvested in the Noto Peninsula, Japan. J. Food Compost. Anal. 2005, 18, 625–633. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Laroque, D.; Chabeaud, A.; Guérard, F. Antioxidant Capacity of Marine Protein Hydrolysates. In Added Value of Fisheries Waste; Bergé, J.-P., Ed.; Transworld Research Network Publisher: Kerala, India, 2008; pp. 147–162. [Google Scholar]
- Bashir, K.M.I.; Mohibbullah, M.; An, J.H.; Choi, J.-Y.; Hong, Y.-K.; Sohn, J.H.; Kim, J.-S.; Choi, J.-S. In vivo antioxidant activity of mackerel (Scomber japonicus) muscle protein hydrolysate. PeerJ 2018, 6, e6181. [Google Scholar] [CrossRef]
- Forghani, B.; Ebrahimpour, A.; Bakar, J.; Abdul Hamid, A.; Hassan, Z.; Saari, N. Enzyme hydrolysates from Stichopus horrens as a new source for angiotensin-converting enzyme inhibitory peptides. Evid.-Based Complement. Altern. Med. 2012, 2012, 236384. [Google Scholar]
- Islam, M.S.; Wang, H.; Noman, A.; Admassu, H.; Ma, C.; Anwei, F. Degree of hydrolysis, functional and antioxidant properties of protein hydrolysates from Grass Turtle (Chinemys reevesii) as influenced by enzymatic hydrolysis conditions. Food Sci. Nutr. 2021, 9, 4031–4047. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.M.; Sylla, K.S.B.; Randriamahatody, Z.; Donnay-Moreno, C.; Moreau, J.; Tran, L.T.; Bergé, J.P. Enzymatic hydrolysis of yellowfin tuna (Thunnus albacares) by-products using Protamex protease. Food Technol. Biotechnol. 2011, 49, 48–55. [Google Scholar]
- Awuor, O.L.; Kirwa, M.E.; Betty, M.; Jackim, M.F. Optimization of Alcalase hydrolysis conditions for production of Dagaa (Rastrineobola argentea) protein hydrolysate with antioxidative properties. Ind. Chem. 2017, 3, 122. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Ghaly, A.E.; Budge, S.M. Extraction of proteins from mackerel fish processing waste using Alcalase enzyme. J. Bioprocess. Biotech. 2013, 3, 2. [Google Scholar]
- García-Moreno, P.J.; Batista, I.; Pires, C.; Bandarra, N.M.; Espejo-Carpio, F.J.; Guadix, A.; Guadix, E.M. Antioxidant activity of protein hydrolysates obtained from discarded Mediterranean fish species. Food Res. Int. 2014, 65, 469–476. [Google Scholar] [CrossRef]
- Peñta-Ramos, E.A.; Xiong, Y.L. Antioxidant activity of soy protein hydrolysates in a liposomal system. J. Food Sci. 2002, 67, 2952–2956. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiong, H.; Selomulya, C.; Chen, X.D.; Zhong, H.; Wang, S.; Sun, W.; Zhou, Q. Enzymatic hydrolysis of rice dreg protein: Effects of enzyme type on the functional properties and antioxidant activities of recovered proteins. Food Chem. 2012, 134, 1360–1367. [Google Scholar] [CrossRef]
- Rueda, J.; Lobo, M.O.; Sammán, N. Changes in the antioxidant activity of peptides released during the hydrolysis of quinoa (Chenopodium quinoa willd) protein concentrate. Proceedings 2020, 53, 12. [Google Scholar] [CrossRef]
- Kim, S.Y.; Je, J.Y.; Kim, S.K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef]
- Ko, J.-Y.; Lee, J.-H.; Samarakoon, K.; Kim, J.-S.; Jeon, Y.-J. Purification and determination of two novel antioxidant peptides from flounder fish (Paralichthys olivaceus) using digestive proteases. Food Chem. Toxicol. 2013, 52, 113–120. [Google Scholar] [CrossRef]
- Ryu, B.; Shin, K.H.; Kim, S.K. Muscle protein hydrolysates and amino acid composition in fish. Mar. Drugs 2021, 19, 377. [Google Scholar] [CrossRef] [PubMed]

 ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
 ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.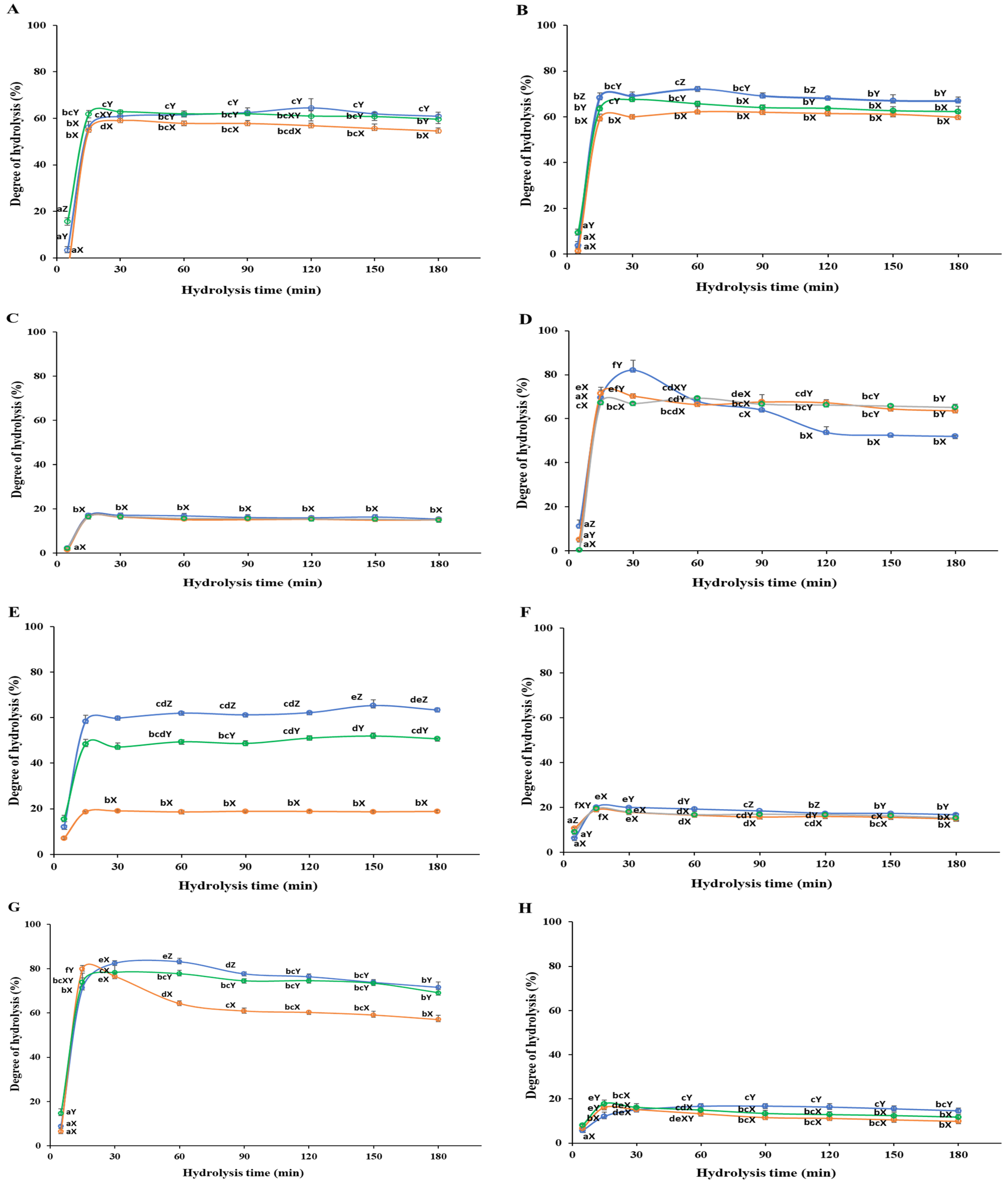
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.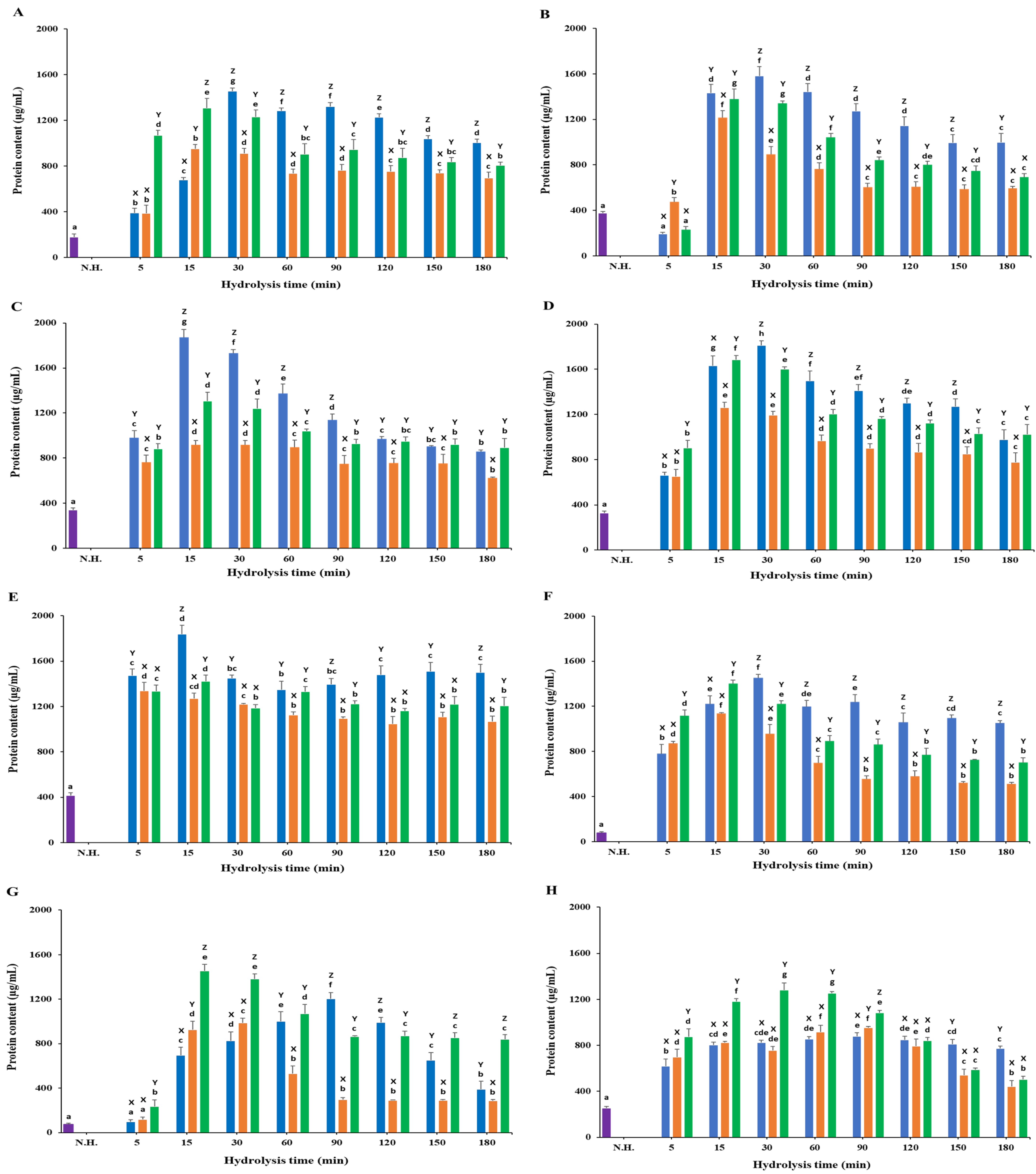
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (7.81 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (7.81 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (7.81 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (7.81 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.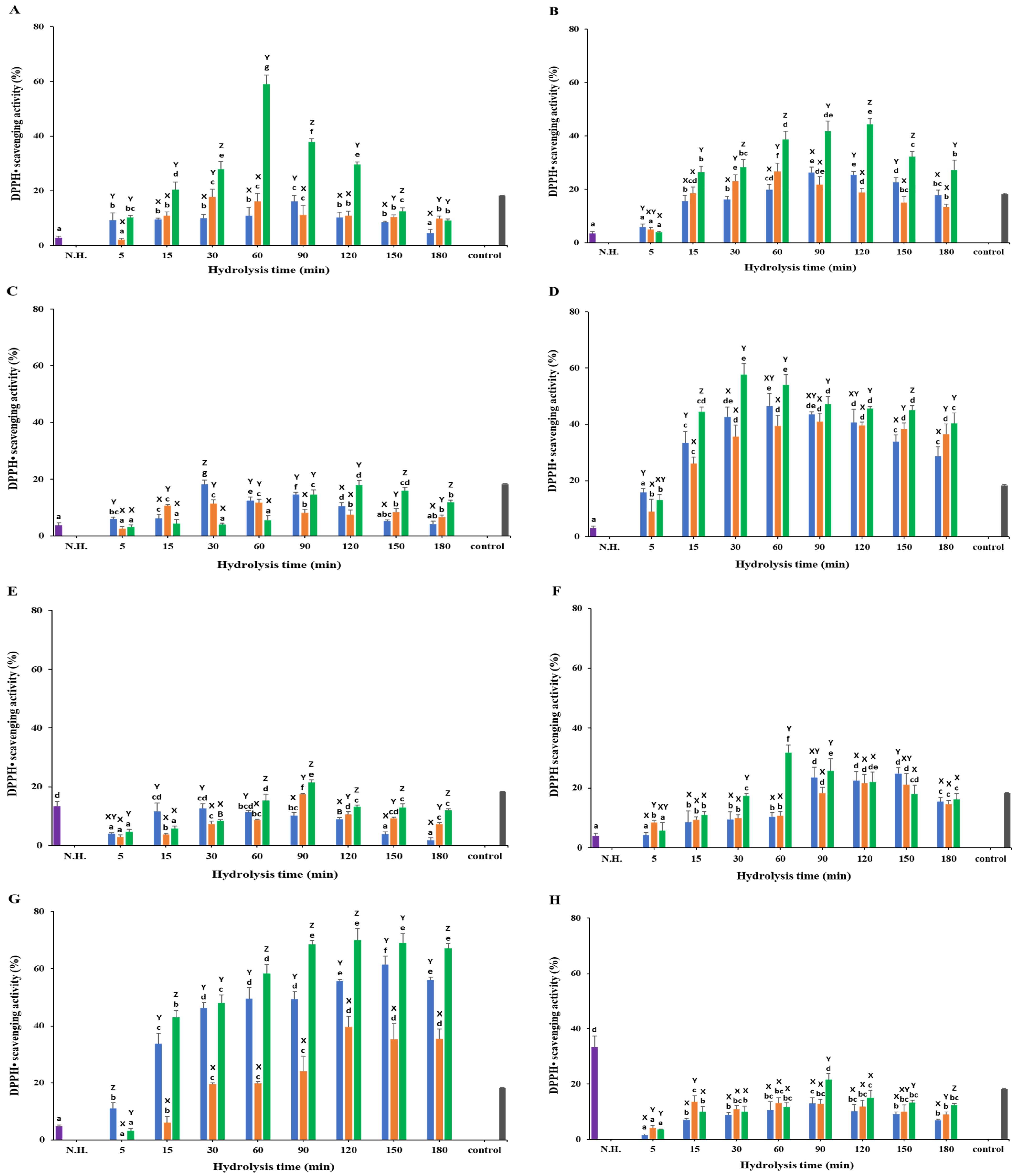
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (31.25 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (31.25 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (31.25 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-ascorbic acid (31.25 µg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–f) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.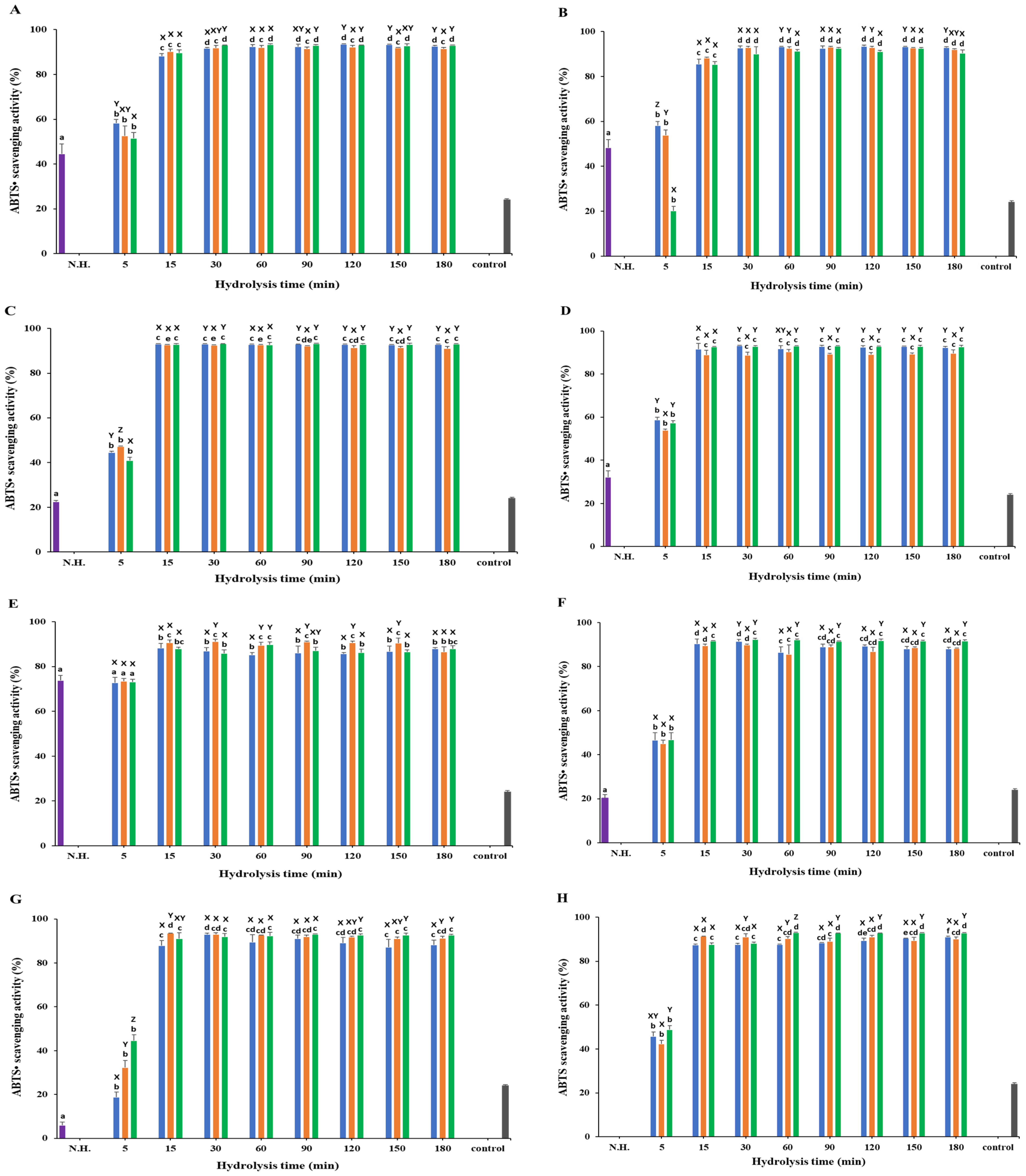
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-Ascorbic acid (200 μg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-Ascorbic acid (200 μg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
 ) Non-hydrolyzed; (
) Non-hydrolyzed; ( ) Neutrase; (
) Neutrase; ( ) Alcalase; (
) Alcalase; ( ) Protamex. N.H.: Non-hydrolyzed sample. L-Ascorbic acid (200 μg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.
) Protamex. N.H.: Non-hydrolyzed sample. L-Ascorbic acid (200 μg/mL) was used as a positive control. Data are means ± S.D., with n = 3. Means with different letters (a–g) among samples at different hydrolysis times for the same body part and using the specific enzyme, or with different letters (X–Z) among different enzymes at the same hydrolysis time for the same body part are significantly different at p < 0.05, as determined by Duncan’s multiple range test.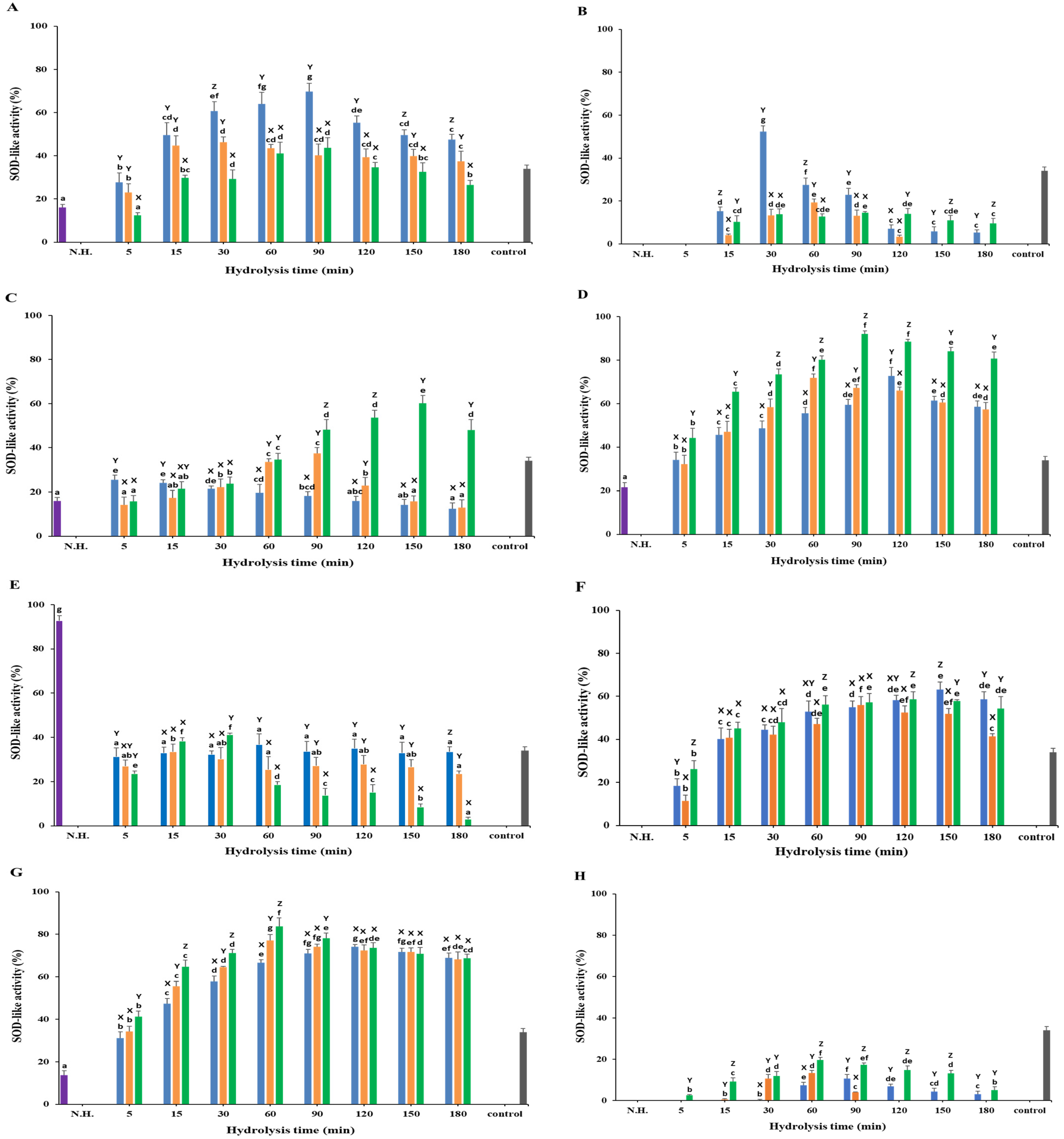
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashir, K.M.I.; Chakniramol, S.; Mansoor, S.; Jahn, A.; Cho, M.-G.; Choi, J.-S. Antioxidant Activity of Protein Hydrolysates from Redlip Mullet (Chelon haematocheilus) Muscle and Byproducts. Foods 2024, 13, 3009. https://doi.org/10.3390/foods13183009
Bashir KMI, Chakniramol S, Mansoor S, Jahn A, Cho M-G, Choi J-S. Antioxidant Activity of Protein Hydrolysates from Redlip Mullet (Chelon haematocheilus) Muscle and Byproducts. Foods. 2024; 13(18):3009. https://doi.org/10.3390/foods13183009
Chicago/Turabian StyleBashir, Khawaja Muhammad Imran, Sukwasa Chakniramol, Sana Mansoor, Alexander Jahn, Man-Gi Cho, and Jae-Suk Choi. 2024. "Antioxidant Activity of Protein Hydrolysates from Redlip Mullet (Chelon haematocheilus) Muscle and Byproducts" Foods 13, no. 18: 3009. https://doi.org/10.3390/foods13183009






