Diversity of ‘Cabernet Sauvignon’ Grape Epidermis and Environmental Bacteria in Wineries from Different Sub-Regions of the Eastern Foothills of Helan Mountain, Ningxia
Abstract
1. Introduction
2. Materials and Methods
2.1. Location Description and Sampling
2.2. Grape Epidermal Sampling
2.3. Sampling of Winery Environment
2.4. DNA Extraction and PCR Amplification
2.5. Illumina MiSeq Sequencing
2.6. Processing of Sequencing Data
3. Results
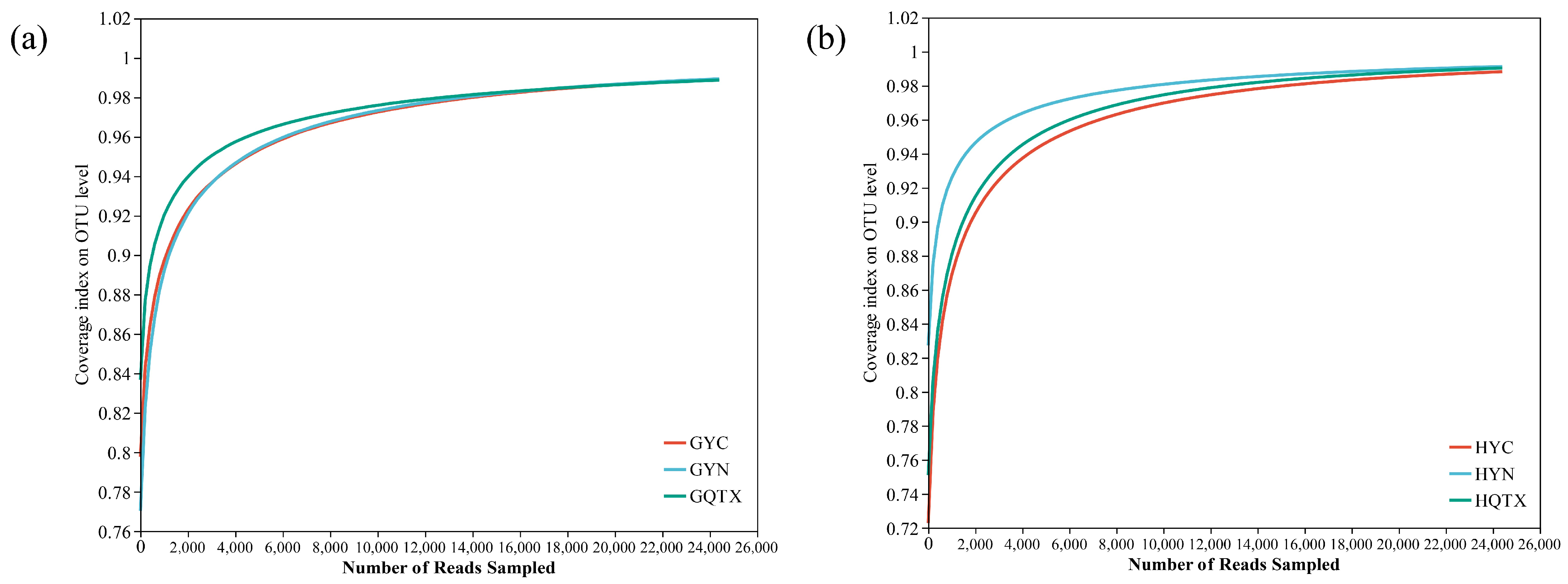
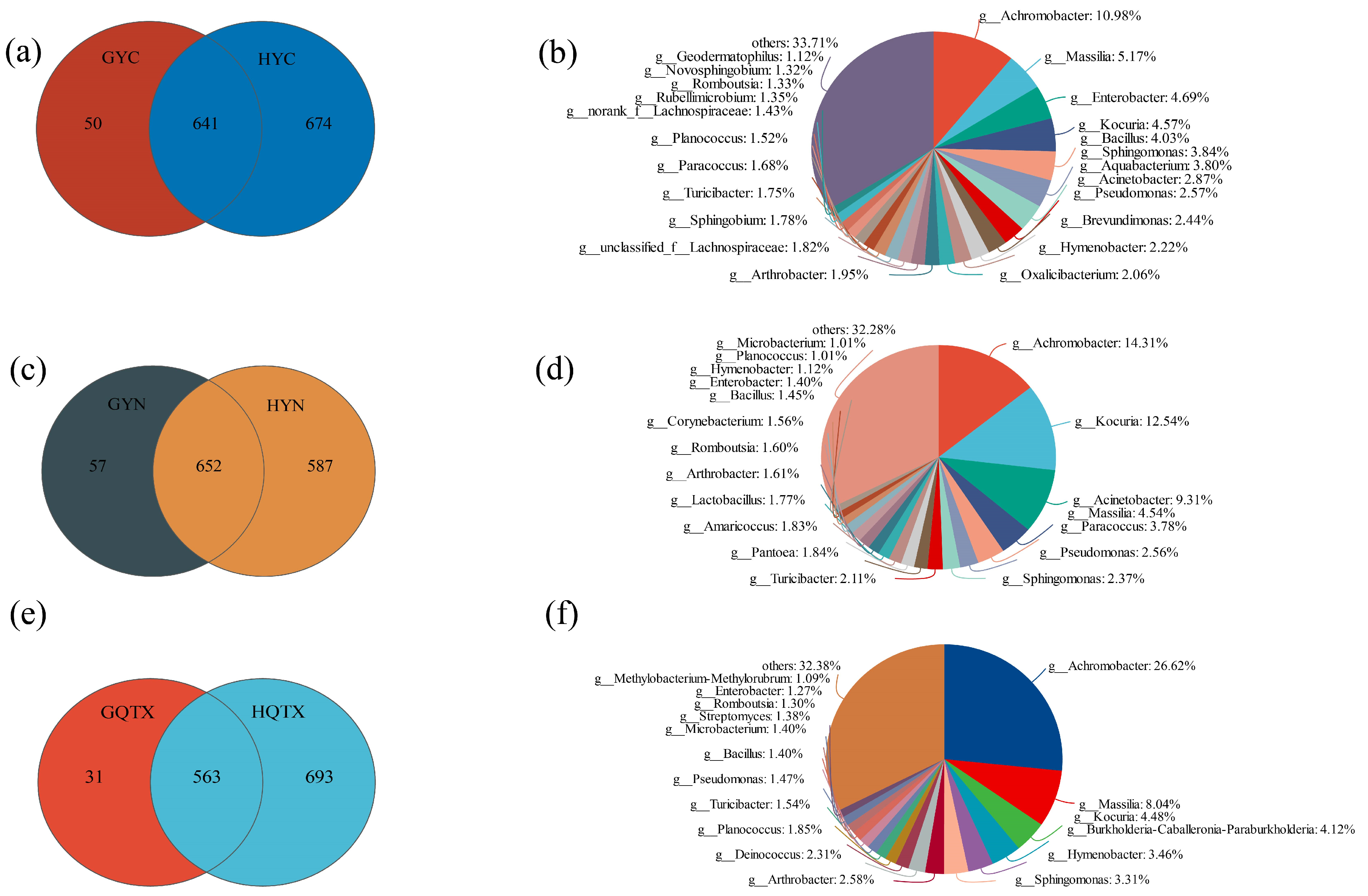
3.1. Evaluation of Bacterial Community Diversity on the Grape Epidermis and in Winery Environments
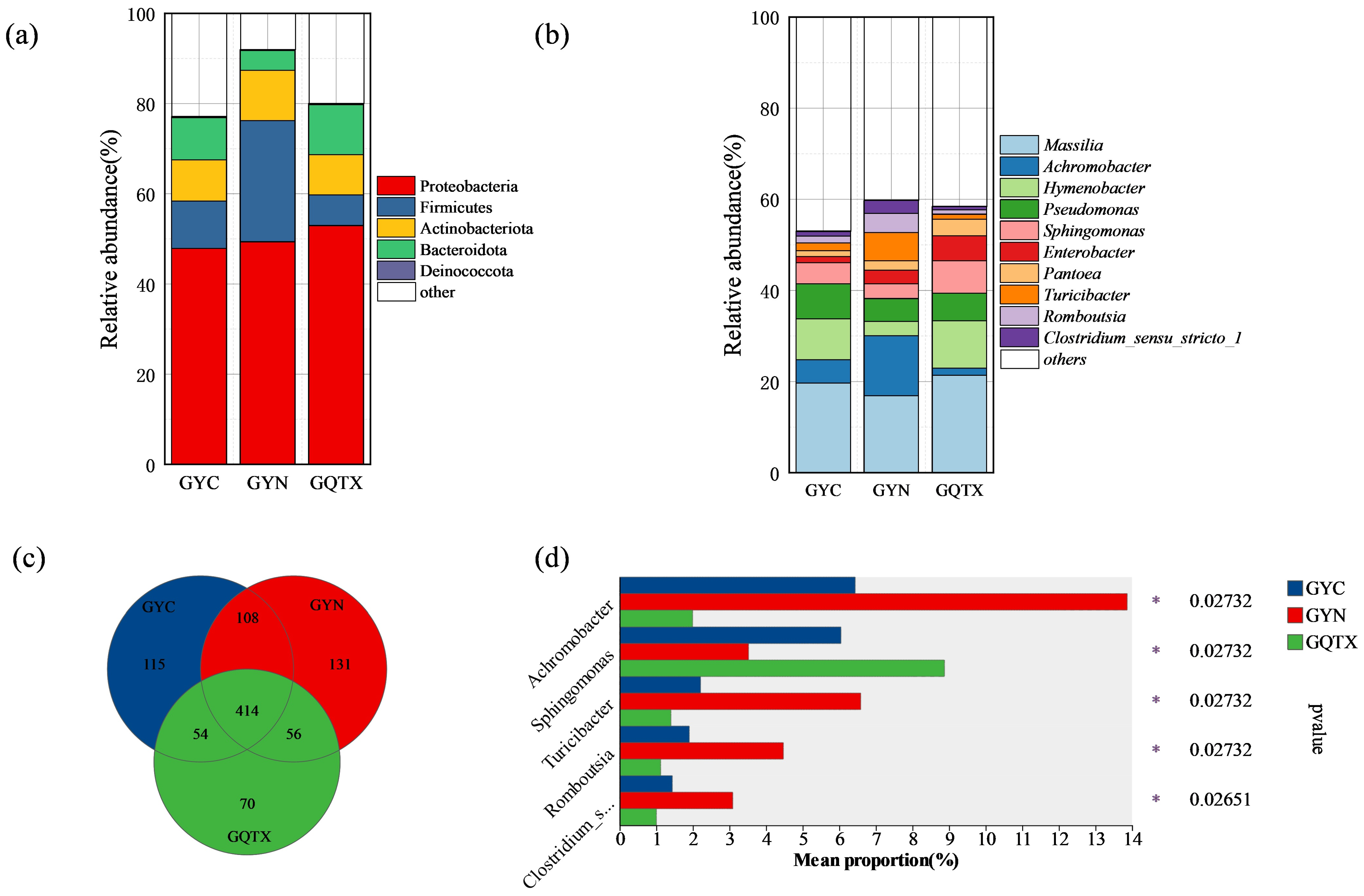
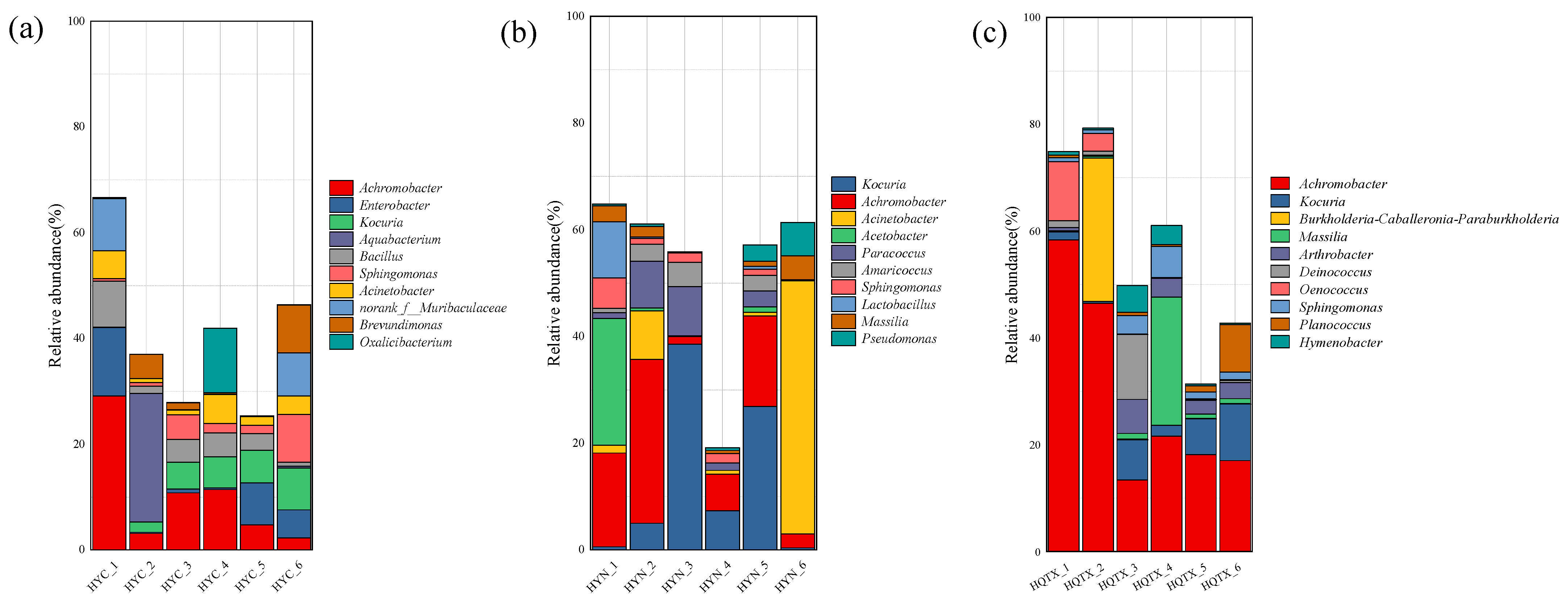
3.2. Analyze of Bacterial Community Consisting of Grape Epidermis in Various Sub-Producing areas
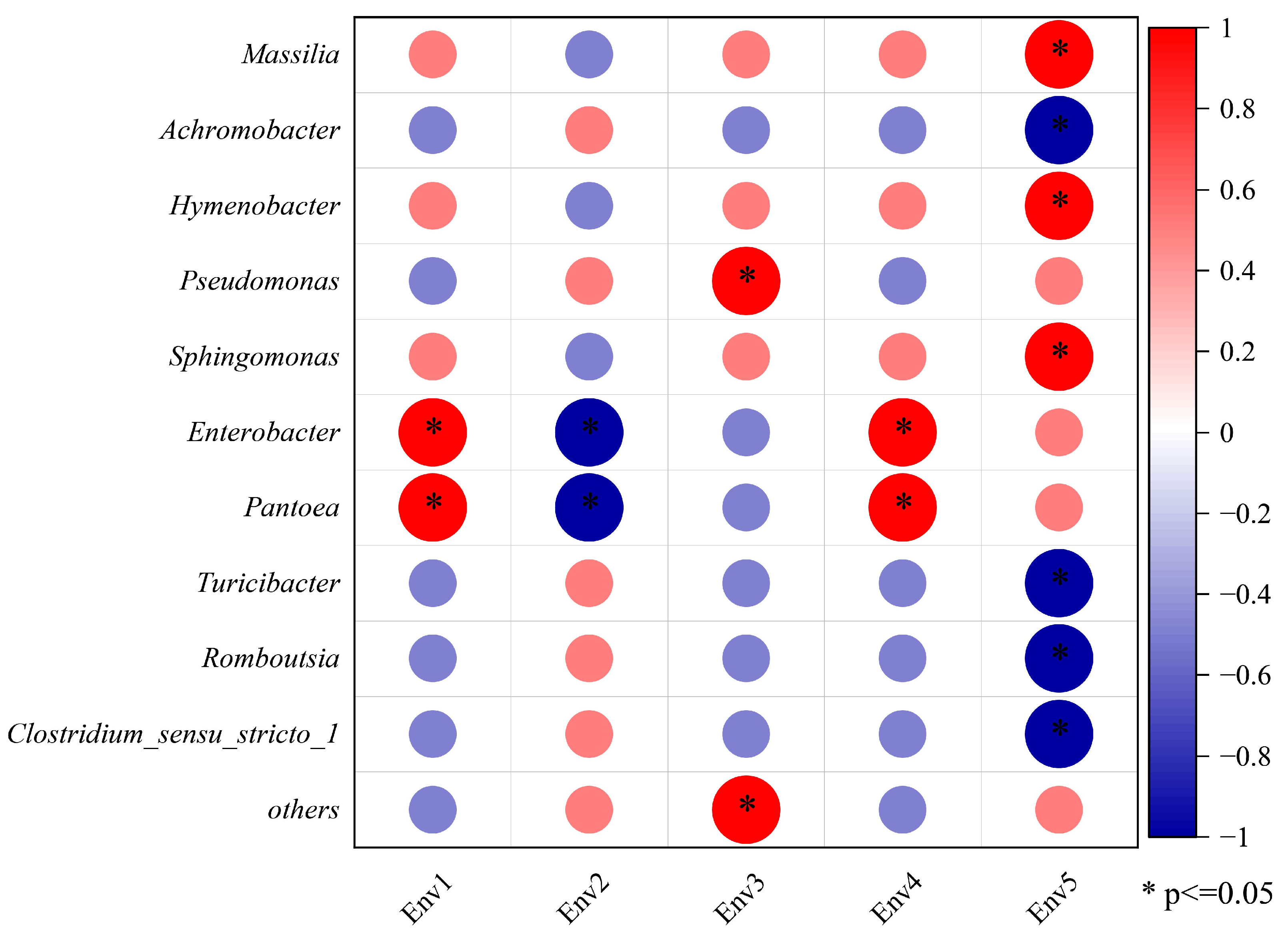
3.3. Correlation between Vineyard Climate and Bacterial Communities on the Grape Epidermis
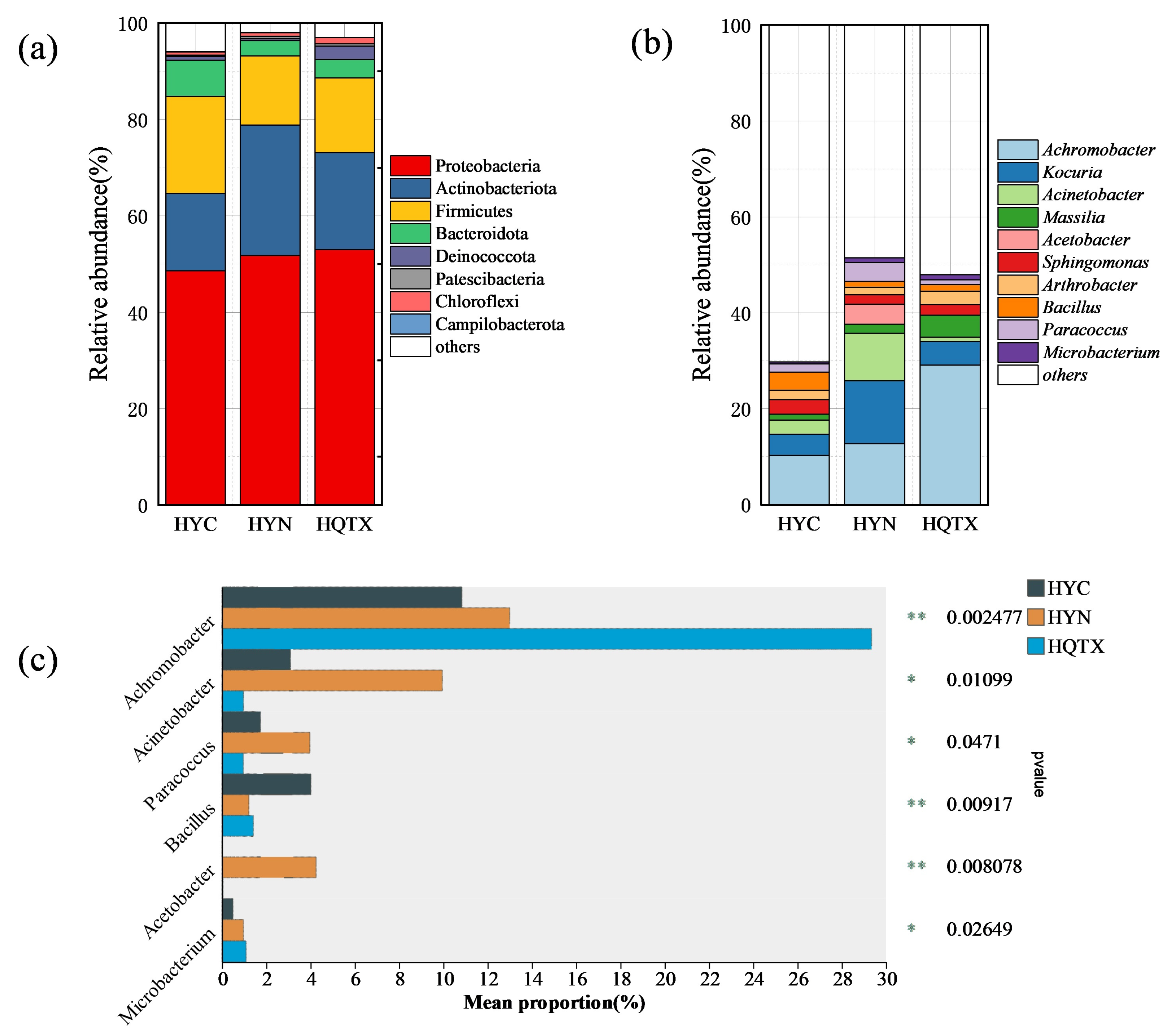
3.4. Environmental Bacterial Diversity of Wineries in Different Sub-Regions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, G.V. The Climate Component of the Terroir. Elements 2018, 14, 167–172. [Google Scholar] [CrossRef]
- Zhou, J.; Cavagnaro, T.R.; De Bei, R.; Nelson, T.M.; Stephen, J.R.; Metcalfe, A.; Gilliham, M.; Breen, J.; Collins, C.; López, C.M.R. Wine Terroir and the Soil Bacteria: An Amplicon Sequencing-Based Assessment of the Barossa Valley and Its Sub-Regions. Front. Microbiol. 2021, 11, 597944. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “terroir” Concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.C.; Yang, H.; Sun, L.J.; Xia, H.C.; Sun, W.J.; Wang, Z.; Zhang, J.X. Bacterial Communities Related to Aroma Formation during Spontaneous Fermentation of ‘Cabernet Sauvignon’ Wine in Ningxia, China. Foods 2022, 11, 2775. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.Z.; Chen, D.L.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef]
- Ma, W.R.; Wu, Y.; Wei, Y.J.; Zou, W.; Yan, Y.Z.; Xue, J.; Tian, G.; Wang, L.Y.; Wang, W.; Pan, H.Y. Microbial diversity analysis of vineyards in the Xinjiang region using high-throughput sequencing. J. Inst. Brew. 2018, 124, 276–283. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, E.T.; Singh, R.P.; Guo, C.; Shang, Y.; Chen, J.; Liu, C. Grape berry surface bacterial microbiome: Impact from the varieties and clones in the same vineyard from central China. J. Appl. Microbiol. 2019, 126, 204–214. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, 12. [Google Scholar] [CrossRef]
- Morgan, H.H.; Du Toit, M.; Setati, M.E. The Grapevine and Wine Microbiome: Insights from High-Throughput Amplicon Sequencing. Front. Microbiol. 2017, 8, 820. [Google Scholar] [CrossRef]
- Liang, H.B.; Wang, X.W.; Yan, J.W.; Luo, L.X. Characterizing the Intra-Vineyard Variation of Soil Bacterial and Fungal Communities. Front. Microbiol. 2019, 10, 1239. [Google Scholar]
- Wei, Y.J.; Wu, Y.; Yan, Y.Z.; Zou, W.; Xue, J.; Ma, W.R.; Wang, W.; Tian, G.; Wang, L.Y. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE 2018, 13, e0193097. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.N.; Kluepfel, D.A.; Strauss, S.L.; Bokulich, N.A.; Cantu, D.; Steenwerth, K.L. Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: Differentiation by geographic features. Soil Biol. Biochem. 2015, 91, 232–247. [Google Scholar] [CrossRef]
- Guzzon, R.; Bertoldi, D.; Roman, T.; Zanzotti, R.; Franciosi, E. Spatial and Seasonal Structure of Bacterial Communities Within Alpine Vineyards: Trentino as a Case Study. Microb. Ecol. 2023, 85, 108–120. [Google Scholar] [CrossRef]
- Portillo, M.D.; Franquès, J.; Araque, I.; Reguant, C.; Bordons, A. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 2016, 219, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Miao, Y.Y.; Wang, H.; Du, J.; Wang, C.Q.; Shi, X.W.; Wang, B. Analysis of Microbial Community Diversity on the Epidermis of Wine Grapes in Manasi’s Vineyard, Xinjiang. Foods 2022, 11, 3174. [Google Scholar] [CrossRef]
- Ding, Y.T.; Wei, R.T.; Wang, L.; Wang, W.N.; Wang, H.; Li, H. Exploring the ecological characteristics of natural microbial communities along the continuum from grape berries to winemaking. Food Res. Int. 2023, 167, 112718. [Google Scholar] [CrossRef]
- Wei, R.T.; Ding, Y.T.; Chen, N.; Wang, L.; Gao, F.F.; Zhang, L.; Song, R.; Liu, Y.H.; Li, H.; Wang, H. Diversity and dynamics of microbial communities during spontaneous fermentation of Cabernet Sauvignon (Vitis vinifera L.) from different regions of China and their relationship with the volatile components in the wine. Food Res. Int. 2022, 156, 111372. [Google Scholar] [CrossRef]
- Gao, F.F.; Chen, J.L.; Xiao, J.; Cheng, W.D.; Zheng, X.J.; Wang, B.; Shi, X.W. Microbial community composition on grape surface controlled by geographical factors of different wine regions in Xinjiang, China. Food Res. Int. 2019, 122, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Howell, K. Community succession of the grapevine fungal microbiome in the annual growth cycle. Environ. Microbiol. 2021, 23, 1842–1857. [Google Scholar] [CrossRef]
- Wei, R.T.; Ding, Y.T.; Gao, F.F.; Zhang, L.; Wang, L.; Li, H.; Wang, H. Community succession of the grape epidermis microbes of cabernet sauvignon (Vitis vinifera L.) from different regions in China during fruit development. Int. J. Food Microbiol. 2022, 362, 109475. [Google Scholar] [CrossRef] [PubMed]
- Nisiotou, A.A.; Rantsiou, K.; Iliopoulos, V.; Cocolin, L.; Nychas, G.J.E. Bacterial species associated with sound and Botrytis-infected grapes from a Greek vineyard. Int. J. Food Microbiol. 2011, 145, 432–436. [Google Scholar] [CrossRef]
- Garijo, P.; Santamaría, P.; López, R.; Sanz, S.; Olarte, C.; Gutiérrez, A.R. The occurrence of fungi, yeasts and bacteria in the air of a Spanish winery during vintage. Int. J. Food Microbiol. 2008, 125, 141–145. [Google Scholar] [CrossRef]
- Kecskeméti, E.; Berkelmann-Löhnertz, B.; Reineke, A. Are Epiphytic Microbial Communities in the Carposphere of Ripening Grape Clusters (Vitis vinifera L.) Different be-tween Conventional, Organic, and Biodynamic Grapes? PLoS ONE 2016, 11, e0160852. [Google Scholar] [CrossRef] [PubMed]
- Bubeck, A.M.; Preiss, L.; Jung, A.N.; Dörner, E.; Podlesny, D.; Kulis, M.; Maddox, C.; Arze, C.; Zörb, C.; Merkt, N. Bacterial microbiota diversity and composition in red and white wines correlate with plant-derived DNA contributions and botrytis infection. Sci. Rep. 2020, 10, 13. [Google Scholar] [CrossRef]
- Carpena, M.; Fraga-Corral, M.; Otero, P.; Nogueira, R.A.; Garcia-Oliveira, P.; Prieto, M.A.; Simal-Gandara, J. Secondary Aroma: Influence of Wine Microorganisms in Their Aroma Profile. Foods 2021, 10, 51. [Google Scholar] [CrossRef]
- Pérez-Martín, F.; Seseña, S.; Fernández-González, M.; Arévalo, M.; Palop, M.L. Microbial communities in air and wine of a winery at two consecutive vintages. Int. J. Food Microbiol. 2014, 190, 44–53. [Google Scholar] [CrossRef]
- Mortimer, R.; Polsinelli, M. On the origin of wine yeast. Res. Microbiol. 1999, 150, 199–204. [Google Scholar] [CrossRef]
- Bao, L.J.; Gu, L.K.; Sun, B.; Cai, W.Y.; Zhang, S.W.; Zhuang, G.Q.; Bai, Z.H.; Zhuang, X.L. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiol. Ecol. 2020, 96, 13. [Google Scholar] [CrossRef]
- Jia, C.B.; An, Y.R.; Du, Z.Y.; Gao, H.H.; Su, J.Y.; Xu, C.Y. Differences in Soil Microbial Communities between Healthy and Diseased Lycium barbarum cv. Ningqi-5 Plants with Root Rot. Microorganisms 2023, 11, 694. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Gayevskiy, V.; Goddard, M.R. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 2012, 6, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lei, X.M.; Jiang, J.; Qin, Y.; Jiang, L.; Liu, Y.L. Microbial diversity on grape epidermis and wine volatile aroma in spontaneous fermentation comprehensively driven by geography, subregion, and variety. Int. J. Food Microbiol. 2023, 404, 110315. [Google Scholar] [CrossRef]
- GB/T 15038-2006; General Analysis Methods for Wine and Fruit Wine. China Standard Press: Beijing, China, 2006; pp. 5–8.
- Sun, D.Y.; Qu, J.J.; Huang, Y.; Lu, J.; Yin, L. Analysis of microbial community diversity of muscadine grape skins. Food Res. Int. 2021, 145, 110417. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Manzari, C.; Grieco, F.; Intranuovo, M.; Cozzi, G.; Mulè, G.; Scioscia, G.; Valiente, G.; Tullo, A. Complexity and Dynamics of the Winemaking Bacterial Communities in Berries, Musts, and Wines from Apulian Grape Cultivars through Time and Space. PLoS ONE 2016, 11, e0157383. [Google Scholar] [CrossRef]
- Vitulo, N.; Lemos, W.J.F.; Calgaro, M.; Confalone, M.; Felis, G.E.; Zapparoli, G.; Nardi, T. Bark and Grape Microbiome of Vitis vinifera: Influence of Geographic Patterns and Agronomic Management on Bacterial Diversity. Front. Microbiol. 2019, 9, 3203. [Google Scholar] [CrossRef]
- Ocón, E.; Garijo, P.; Sanz, S.; Olarte, C.; López, R.; Santamaría, P.; Gutiérrez, A.R. Analysis of airborne yeast in one winery over a period of one year. Food Control 2013, 30, 585–589. [Google Scholar] [CrossRef]
- Leveau, J.H.J.; Tech, J.J. Grapevine Microbiomics: Bacterial Diversity on Grape Leaves and Berries Revealed by High-Throughput Sequence Analysis of 16S rRNA Amplicons. In Proceedings of the International Symposium on Biological Control of Postharvest Diseases: Challenges and Opportunitie, Leesburg, VA, USA, 25–28 October 2011; Volume 95, pp. 31–42. [Google Scholar]
- Martins, G.; Lauga, B.; Miot-Sertier, C.; Mercier, A.; Lonvaud, A.; Soulas, M.L.; Soulas, G.; Masneuf-Pomarède, I. Characterization of Epiphytic Bacterial Communities from Grapes, Leaves, Bark and Soil of Grapevine Plants Grown, and Their Relations. PLoS ONE 2013, 8, e73013. [Google Scholar] [CrossRef]
- Mezzasalma, V.; Sandionigi, A.; Guzzetti, L.; Galimberti, A.; Grando, M.S.; Tardaguila, J.; Labra, M. Geographical and Cultivar Features Differentiate Grape Microbiota in Northern Italy and Spain Vineyards. Front. Microbiol. 2018, 9, 946. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Santoni, S.; This, P.; Péros, J.P. Genotype-Environment Interaction Shapes the Microbial Assemblage in Grapevine’s Phyllosphere and Carposphere: An NGS Approach. Microorganisms 2018, 6, 96. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P.; Afonso, I.M.; Pereira, J.; Rocha, R.; Rodrigues, A.S. Epiphitic Microbiome of Alvarinho Wine Grapes from Different Geographic Regions in Portugal. Biology 2023, 12, 146. [Google Scholar] [CrossRef]
- Ye, X.F.; Zhang, X.Y.; Hao, L.F.; Lin, Q.; Bao, Y.Y. Effects of Different Fermentation Methods on the Quality and Microbial Diversity of Passion Fruit Wine. Fermentation 2023, 9, 439. [Google Scholar] [CrossRef]
- Schmid, F.; Moser, G.; Müller, H.; Berg, G. Functional and Structural Microbial Diversity in Organic and Conventional Viticulture: Organic Farming Benefits Natural Biocontrol Agents. Appl. Environ. Microbiol. 2011, 77, 2188–2191. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Verginer, M.; Leitner, E.; Berg, G. Production of Volatile Metabolites by Grape-Associated Microorganisms. J. Agric. Food Chem. 2010, 58, 8344–8350. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Wang, Z.; Zhang, Z.; Shu, C.; Zhu, J.; Li, Y.; Zhang, J. Diversity of ‘Cabernet Sauvignon’ Grape Epidermis and Environmental Bacteria in Wineries from Different Sub-Regions of the Eastern Foothills of Helan Mountain, Ningxia. Foods 2024, 13, 252. https://doi.org/10.3390/foods13020252
Yang H, Wang Z, Zhang Z, Shu C, Zhu J, Li Y, Zhang J. Diversity of ‘Cabernet Sauvignon’ Grape Epidermis and Environmental Bacteria in Wineries from Different Sub-Regions of the Eastern Foothills of Helan Mountain, Ningxia. Foods. 2024; 13(2):252. https://doi.org/10.3390/foods13020252
Chicago/Turabian StyleYang, Hui, Zheng Wang, Zhong Zhang, Chao Shu, Jiaqi Zhu, Ying Li, and Junxiang Zhang. 2024. "Diversity of ‘Cabernet Sauvignon’ Grape Epidermis and Environmental Bacteria in Wineries from Different Sub-Regions of the Eastern Foothills of Helan Mountain, Ningxia" Foods 13, no. 2: 252. https://doi.org/10.3390/foods13020252
APA StyleYang, H., Wang, Z., Zhang, Z., Shu, C., Zhu, J., Li, Y., & Zhang, J. (2024). Diversity of ‘Cabernet Sauvignon’ Grape Epidermis and Environmental Bacteria in Wineries from Different Sub-Regions of the Eastern Foothills of Helan Mountain, Ningxia. Foods, 13(2), 252. https://doi.org/10.3390/foods13020252




