Growth Control of Listeria monocytogenes in Raw Sausage via Bacteriocin-Producing Leuconostoc carnosum DH25
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of LAB Strains
2.2. Identification of Presumptive LAB Isolates
2.3. Screening for Antimicrobial Activity
2.3.1. Antimicrobial Screening by Agar Spot Assay
2.3.2. Anti-Listeria Screening by Well Diffusion Assay
2.4. Characterization of Anti-Listeria Substances Produced by Selected LAB
2.4.1. Sensitivity of Anti-Listeria Metabolites to Proteolytic Enzymes
2.4.2. Determination of the Presence of Structural Genes for Bacteriocins
2.5. Challenge Test in a Meat Model and in Raw Sausage
2.6. Characterization of the Bacteriocin Produced by Leuconostoc carnosum DH25
2.6.1. Stability and Activity of the Anti-Listeria Substance of Leuconostoc carnosum DH25 in Various Conditions
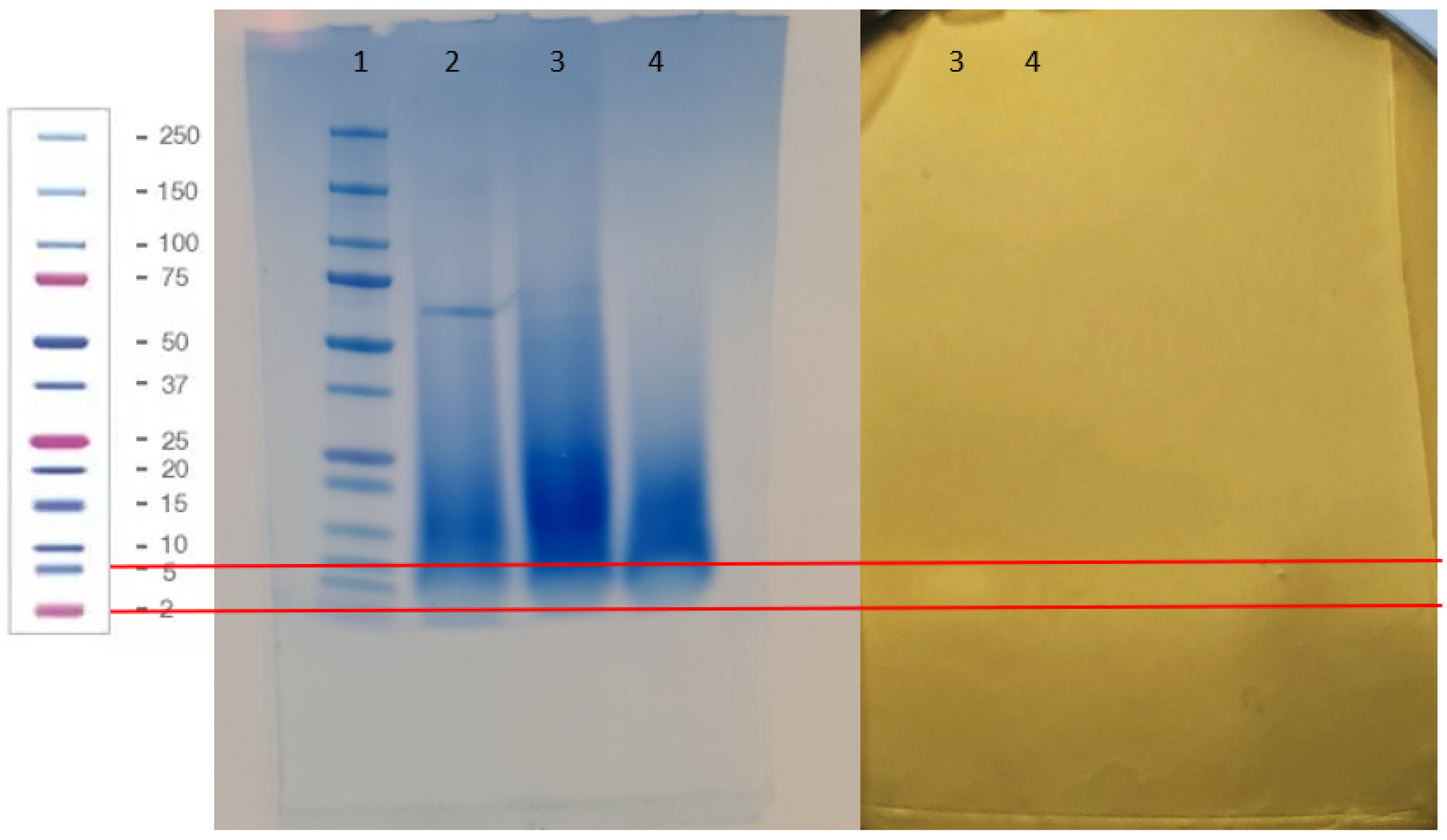
2.6.2. Size Estimation of the Proteinacoeus Anti-Listeria Substance of Lc. carnosum DH25
2.7. Safety Assessement of Lc. carnosum DH25
2.7.1. Phenotypic Detection of Biogenic Amine Formation by the Decarboxylase Assay
2.7.2. Genome Sequencing of Lc. carnosum DH25
2.8. Statistical Analysis
3. Results
3.1. Isolation and Identification of LAB Strains
3.2. Screening of Anti-Listeria Activity
3.3. Sensitivity of Anti-Listeria Active Substances to Proteolytic Enzymes and the Presence of Structural Genes of Known Bacteriocins
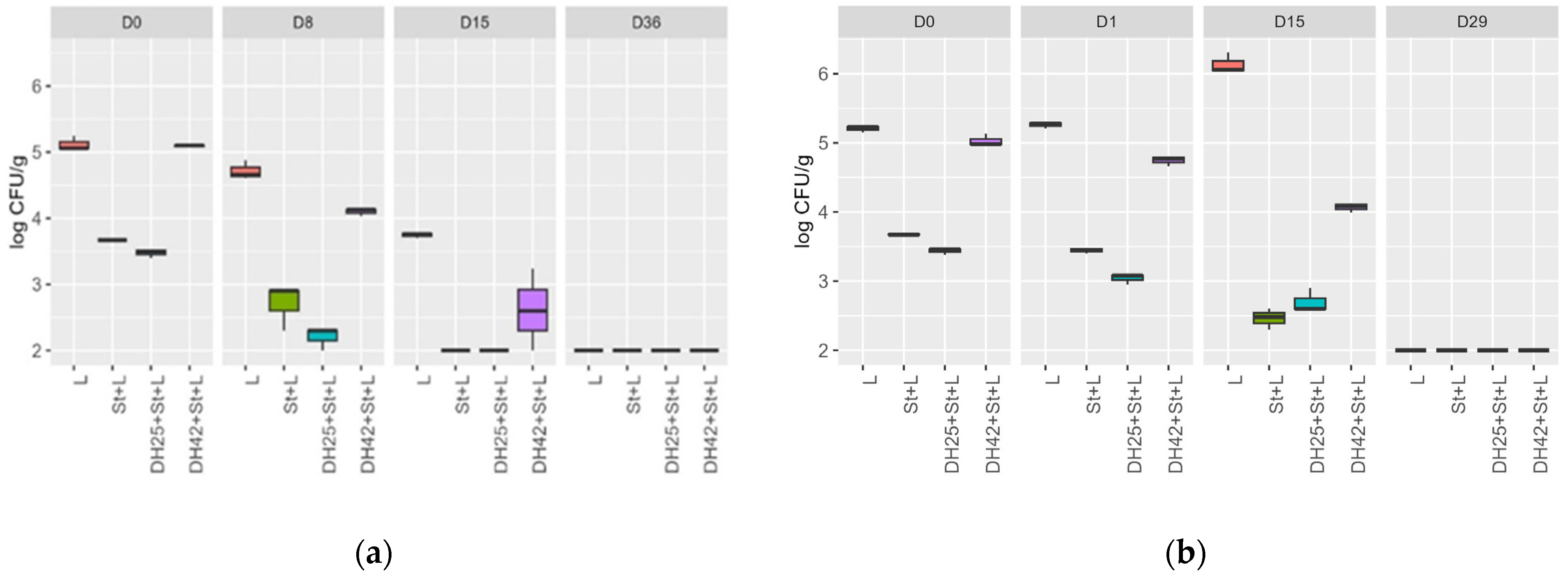
3.4. Challenge Test with a Meat Model and Raw Sausages
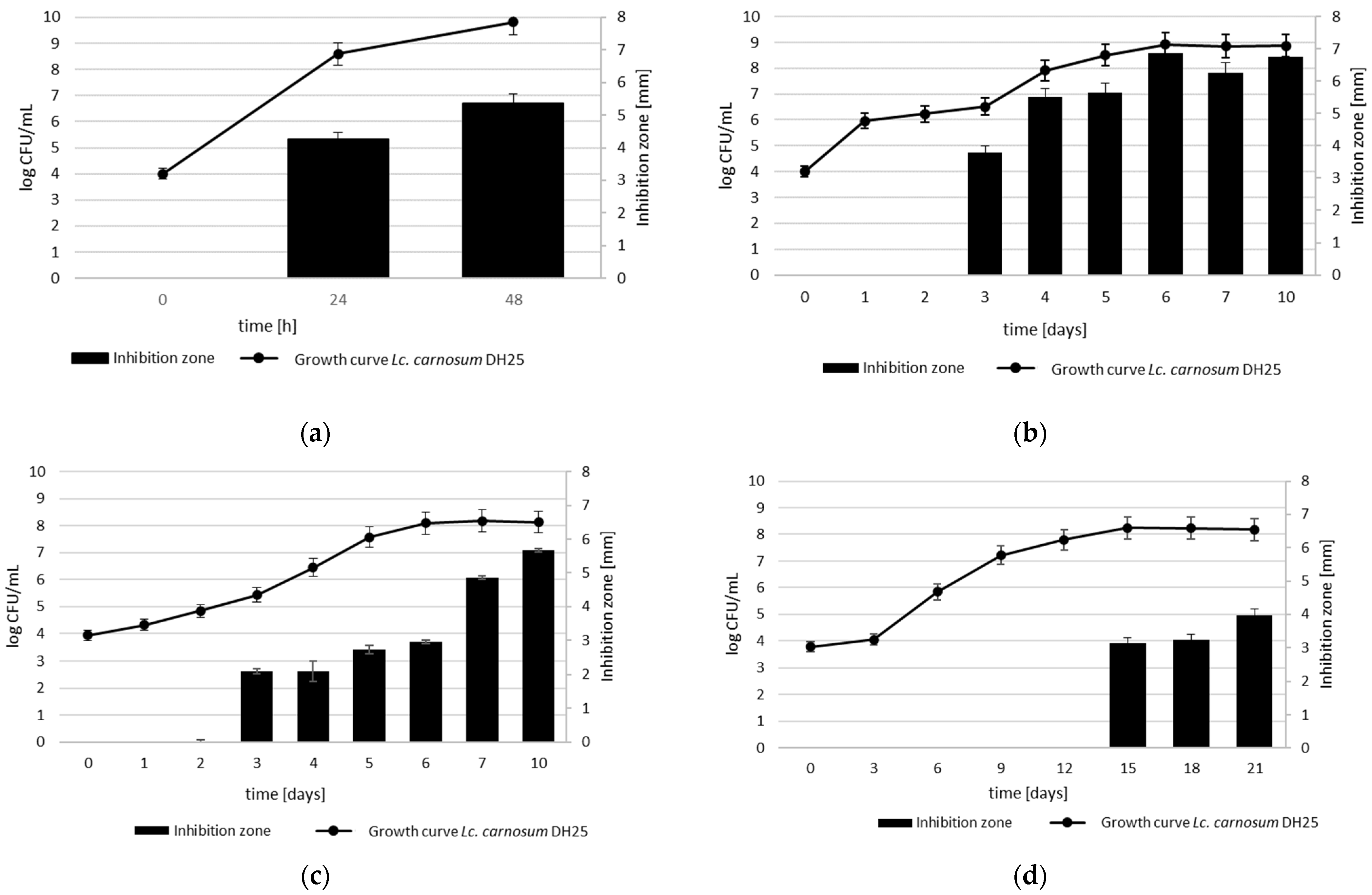
3.5. Growth-Dependent Production of Anti-Listeria Metabolites at Different Temperatures and Temperature and pH Stability of CFS
3.6. Characterization of the Proteinaocus Anti-Listeria Metabolites Produced by Lc. carnosum DH25
3.7. Safety Assessment by an Evaluation of Biogenic Amine Formation and Genome Sequencing of Lc. Carnosum DH25
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Listeriosis. Available online: https://www.who.int (accessed on 2 October 2023).
- European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report|EFSA. Available online: https://european-union.europa.eu/index_en (accessed on 2 October 2023).
- Osek, J.; Wieczorek, K. Listeria moznocytogenes—How This Pathogen Uses Its Virulence Mechanisms to Infect the Hosts. Pathogens 2022, 11, 1491. [Google Scholar] [CrossRef]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a Food-Borne Pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [CrossRef]
- Degenhardt, R.; Sant’Anna, E.S. Survival of Listeria monocytogenes in Low Acid Italian Sausage Produced under Brazilian Conditions. Braz. J. Microbiol. 2007, 38, 309–314. [Google Scholar] [CrossRef]
- Barcenilla, C.; Ducic, M.; López, M.; Prieto, M.; Álvarez-Ordóñez, A. Application of Lactic Acid Bacteria for the Biopreservation of Meat Products: A Systematic Review. Meat Sci. 2022, 183, 108661. [Google Scholar] [CrossRef] [PubMed]
- Ammor, M.S.; Mayo, B. Selection Criteria for Lactic Acid Bacteria to Be Used as Functional Starter Cultures in Dry Sausage Production: An Update. Meat Sci. 2007, 76, 138–146. [Google Scholar] [CrossRef]
- Pragalaki, T.; Bloukas, J.G.; Kotzekidou, P. Inhibition of Listeria monocytogenes and Escherichia coli O157:H7 in Liquid Broth Medium and during Processing of Fermented Sausage Using Autochthonous Starter Cultures. Meat Sci. 2013, 95, 458–464. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; López, R.L.; Omar, N.B. Bacteriocin-Based Strategies for Food Biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, T.M.; Tang, S.; Wiedmann, M.; Boor, K.J. Nisin Resistance of Listeria monocytogenes Is Increased by Exposure to Salt Stress and Is Mediated via LiaR. Appl. Environ. Microbiol. 2013, 79, 5682–5688. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing Innate Immunity for Food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Kheadr, E.; Lacroix, C.; Fliss, I. Class I/Class IIa Bacteriocin Cross-Resistance Phenomenon in Listeria monocytogenes. Food Microbiol. 2007, 24, 718–727. [Google Scholar] [CrossRef]
- Hammes, W.P.; Hertel, C. The Genera Lactobacillus and Carnobacterium. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 320–403. ISBN 978-0-387-25494-4. [Google Scholar]
- Miescher Schwenninger, S.; Freimüller Leischtfeld, S.; Gantenbein-Demarchi, C. High-Throughput Identification of the Microbial Biodiversity of Cocoa Bean Fermentation by MALDI-TOF MS. Lett. Appl. Microbiol. 2016, 63, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Inglin, R.C.; Stevens, M.J.A.; Meile, L.; Lacroix, C.; Meile, L. High-Throughput Screening Assays for Antibacterial and Antifungal Activities of Lactobacillus Species. J. Microbiol. Methods 2015, 114, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic Acid Bacteria as Functional Starter Cultures for the Food Fermentation Industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Tagg, J.R.; McGiven, A.R. Assay System for Bacteriocins. Appl. Environ. Microbiol. 1971, 21, 943. [Google Scholar] [CrossRef] [PubMed]
- Miescher, S.; Stierli, M.P.; Teuber, M.; Meile, L. Propionicin SM1, a Bacteriocin from Propionibacterium Jensenii DF1: Isolation and Characterization of the Protein and Its Gene. Syst. Appl. Microbiol. 2000, 23, 174–184. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Stevens, M.J.A.; Cernela, N.; Corti, S.; Stephan, R. Draft Genome Sequence of Streptococcus Parasuis 4253, the First Available for the Species. Microbiol. Resour. Announc. 2019, 8, e00203–e00219. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Shovill—Assemble Bacterial Isolate Genomes from Illumina Paired-End Reads. Available online: https://github.com/tseemann/shovill (accessed on 10 November 2023).
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Nie, R.; Liu, Y.; Li, X.; Duan, J.; Hao, X.; Shan, Y.; Zhang, J. Bacillus subtilis BS-15 Effectively Improves Plantaricin Production and the Regulatory Biosynthesis in Lactiplantibacillus plantarum RX-8. Front. Microbiol. 2022, 12, 772546. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yoshida, Y.; Cisar, J.O. Genetic Basis of Coaggregation Receptor Polysaccharide Biosynthesis in Streptococcus Sanguinis and Related Species. Mol. Oral Microbiol. 2014, 29, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Elidrissi, A.; Ezzaky, Y.; Boussif, K.; Achemchem, F. Isolation and Characterization of Bioprotective Lactic Acid Bacteria from Moroccan Fish and Seafood. Braz. J. Microbiol. 2023, 54, 2117–2127. [Google Scholar] [CrossRef]
- Todorov, S.D.; Rachman, C.; Fourrier, A.; Dicks, L.M.T.; van Reenen, C.A.; Prévost, H.; Dousset, X. Characterization of a Bacteriocin Produced by Lactobacillus Sakei R1333 Isolated from Smoked Salmon. Anaerobe 2011, 17, 23–31. [Google Scholar] [CrossRef]
- Todorov, S.D.; Vaz-Velho, M.; de Melo Franco, B.D.G.; Holzapfel, W.H. Partial Characterization of Bacteriocins Produced by Three Strains of Lactobacillus Sakei, Isolated from Salpicao, a Fermented Meat Product from North-West of Portugal. Food Control 2013, 30, 111–121. [Google Scholar] [CrossRef]
- Andrighetto, C.; Zampese, L.; Lombardi, A. RAPD-PCR Characterization of Lactobacilli Isolated from Artisanal Meat Plants and Traditional Fermented Sausages of Veneto Region (Italy). Lett. Appl. Microbiol. 2001, 33, 26–30. [Google Scholar] [CrossRef]
- Castellano, P.; Vignolo, G. Inhibition of Listeria innocua and Brochothrix thermosphacta in Vacuum-Packaged Meat by Addition of Bacteriocinogenic lactobacillus Curvatus CRL705 and Its Bacteriocins. Lett. Appl. Microbiol. 2006, 43, 194–199. [Google Scholar] [CrossRef]
- Gao, Y.; Li, D.; Liu, X. Bacteriocin-Producing Lactobacillus sakei C2 as Starter Culture in Fermented Sausages. Food Control. 2014, 35, 1–6. [Google Scholar] [CrossRef]
- Martinez, R.C.R.; Staliano, C.D.; Vieira, A.D.S.; Villarreal, M.L.M.; Todorov, S.D.; Saad, S.M.I.; Franco, B.D.G.d.M. Bacteriocin Production and Inhibition of Listeria monocytogenes by Lactobacillus sakei Subsp. sakei 2a in a Potentially Synbiotic Cheese Spread. Food Microbiol. 2015, 48, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Koep, K.S.C.; Van Reenen, C.A.; Hoffman, L.C.; Slinde, E.; Dicks, L.M.T. Production of Salami from Beef, Horse, Mutton, Blesbok (Damaliscus dorcas phillipsi) and Springbok (Antidorcas marsupialis) with Bacteriocinogenic Strains of Lactobacillus plantarum and Lactobacillus curvatus. Meat Sci. 2007, 77, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Drider, D.; Fimland, G.; Héchard, Y.; McMullen, L.M.; Prévost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef]
- Guyonnet, D.; Fremaux, C.; Cenatiempo, Y.; Berjeaud, J.M. Method for Rapid Purification of Class IIa Bacteriocins and Comparison of Their Activities. Appl. Environ. Microbiol. 2000, 66, 1744–1748. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, H.A.; Wilke, T.; Erdmann, R. Efficacy of Bacteriocin-Containing Cell-Free Culture Supernatants from Lactic Acid Bacteria to Control Listeria monocytogenes in Food. Int. J. Food Microbiol. 2011, 146, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Katla, T.; Naterstad, K.; Vancanneyt, M.; Swings, J.; Axelsson, L. Differences in Susceptibility of Listeria monocytogenes Strains to Sakacin P, Sakacin A, Pediocin PA-1, and Nisin. Appl. Environ. Microbiol. 2003, 69, 4431–4437. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Sullivan, G.A.; Jackson, A.L.; Zhou, G.H.; Sebranek, J.G. Use of Natural Antimicrobials to Improve the Control of Listeria monocytogenes in a Cured Cooked Meat Model System. Meat Sci. 2011, 88, 503–511. [Google Scholar] [CrossRef]
- Chen, N.; Shelef, L.A. Relationship Between Water Activity, Salts of Lactic Acid, and Growth of Listeria monocytogenes in a Meat Model System. J. Food Prot. 1992, 55, 574–578. [Google Scholar] [CrossRef]
- Dussault, D.; Vu, K.D.; Lacroix, M. Development of a Model Describing the Inhibitory Effect of Selected Preservatives on the Growth of Listeria monocytogenes in a Meat Model System. Food Microbiol. 2016, 53, 115–121. [Google Scholar] [CrossRef]
- Thévenot, D.; Delignette-Muller, M.L.; Christieans, S.; Vernozy-Rozand, C. Fate of Listeria monocytogenes in Experimentally Contaminated French Sausages. Int. J. Food Microbiol. 2005, 101, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Chaves-López, C.; Paparella, A.; Tofalo, R.; Suzzi, G. Proteolytic Activity of Saccharomyces cerevisiae Strains Associated with Italian Dry-Fermented Sausages in a Model System. Int. J. Food Microbiol. 2011, 150, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Austrich-Comas, A.; Serra-Castelló, C.; Jofré, A.; Gou, P.; Bover-Cid, S. Control of Listeria monocytogenes in Chicken Dry-Fermented Sausages with Bioprotective Starter Culture and High-Pressure Processing. Front. Microbiol. 2022, 13, 983265. [Google Scholar] [CrossRef] [PubMed]
- Hugas, M.; Garriga, M.; Aymerich, M.T.; Monfort, J.M. Inhibition of Listeria in Dry Fermented Sausages by the Bacteriocinogenic lactobacillus Sake CTC494. J. Appl. Bacteriol. 1995, 79, 322–330. [Google Scholar] [CrossRef]
- Castro, M.P.; Palavecino, N.Z.; Herman, C.; Garro, O.A.; Campos, C.A. Lactic Acid Bacteria Isolated from Artisanal Dry Sausages: Characterization of Antibacterial Compounds and Study of the Factors Affecting Bacteriocin Production. Meat Sci. 2011, 87, 321–329. [Google Scholar] [CrossRef]
- Budde, B. Leuconostoc carnosum 4010 Has the Potential for Use as a Protective Culture for Vacuum-Packed Meats: Culture Isolation, Bacteriocin Identification, and Meat Application Experiments. Int. J. Food Microbiol. 2003, 83, 171–184. [Google Scholar] [CrossRef]
- Baka, M.; Noriega, E.; Tsakali, E.; Van Impe, J.F.M. Influence of Composition and Processing of Frankfurter Sausages on the Growth Dynamics of Listeria monocytogenes under Vacuum. Food Res. Int. 2015, 70, 94–100. [Google Scholar] [CrossRef]
- Luo, K.; Hong, S.-S.; Oh, D.-H. Modeling the Effect of Storage Temperatures on the Growth of Listeria monocytogenes on Ready-to-Eat Ham and Sausage. J. Food Prot. 2015, 78, 1675–1681. [Google Scholar] [CrossRef]
- Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs (Text with EEA Relevance). 2005. Available online: https://www.legislation.gov.uk (accessed on 3 November 2023).
- Martín, I.; Rodríguez, A.; Alía, A.; Martínez, R.; Córdoba, J.J. Selection and Characterization of Lactic Acid Bacteria with Activity against Listeria monocytogenes from Traditional RTE Ripened Foods. LWT 2022, 163, 113579. [Google Scholar] [CrossRef]
- Kumar, V.; Sheoran, P.; Gupta, A.; Yadav, J.; Tiwari, S.K. Antibacterial Property of Bacteriocin Produced by Lactobacillus Plantarum LD4 Isolated from a Fermented Food. Ann. Microbiol. 2016, 66, 1431–1440. [Google Scholar] [CrossRef]
- Daba, G.M.; Mostafa, F.A.; Saleh, S.A.A.; Elkhateeb, W.A.; Awad, G.; Nomiyama, T.; Zendo, T.; El-Dein, A.N. Purification, Amino Acid Sequence, and Characterization of Bacteriocin GA15, a Novel Class IIa Bacteriocin Secreted by Lactiplantibacillus Plantarum GCNRC_GA15. Int. J. Biol. Macromol. 2022, 213, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Saris, P.E.J.; Takala, T.M. Genetic Characterization and Expression of Leucocin B, a Class IId Bacteriocin from Leuconostoc Carnosum 4010. Res. Microbiol. 2015, 166, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Hill, C. Virulence or Niche Factors: What’s in a Name? J. Bacteriol. 2012, 194, 5725–5727. [Google Scholar] [CrossRef] [PubMed]
- FDA. GRAS-Notice-GRN-936-Leuconostoc carnosum DSM 32756. 2020. Available online: https://www.fda.gov (accessed on 10 November 2023).


| ID 1 | Product Type 2 | Animal Origin 4 | Package 5 | Aliquot Size 6 | Growth Media 7 |
|---|---|---|---|---|---|
| Fish products | |||||
| H1 | Graved fish | Farmed salmon (NO) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H6 | Fresh fish, after thawing (I) 3 | Wild salmon (AK) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H7 | Fresh fish, dry salted (I) | Wild salmon (AK) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H9 | Fresh fish, salted by injection of brine (I) | Wild salmon (AK) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H8 | Fresh fish, salted by injection of brine (I) | Farmed salmon (NO) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H4 | Cold-smoked fish | Farmed salmon (NO) | VP, −20 °C | 20 g | MRS, DMRS, ELK |
| H10 | Cold-smoked fish | Farmed salmon (NO) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| Meat products | |||||
| H2 | Fermented sausage | Beef, pork (CH) | VP, RT | 40 g | MRS, DMRS, ELK |
| H3 | Fermented sausage | Elk (EU), pork (EU) | MAP, RT | 20 g | MRS, DMRS, ELK |
| H5 | Fermented sausage | Boar (CH) | UP, RT | 40 g | MRS, DMRS, ELK |
| H11 | Fresh meat, dry salted (I) | Beef (CH) | UP, 4 °C | 40 g | MRS, DMRS, ELK |
| H12 | Dry-cured meat | Ibex (CH) | VP, RT | 20 g | MRS, DMRS |
| H13 | Fermented sausage, lightly smoked | Beef, pork (CH) | VP, RT | 20 g | MRS, DMRS |
| H14 | Fresh sausage, lightly smoked | Beef, pork (CH) | VP, 4 °C | 40 g | MRS, DMRS |
| H15 | Dry-cured meat | Deer (AT) | VP, RT | 20 g | MRS, DMRS |
| H16 | Dry-cured meat | Pork (CH) | VP, RT | 20 g | MRS, DMRS |
| H17 | Fermented sausage | Chamois, pork (CH) | VP, RT | 20 g | MRS, DMRS |
| H18 | Fresh sausage (I) | Beef (CH) | UP, 4 °C | 40 g | MRS, DMRS |
| H19 | Fermented sausage | Beef (CH) | VP, RT | 20 g | MRS, DMRS |
| H20 | Fresh sausage (I) | Sheep, pork (CH) | UP, 4 °C | 40 g | MRS, DMRS |
| H21 | Fermented sausage | Sheep, pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H22 | Fresh sausage with 9% beetroot (I) | Beef, pork (CH) | UP, 4 °C | 40 g | MRS, DMRS |
| H23 | Fermented sausage with 9% beetroot | Beef, pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H24 | Fermented sausage | Goat (CH) | UP, RT | 20 g | MRS, DMRS |
| H25 | Fermented sausage | Deer (CH/NZ), pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H26 | Fermented sausage | Pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H27 | Fermented sausage | Boar (CH/NZ), pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H28 | Dry-cured meat | Veal (n.i.) | UP, RT | 20 g | MRS, DMRS |
| H29 | Fermented sausage | Deer (CH/AUT), pork (CH) | UP, RT | 20 g | MRS, DMRS |
| H30 | Fermented sausage | Pheasant, pork (EU) | UP, RT | 20 g | MRS, DMRS |
| Time [h] | Temperature [°C] | Relative Humidity [%] |
|---|---|---|
| 28 | 24 | 94 |
| Mettwurst: storage at 2 °C, Salami: follow further ripening steps | ||
| 14 | 22 | 90 |
| 14 | 20 | 88 |
| 12 | 18 | 86 |
| 12 | 16 | 84 |
| 12 | 14 | 82 |
| 12 | 14 | 80 |
| 216 | 14 | 76 |
| Strain | Agar Spot Assay and WDA | Positive PCR Reaction (Primer) | Sensitivity to Proteolytic Enzymes | Biogenic Amine Formation (Tyramine, Histamine, Putrescine, and Cadaverine) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. monocytogenes ATTC 15313 | L. monocytogenes SLCC 27555 | L. monocytogenes ATCC 19115 | |||||||||||||
| 37 °C/mm | 25 °C | 15 °C | 8 °C | 37 °C/mm | 25 °C | 15 °C | 8 °C | 37 °C/mm | 25 °C | 15 °C | 8 °C | ||||
| Lactiplantibacillus plantarum DH2 | 4.55 | ++++S | ++++S | ++++S | 6.9 | ++++S | ++++S | ++++S | 7.2 | ++++S | ++++S | ++++S | Ped, Pln A, PlanW, Pln | Yes | none |
| Lactiplantibacillus plantarum DH3 | 5.1 | ++++S | ++++S | ++++S | 6.95 | ++++S | ++++S | ++++S | 7.5 | ++++S | ++++S | ++++S | Pln A, Pln | Yes | none |
| Lactiplantibacillus plantarum DH9 | 3.6 | ++++S | ++++S | ++++S | 6.85 | ++++S | ++++S | ++++S | 7.6 | ++++S | ++++S | ++++S | Ped, Pln A, PlanS, Pln | Yes | none |
| Leuconostoc carnosum DH25 | 3.5 | ++++S | ++++S | ++++S | 6.25 | ++++S | ++++S | ++++S | 4.85 | ++++S | ++++S | ++++S | Pln A, PlanS, PlanW | Yes | none |
| Latilactobacillus curvatus DH29 | 0 | +++ | ++++S | ++++S | 7.55 | ++++ | ++++S | ++++S | 0 | ++ | ++++ | ++++S | CurA, Sak Q | NA | none |
| Latilactobacillus sakei DH42 | 0 | ++++S | ++++S | ++++S | 3.15 | ++++S | ++++S | ++++S | 4.1 | +++ | ++++S | ++++S | Sak Q, Sak P | Yes | none |
| Latilactobacillus sakei DH45 | 0 | ++++ | ++++S | ++++S | 1.55 | +++ | ++++ | ++++S | 0.6 | +++ | ++++S | ++++S | Sak Q, Sak P | Yes | none |
| Latilactobacillus sakei DH51 | 0 | ++++S | ++++S | ++++S | 2 | ++++S | ++++S | ++++S | ND | ++++ | ++++S | ++++S | skgA2 | NA | none |
| Latilactobacillus sakei DH54 | 0 | ++++S | ++++S | ++++S | 1.85 | ++++S | ++++S | ++++S | ND | ++++ | ++++S | ++++S | PlanW, skgA2 | Yes | none |
| Latilactobacillus sakei DH61 | 0 | ++++S | ++++S | ++++S | 1.65 | ++++ | ++++S | ++++S | ND | ++++ | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DH64 | 0 | ++++S | ++++S | ++++S | 1.6 | ++++ | ++++S | ++++S | ND | ++++ | ++++S | ++++S | skgA2 | NA | none |
| Latilactobacillus sakei DH80 | 0 | ++++S | ++++S | ++++S | 1.15 | ++++ | ++++S | ++++S | ND | ++++ | ++++S | ++++S | PlanW, skgA2 | NA | none |
| Latilactobacillus sakei DH84 | 0 | ++++S | ++++S | ++++S | 1.55 | ++++ | ++++S | ++++S | ND | ++++ | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DH85 | 0 | ++++S | ++++S | ++++S | 2.1 | ++++ | ++++S | ++++S | 3.1 | ++++ | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DH87 | 0 | +++ | +++ | ++++ | 0 | ++ | ++++S | ++++ | 0 | +++ | ++++ | ++++S | Sak P, Sak Q | NA | none |
| Lactiplantibacillus plantarum DH106 | ND | ++++S | ++++S | ND | ND | ++++S | ++++S | ++++S | ND | ++++S | ++++S | ++++S | Pln A, Pln J, Pln K, PlanS, PlanW, Pln | NA | none |
| Lactiplantibacillus plantarum DH108 | ND | +++ | ND | ND | ND | +++ | ND | ++++ | NA | +++ | ND | ++++S | Pln A, Pln J, Pln K, PlanS, PlanW, Pln | NA | none |
| Latilactobacillus sakei DH126 | 0 | + | ++++ | ND | 0 | + | + | ++++S | 0 | ++ | +++ | ++++ | CurA, Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DH134 | 0 | +++ | ++++ | ND | 3.35 | + | + | ++++ | 0 | ++ | ++ | ++++ | none | NA | none |
| Latilactobacillus sakei DH140 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | none | NA | none |
| Latilactobacillus sakei DH150 | 0 | ++++ | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DH151 | 0 | ++++ | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DH152 | 0 | ++++ | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | CurA, Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DH153 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++ | ++++S | ++++S | Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DH165 | 0 | ++++S | ++++S | ++++S | 0 | ++++ | ++++ | ++++S | 0 | ++++ | ++++ | ++++S | Sak P, Sak Q | NA | none |
| Leuconostoc citreum DH173 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++ | ++++S | ++++S | CurA | NA | none |
| Lactiplantibacillus plantarum DH176 | 7.05 | ++++S | ++++S | ++++S | 4.45 | ++++S | ++++S | ++++S | 9.05 | ++++S | ++++S | ++++S | PlanS, PlanW, Pln | Yes | none |
| Latilactobacillus sakei DH192 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 7.65 | ++++S | ++++S | ++++S | none | Yes | none |
| Latilactobacillus sakei DH208 | 5.5 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | none | Yes | none |
| Latilactobacillus sakei DH209 | 0 | ++++S | ++++S | ++++S | 0 | ++++ | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | none | NA | none |
| Latilactobacillus sakei DH210 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | none | NA | none |
| Latilactobacillus sakei DH213 | 0 | ++++ | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 2.8 | ++++ | ++++S | ++++S | none | Yes | Tyramine |
| Latilactobacillus sakei DH228 | 2.25 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 1.4 | ++++S | ++++S | ++++S | none | Yes | none |
| Lactiplantibacillus plantarum DH229 | 7.8 | ++++S | ++++S | ++++S | 4.8 | ++++S | ++++S | ++++S | 9.45 | ++++S | ++++S | ++++S | PlanW | Yes | Tyramine |
| Latilactobacillus sakei DH230 | 0 | ++++S | ++++S | ++++S | 4.35 | ++++S | ++++S | ++++S | 8.15 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DH231 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | NA | none |
| Latilactobacillus sakei DH232 | 1.6 | ++++S | ++++S | ++++S | 2.6 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DH233 | 0.75 | ++++S | ++++S | ++++S | 2.7 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Lactobacillus curvatus DH234 | 0 | ++++S | ++++S | ++++S | 4 | ++++S | ++++S | ++++S | 5.8 | ++++S | ++++S | ++++S | none | Yes | none |
| Latilactobacillus sakei DH235 | 0 | ++++S | ++++S | ++++S | 9.35 | ++++S | ++++S | ++++S | 0.8 | ++++S | ++++S | ++++S | none | Yes | none |
| Latilactobacillus sakei DH250 | 0 | ++++S | − | ++++S | 0.85 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | Sak Q | Yes | Tyramine |
| Latilactobacillus curvatus DH252 | 0 | ++++S | − | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | Sak Q | Yes | Tyramine |
| Latilactobacillus sakei DHN35 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | Sak P, Sak Q | NA | none |
| Latilactobacillus sakei DHN129 | 1.55 | ++++S | ++++S | ++++S | 2.45 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DHN154 | 1.45 | ++++S | ++++S | ++++S | 2.3 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | Tyramine |
| Latilactobacillus sakei DHN158 | 1.9 | ++++S | ++++S | ++++S | 2.5 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | Tyramine |
| Latilactobacillus sakei DHN62 | 0.65 | ++++S | ++++S | ++++S | 2.3 | ++++S | ++++S | ++++S | 3.25 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei DHN81 | 0 | ++++S | ++++S | ++++S | 2.95 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | skgA2 | Yes | none |
| Latilactobacillus sakei PR21-05 | 0 | ++++S | ++++S | ++++S | 10.05 | ++++S | ++++S | ++++S | 1.6 | ++++S | ++++S | ++++S | none | Yes | Tyramine |
| Not identified PR21-08 | 0 | ++++S | ++++S | ++++S | 2.55 | ++++S | ++++S | ++++S | 1 | ++++S | ++++S | ++++S | Pln | Yes | none |
| Lacticaseibacillus paracasei PR25-02 | 0 | ++++S | ++++S | ++++S | 0 | ++++S | ++++S | ++++S | 1.95 | ++++S | ++++S | ++++S | Pln, PlanW | Yes | none |
| Sample | pH Ø | ||
| D0 | D2 | D4 | |
| L+St+DH25 | 5.83 ± 0.01 | 4.93 ± 0.05 | 5.15 ± 0.03 |
| L+St+DH42 | 5.76 ± 0.01 | 4.95 ± 0.10 | 5.07 ± 0.14 |
| L+St+DH54 | 5.70 ± 0.02 | 5.01 ± 0.02 | 4.99 ± 0.22 |
| L+St+DH61 | 5.63 ± 0.00 | 5.14 ± 0.06 | 5.59 ± 0.63 |
| L+St+DH64 | 5.68 ± 0.01 | 4.81 ± 0.03 | 5.11 ± 0.13 |
| L+St+DH106 | 5.73 ± 0.01 | 4.91 ± 0.06 | 5.01 ± 0.09 |
| L+St+DH108 | 5.76 ± 0.04 | 4.89 ± 0.08 | 5.22 ± 0.02 |
| L | 5.77 ± 0.06 | 5.06 ± 0.27 | 5.16 ± 0.27 |
| L+St | 5.74 ± 0.10 | 4.91 ± 0.13 | 5.18 ± 0.26 |
| aw Ø | |||
| Control | 0.99 ± 0.00 | 0.99 ± 0.00 | 0.99 ± 0.00 |
| Sample | pH | |||
| D0 | D4 | D7 | D14 | |
| DH25+B+L | 5.86 | 5.28 | 5.54 | 7.28 |
| DH25+P+L | 5.74 | 5.24 | 5.51 | 6.05 |
| B+L | 5.80 | 5.29 | 5.60 | 7.21 |
| P+L | 5.86 | 5.30 | 5.62 | 6.44 |
| L | 5.90 | 5.21 | 5.67 | 5.90 |
| aw Ø | ||||
| Control | 0.98 ± 0.01 | 0.99 ± 0.00 | 0.97 ± 0.00 | 0.99 ± 0.00 |
| Sample | pH | ||
| D0 | D4 | D7 | |
| DH25+B+L | 6.06 | 5.65 | 5.29 |
| DH25+P+L | 6.05 | 5.25 | 5.43 |
| B+L | 6.03 | 5.69 | 5.24 |
| P+L | 6.11 | 5.34 | 5.04 |
| L | 6.07 | 5.93 | 5.29 |
| aw Ø | |||
| Control | 0.96 ± 0.00 | 0.95 ± 0.00 | 0.95 ± 0.00 |
| Sample | pH Ø | |||||
| Salami | D0 | D1 | D8 | D15 | D29 | D36 |
| DH25+St+L | 5.58 ± 0.00 | - | 4.76 ± 0.02 | 4.71 ± 0.04 | - | 4.89 ± 0.00 |
| DH42+St+L | 5.53 ± 0.02 | - | 4.80 ± 0.00 | 4.62 ± 0.01 | - | 4.82 ± 0.02 |
| St+L | 5.56 ± 0.00 | - | 4.85 ± 0.01 | 4.72 ± 0.02 | - | 4.90 ± 0.01 |
| L | 5.55 ± 0.00 | - | 5.20 ± 0.01 | 4.70 ± 0.00 | - | 4.91 ± 0.01 |
| Mettwurst | ||||||
| DH25+St+L | 5.55 ± 0.01 | 5.41 ± 0.00 | - | 4.89 ± 0.00 | 4.82 ± 0.01 | - |
| DH42+St+L | 5.56 ± 0.00 | 5.40 ± 0.00 | - | 4.87 ± 0.02 | 4.86 ± 0.00 | - |
| St+L | 5.56 ± 0.00 | 5.29 ± 0.01 | - | 4.74 ± 0.01 | 4.91 ± 0.00 | - |
| L | 5.55 ± 0.00 | 5.30 ± 0.01 | - | 4.79 ± 0.00 | 4.81 ± 0.01 | - |
| aw Ø | ||||||
| Salami | 0.96 ± 0.00 | - | 0.95 ± 0.00 | 0.94 ± 0.00 | - | 0.95 ± 0.00 |
| Mettwurst | 0.97 ± 0.00 | 0.96 ± 0.00 | - | 0.96 ± 0.00 | 0.95 ± 0.00 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tönz, A.; Freimüller Leischtfeld, S.; Stevens, M.J.A.; Glinski-Häfeli, D.; Ladner, V.; Gantenbein-Demarchi, C.; Miescher Schwenninger, S. Growth Control of Listeria monocytogenes in Raw Sausage via Bacteriocin-Producing Leuconostoc carnosum DH25. Foods 2024, 13, 298. https://doi.org/10.3390/foods13020298
Tönz A, Freimüller Leischtfeld S, Stevens MJA, Glinski-Häfeli D, Ladner V, Gantenbein-Demarchi C, Miescher Schwenninger S. Growth Control of Listeria monocytogenes in Raw Sausage via Bacteriocin-Producing Leuconostoc carnosum DH25. Foods. 2024; 13(2):298. https://doi.org/10.3390/foods13020298
Chicago/Turabian StyleTönz, Andrea, Susette Freimüller Leischtfeld, Marc J. A. Stevens, Deborah Glinski-Häfeli, Valentin Ladner, Corinne Gantenbein-Demarchi, and Susanne Miescher Schwenninger. 2024. "Growth Control of Listeria monocytogenes in Raw Sausage via Bacteriocin-Producing Leuconostoc carnosum DH25" Foods 13, no. 2: 298. https://doi.org/10.3390/foods13020298
APA StyleTönz, A., Freimüller Leischtfeld, S., Stevens, M. J. A., Glinski-Häfeli, D., Ladner, V., Gantenbein-Demarchi, C., & Miescher Schwenninger, S. (2024). Growth Control of Listeria monocytogenes in Raw Sausage via Bacteriocin-Producing Leuconostoc carnosum DH25. Foods, 13(2), 298. https://doi.org/10.3390/foods13020298







