Tanshinone Content Prediction and Geographical Origin Classification of Salvia miltiorrhiza by Combining Hyperspectral Imaging with Chemometrics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Pretreatment
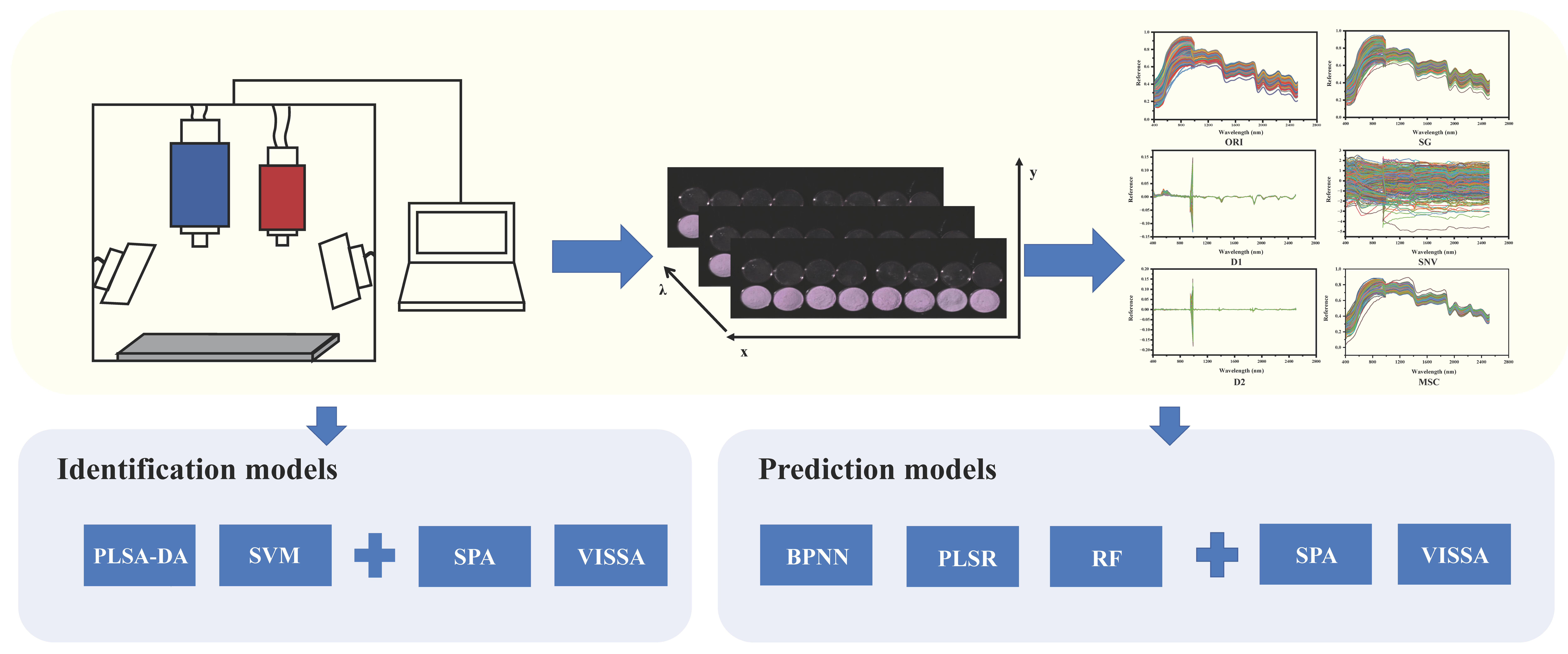
2.2. Data Collection Using an HSI System and Extraction of Interest Origins
2.3. Chemometrics Analysis
2.3.1. Pretreatment
2.3.2. Classification Models
2.3.3. Prediction Models
2.3.4. Effective Wavelength Screening Algorithms
2.4. Reference Determination of Four Tanshinone Content
2.5. Statistical Analysis
3. Results
3.1. Statistical Analysis of Tanshinone Content in S. miltiorrhiza from Different Origins
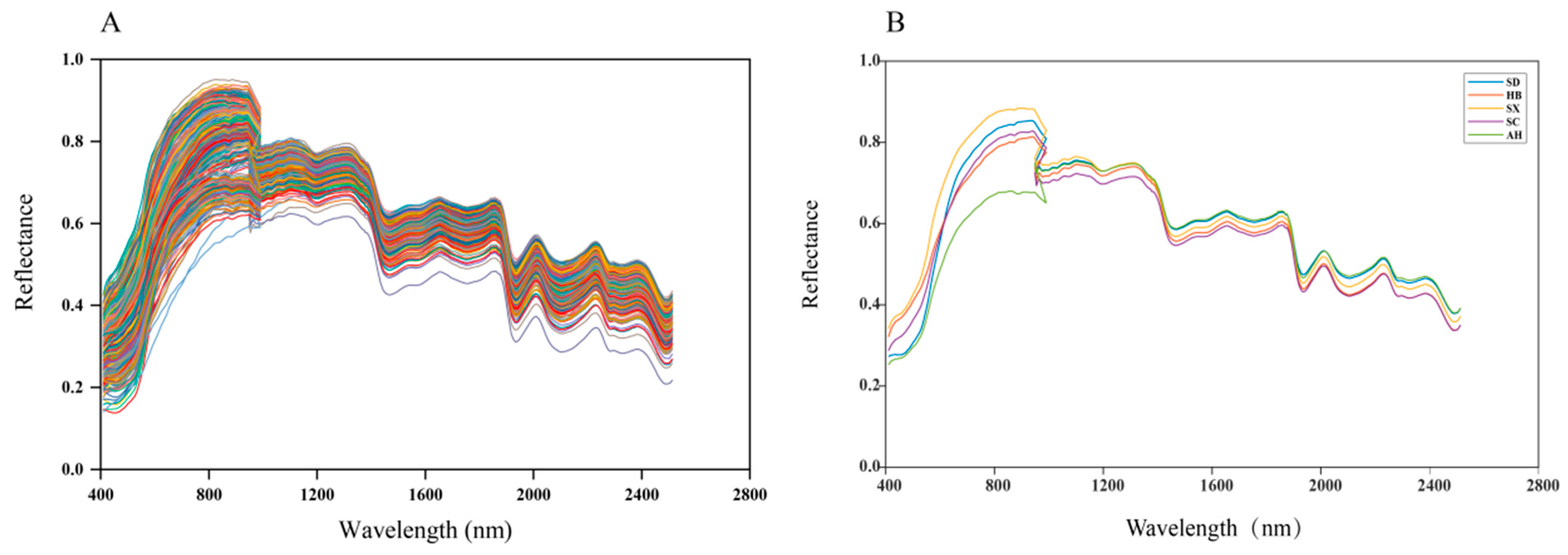
3.2. Raw spectra Characteristics of S. miltiorrhiza
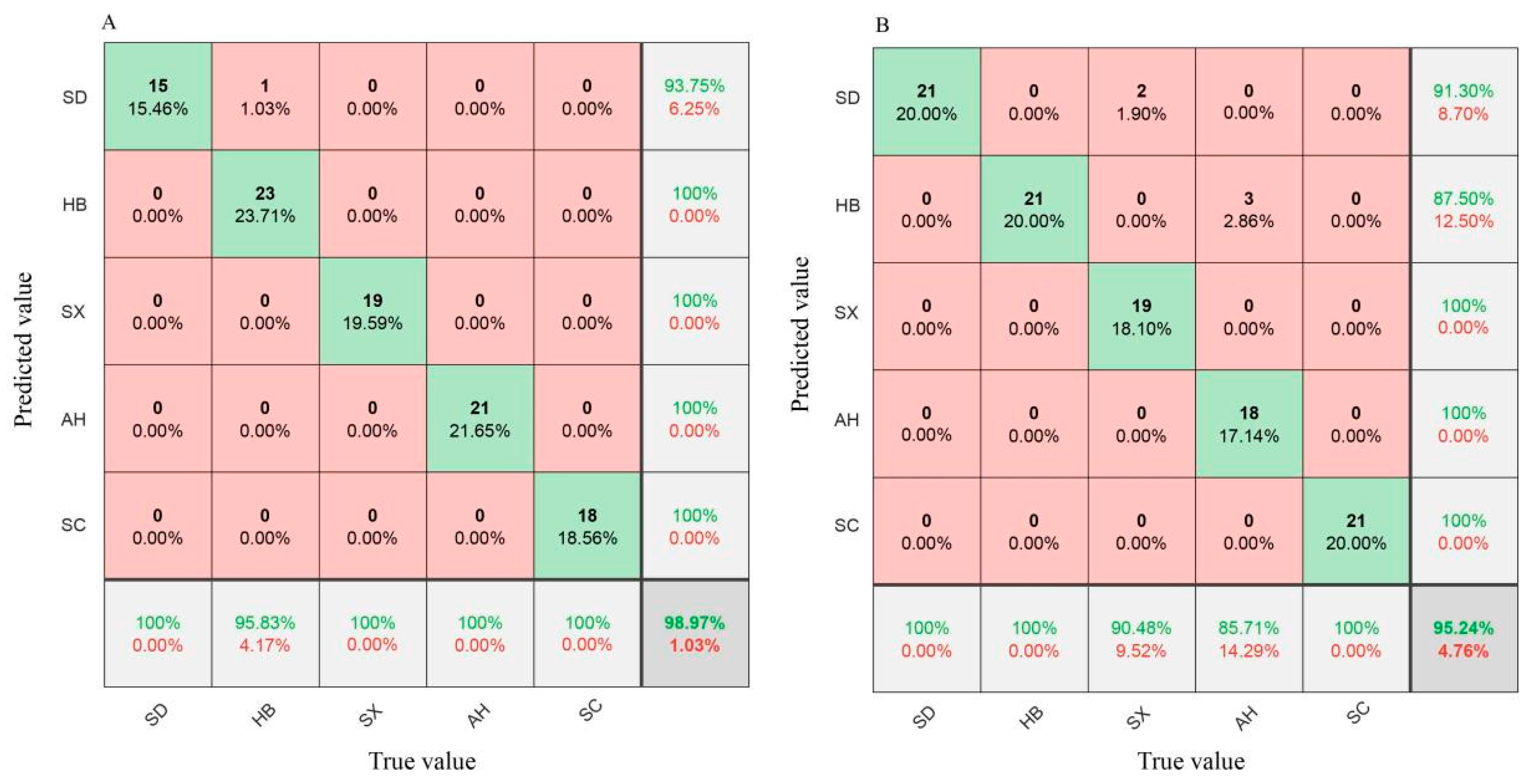
3.3. Classification of the Geographical Origins of S. miltiorrhiza Based on Hyperspectral Imaging Full Wavelengths
3.4. Prediction of Chemical Indicators Based on HSI Full Wavelength
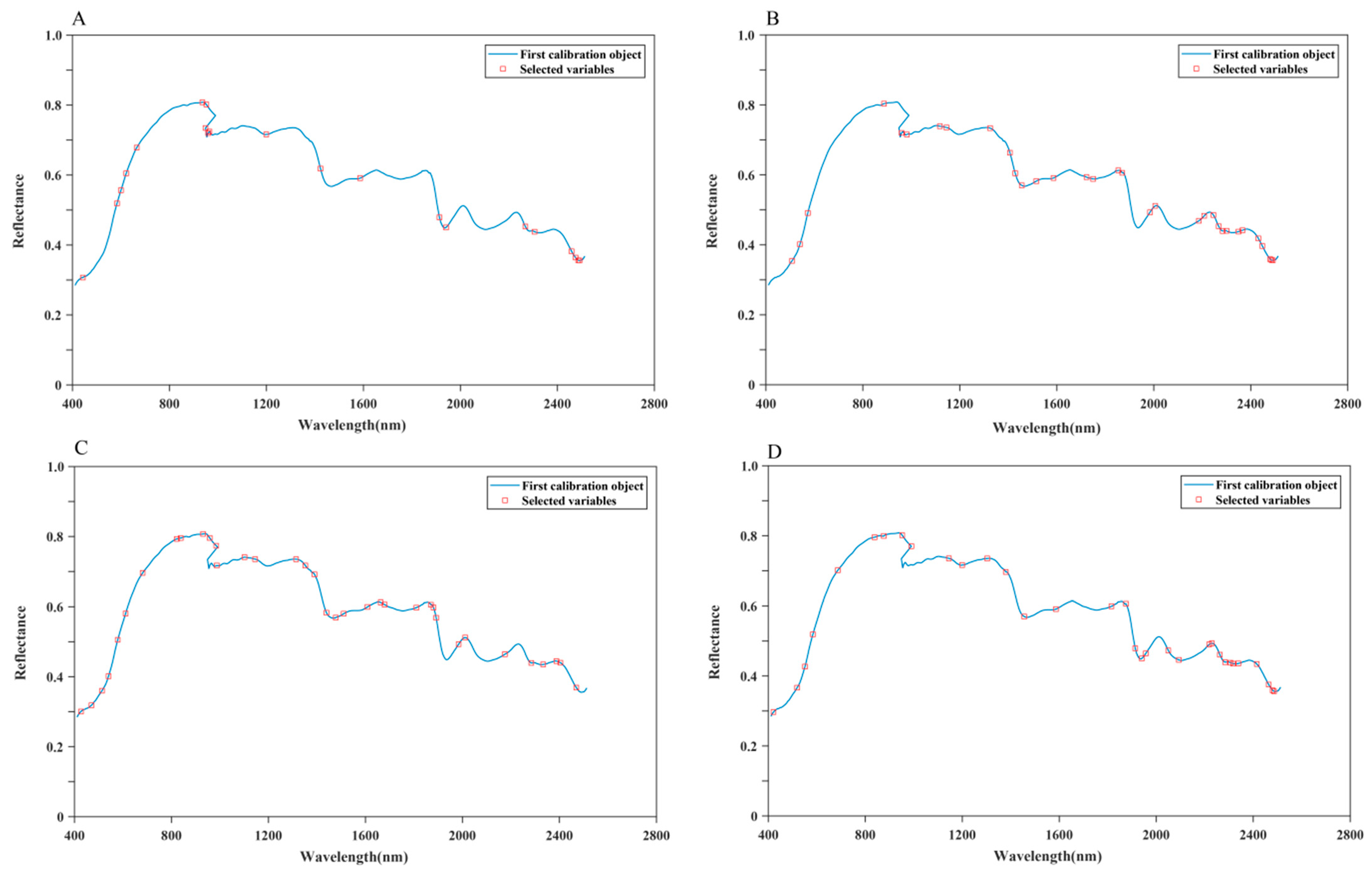
3.5. Classification and Prediction of Chemical Indicators Based on Selected Wavelengths
3.5.1. Classification of the Geographical Origins of S. miltiorrhiza Based on Selected Wavelengths
3.5.2. Prediction of Chemical Indicators Based on HSI Selected Wavelengths
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Deng, A.P.; Guo, L.P.; Zhan, Z.L.; Huang, L.Q. Decipherment of ancient literature about Danshen. Zhongguo Zhong Yao Za Zhi 2016, 41, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Kong, Y.; Chen, W.; Wang, X.; Jia, Z.; Dai, Y.; Yang, X. The quality of wild Salvia miltiorrhiza from Dao Di area in China and its correlation with soil parameters and climate factors. Phytochem Anal. 2021, 32, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Feng, H.; Guo, S.; Han, Y.; Chen, X. Danshenol A inhibits TNF-α-induced expression of intercellular adhesion molecule-1 (ICAM-1) mediated by NOX4 in endothelial cells. Sci. Rep. 2017, 7, 12953. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Liu, H. Research Progress on Structural Optimization on the Lipid-Soluble Ingredients of Salvia miltiorrhiza. Chin. J. Org. Chem. 2023, 43, 471–490. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Z. Salvianolic acid B from Salvia miltiorrhiza bunge: A potential antitumor agent. Front. Pharmacol. 2022, 13, 1042745. [Google Scholar] [CrossRef]
- Guo, Y.; Li, Y.; Xue, L.; Severino, R.P.; Gao, S.; Niu, J.; Qin, L.; Zhang, D.; Brömme, D. Salvia miltiorrhiza: An ancient Chinese herbal medicine as a source for anti-osteoporotic drugs. J. Ethnopharmacol. 2014, 155, 1401–1416. [Google Scholar] [CrossRef]
- Yuan, J.; Du, H.; Wan, M.X.; Li, Z.; Zhang, Y.X.; Li, D.K.; Zhuang, P.W.; Ju, A.C. Research progress on anti-inflammatory pharmacological action of components of Salvia miltiorrhiza and its preparations. Drug Eval. Res. 2021, 44, 2322–2332. [Google Scholar] [CrossRef]
- Lobina, C.; Colombo, G.; Gessa, G.L.; Carai, M.A.M.; Allegrini, P.; Morazzoni, P.; Riva, A. Anxiolytic effect of an extract of Salvia miltiorrhiza roots in rats. J. Chin. Med. Assoc. 2018, 81, 390–397. [Google Scholar] [CrossRef]
- Wu, J.J.; Li, S.L.; Huang, Z.Q.; Gao, R.H.; Wang, H.L.; Peng, Y.; Zhang, C.; Zhao, H.G.; Huang, F. Study on Antioxidative Pharmacodynamics of Aqueous Extract of Salvia miltiorrhiza Bge Leaves in Mice. Strait Pharm. J. 2023, 35, 25–32. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2020.
- Zhan, Z.L.; Deng, A.P.; Peng, H.S.; Zhang, X.B.; Guo, L.P.; Huang, L.Q. Study of genuineness based on changes of ancient herbal origin--taking Astragalus membranaceus and Salvia miltiorrhiza as examples. Zhongguo Zhong Yao Za Zhi 2016, 41, 3202–3208. [Google Scholar] [CrossRef]
- Li, Y.; You, G.; Duan, R.; Liu, K.; Wang, D. Simultaneous Determination of Danshensu, Protocatechuic Acid, Protocatechuic Aldehyde, Rosmarinic Acid and Salvianolic Acid B in Three Processing Products of White Flower Salvia miltiorrhiza by HPLC. Asian J. Chem. 2013, 25, 9558–9560. [Google Scholar] [CrossRef]
- Zhang, L.G.; Hu, T.T.; Zhang, F.F.; Luan, S.R.; Li, W.; Deng, H.X.; Lan, Z.H.; Luo, X.F.; Wu, Z.X.; Leslaw, M. Analysis of lipophilic components of Salvia miltiorrhiza roots and S. yunnanensis roots by UPLC and LC-MS/MS. Zhongguo Zhong Yao Za Zhi 2019, 44, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, Y.; Cen, Y.; Yang, D.; Qi, Z.; Hou, Z.; Han, S.; Cai, Z.; Liu, K. Bivariate Correlation Analysis of the Chemometric Profiles of Chinese Wild Salvia miltiorrhiza Based on UPLC-Qqq-MS and Antioxidant Activities. Molecules 2018, 23, 538. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Gong, S.; Liu, Y.; Li, X.; Li, M.; Zeng, D.; Li, J.; Guo, Y.; Guo, L. Rapid discrimination of the authenticity and geographical origin of bear bile powder using stable isotope ratio and elemental analysis. Anal. Bioanal. Chem. 2023, 415, 345–356. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.Z.; Liu, Z.; Jiang, X.; Zhang, D.L. Identification of Honey Adulteration by Isotope Ratio Mass Spectrometry. Mod. Food Sci. Technol. 2013, 29, 867–871. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, K.; Wang, L.; Han, D.; Yin, G.; Wang, J.; Qin, B.; Li, S.; Wang, T. Identification of Salvia species using high-performance liquid chromatography combined with chemical pattern recognition analysis. J. Sep. Sci. 2018, 41, 609–617. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, J.; Li, M.; Chen, Y.; Cui, Q.; Lu, C.; Wang, Y.; Li, L.; Xu, Z.; Zhong, Y.; et al. Rapid identification of the green tea geographical origin and processing month based on near-infrared hyperspectral imaging combined with chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120537. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Yu, S.; Fu, H.; He, S.; Yang, B.; Nan, T.; Yuan, Y.; Huang, L. Prediction of chemical indicators for quality of Zanthoxylum spices from multi-regions using hyperspectral imaging combined with chemometrics. Front. Sustain. Food Syst. 2022, 6, 1036892. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Yuan, Y.; Zhao, Y.; Nie, J.; Nan, T.; Huang, L.; Yang, J. Nutrient content prediction and geographical origin identification of red raspberry fruits by combining hyperspectral imaging with chemometrics. Front. Nutr. 2022, 9, 980095. [Google Scholar] [CrossRef]
- Gehlot, S.; Ansari, N.; Gupta, A. WTL-I: Mutual Information-Based Wavelet Transform Learning for Hyperspectral Imaging. Front. Signal Process. 2022, 2, 854207. [Google Scholar] [CrossRef]
- Liu, L.L.; Wang, Y.Y.; Yang, J.; Zhang, X.B. Rapid detection technology of chemical component content in Lycii Fructus based on hyperspectral technology. China J. Chin. Mater. Medica 2023, 48, 4328–4336. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, C.; Wang, S.; Yuan, Y.; Bai, R.; Nan, T.; Yang, J. Ginsenoside Rg2 content prediction in Panax ginseng based on the fusion of hyperspectral wavelengths combined with chemometric analysis. J. Food Compos. Anal. 2023, 123, 105619. [Google Scholar] [CrossRef]
- Zijian, L.; Jiacheng, G.; Cong, Z.; Youyou, W.; Jian, Y.; Jun, H.; Hongpeng, W.; Ruibin, B. Identification of Geographical Origin for Hawthorn Based on Hyperspectral Imaging Technology. Sci. Technol. Food Ind. 2023, 45, 282–291. [Google Scholar] [CrossRef]
- Xuan, G.; Gao, C.; Shao, Y. Spectral and image analysis of hyperspectral data for internal and external quality assessment of peach fruit. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 272, 121016. [Google Scholar] [CrossRef]
- Liu, Y.L.; Lyu, Q.; He, S.L.; Yi, S.L.; Liu, X.F.; Xie, R.J.; Zheng, Y.Q.; Deng, L. Prediction of nitrogen and phosphorus contents in citrus leaves based on hyperspectral imaging. Int. J. Agric. Biol. Eng. 2015, 8, 80–88. [Google Scholar] [CrossRef]
- Zulfiqar, M.; Ahmad, M.; Sohaib, A.; Mazzara, M.; Distefano, S. Hyperspectral Imaging for Bloodstain Identification. Sensors 2021, 21, 3045. [Google Scholar] [CrossRef]
- Ning, H.; Wang, J.; Jiang, H.; Chen, Q. Quantitative detection of zearalenone in wheat grains based on near-infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 280, 121545. [Google Scholar] [CrossRef]
- Xu, J.; Sun, D. Identification of freezer burn on frozen salmon surface using hyperspectral imaging and computer vision combined with machine learning algorithm. Int. J. Refrig. 2017, 74, 151–164. [Google Scholar] [CrossRef]
- Chaudhary, S.; Ninsawat, S.; Nakamura, T. Non-Destructive Trace Detection of Explosives Using Pushbroom Scanning Hyperspectral Imaging System. Sensors 2019, 19, 97. [Google Scholar] [CrossRef]
- Feng, J.; Liu, Y.; Shi, X.; Wang, Q. Potential of hyperspectral imaging for rapid identification of true and false honeysuckle tea leaves. J. Food Meas. Charact. 2018, 12, 2184–2192. [Google Scholar] [CrossRef]
- Ong, P.; Chen, S.; Tsai, C.; Chuang, Y. Prediction of tea theanine content using near-infrared spectroscopy and flower pollination algorithm. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 255, 119657. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.; Zhu, S.; He, Y.; Zhang, C. Grading of Chinese Cantonese Sausage Using Hyperspectral Imaging Combined with Chemometric Methods. Sensors 2017, 17, 1706. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xu, M.; Zhang, W.; Liu, G.; Tong, L. Identification of zinc pollution in rice plants based on two characteristic variables. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120043. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, Y.J.; Fu, H.Y.; Li, X.; Yang, J. Quality Difference Analysis of Salviae Miltiorrhizae Radix et Rhizoma from Different Origins Based on Multi-index Content Determination Combined with Chemometrics. J. Instrum. Anal. 2023, 42, 389–401. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, L.H.; Yang, G.J.; Ye, Z.L. Rapid Determination of Multi-maker Ingredients in Salvia miltiorrhiza by Near Infrared Diffused Reflection Spectroscopy. China Pharm. 2017, 28, 4247–4251. [Google Scholar] [CrossRef]
- Yuan, W.D.; Jiang, H.Z.; Yang, S.Y.; Zhang, C.; Zhou, Y.; Zhou, H.P. Geographical Origin Identification of Ningxia Lycium Barbarum Using Hyperspectral Imaging Technology. Food Sci. 2024, 45, 254–260. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, F.; Zhang, Y.; Wang, S.; Yuan, Y.; Lu, C.; Nie, J.; Nan, T.; Yang, B.; Huang, L.; et al. Application of hyperspectral imaging assisted with integrated deep learning approaches in identifying geographical origins and predicting nutrient contents of Coix seeds. Food Chem. 2023, 404, 134503. [Google Scholar] [CrossRef]
- Peng, Y.F.; Wang, J.; Wu, Z.S.; Liu, X.N.; Qiao, Y.J. NIR Band Assignment of Tanshinone IIA and Cryptotanshinone by 2D-COS Technology and Model Application Tanshinone Extract. Spectrosc. Spect. Anal. 2022, 42, 1781–1785. [Google Scholar] [CrossRef]
- Fang, W.T.; Deng, A.P.; Ren, Z.L.; Nan, T.G.; Kang, L.P.; Guo, L.P.; Huang, L.Q.; Zhan, Z.L. Research progress on quality evaluation of Salviae Miltiorrhizae Radix et Rhizoma (Danshen). Zhongguo Zhong Yao Za Zhi 2018, 43, 1077–1085. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, D.; Liang, Z.; Liu, J.; Yan, K.; Zhu, Y.; Yang, S. Climatic factors control the geospatial distribution of active ingredients in Salvia miltiorrhiza Bunge in China. Sci. Rep. 2019, 9, 904. [Google Scholar] [CrossRef]
- Zhang, X.D.; Yu, Y.G.; Yang, D.F.; Qi, Z.C.; Liu, R.Z.; Deng, F.T.; Cai, Z.X.; Li, Y.; Sun, Y.F. Chemotaxonomic variation in secondary metabolites contents and their correlation between environmental factors in Salvia miltiorrhiza Bunge from natural habitat of China. Ind. Crop Prod. 2018, 113, 335–347. [Google Scholar] [CrossRef]
- Pan, X.M.; Wei, H.; Liu, Y.; Liu, S.X.; Zhang, T.J.; Ma, X.W.; Han, F.N. Study on quality of Salvia miltiorrhiza from different habitats. Chin. Tradit. Herb. Drugs 2011, 42, 1833–1836. [Google Scholar]
- Wu, R.Q.; Xie, Y.; Zhang, Y.Q.; Chen, X.F.; Wu, X.Q.; Gao, Y.M. Study on the Quality Consistency of Salvia miltiorrhiza for Different Districts from Sichuan Province. Chin. J. Ethmed. Ethnopharacy 2023, 32, 27–33. [Google Scholar]
- Li, J.; Fan, G.; He, Y. Predicting the current and future distribution of three Coptis herbs in China under climate change conditions, using the MaxEnt model and chemical analysis. Sci. Total Environ. 2020, 698, 134141. [Google Scholar] [CrossRef] [PubMed]
- Uchimiya, M. Aromaticity of secondary products as the marker for sweet sorghum [Sorghum bicolor (L.) Moench] genotype and environment effects. J. Agric. Food Res. 2022, 9, 100338. [Google Scholar] [CrossRef]
- Lee, J.; Jung, Y.; Shin, J.; Kim, H.K.; Moon, B.C.; Ryu, D.H.; Hwang, G. Secondary Metabolite Profiling of Curcuma Species Grown at Different Locations Using GC/TOF and UPLC/Q-TOF MS. Molecules 2014, 19, 9535–9551. [Google Scholar] [CrossRef]
- Sun, Y.C.; Jiao, L. Identification of Salvia miltiorrhiza Regions by Hyperspectrum and Support Vector Machine. Fujian Anal. Test. 2023, 32, 11–15. [Google Scholar] [CrossRef]
- Zhang, S.S.; Wang, D.Q.; Zhu, J.J.; Li, C.Y.; Wang, Z.M.; Kuang, Y.H. Process Quality Control of Tanshinones in Salvia miltiorrhiza with the extraction and concertration of Fufangdanshen Tabletsby Near Infrared Spectroscopy. Ginsheng Res. 2021, 33, 15–20. [Google Scholar] [CrossRef]
- Xia, Z.; Sun, Y.; Cai, C.; He, Y.; Nie, P. Rapid Determination of Chlorogenic Acid, Luteoloside and 3,5-O-dicaffeoylquinic Acid in Chrysanthemum Using Near-Infrared Spectroscopy. Sensors 2019, 19, 1981. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; He, Y. Application of Near-Infrared Hyperspectral Imaging to Detect Sulfur Dioxide Residual in the Fritillaria thunbergii Bulbus Treated by Sulfur Fumigation. Appl. Sci. 2017, 7, 77. [Google Scholar] [CrossRef]
- Lu, Z.; Yu, H.; Yin, Y.; Yuan, Y.; Liang, H.; Li, F.; Li, Z. Determination of the Acid and Peroxide Values of Vegetable Oils by Raman Spectroscopy with Competitive Adaptive Reweighted Sampling (CARS) and Back Propagation Neural Network (BPNN). Anal. Lett. 2024, 57, 2289–2306. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, X.; Li, Q.; Shi, Y.; Guo, G.; Zhao, L.; Wang, J.; Su, Y.; Zhang, C. Spatial distribution prediction of soil As in a large-scale arsenic slag contaminated site based on an integrated model and multi-source environmental data. Environ. Pollut. 2020, 267, 115631. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, C.; Song, S.; Xie, S.; Kang, F. Study on starch content detection and visualization of potato based on hyperspectral imaging. Food Sci. Nutr. 2021, 9, 4420–4430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.Y.; Zhang, T.; Qin, Y.C.; Nan, T.G.; Yang, J.; Lu, Y.J. Identification of Citri Reticulatae Pericarpium Form Different Scales Geographical Origin by Hyperspectral Imaging Combined with Image Segmentation Algorithm. Chem. Reag. 2023, 45, 136–143. [Google Scholar] [CrossRef]
- Xiao, D.; Wang, S.M.; Zhang, Y.; Liu, D.F.; Hao, Q.X.; Bai, R.B.; Yang, J. Research on the detection of andrographolide components in Andrographis paniculata based on hyperspectral technology. Chem. Reag. 2024, 46, 89–98. [Google Scholar] [CrossRef]
- Haruna, S.A.; Li, H.; Wei, W.; Geng, W.; Luo, X.; Zareef, M.; Yao-Say Solomon Adade, S.; Ivane, N.M.A.; Isa, A.; Chen, Q. Simultaneous quantification of total flavonoids and phenolic content in raw peanut seeds via NIR spectroscopy coupled with integrated algorithms. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 285, 121854. [Google Scholar] [CrossRef]


| Collection Origins | Tan I (mg/g) | Tan IIA (mg/g) | CTS (mg/g) | Total (mg/g) |
|---|---|---|---|---|
| SD (n = 84) | 1.090 ± 0.373 d | 2.807 ± 0.983 d | 3.616 ± 2.070 d | 7.469 ± 3.126 d |
| HB (n = 84) | 0.316 ± 0.268 ab | 0.826 ± 0.633 a | 0.451 ± 0.379 a | 1.593 ± 1.231 a |
| SX (n = 84) | 0.132 ± 0.058 c | 1.142 ± 0.608 b | 0.640 ± 0.420 b | 1.914 ± 1.014 a |
| SC (n = 84) | 0.271 ± 0.186 a | 1.663 ± 1.317 c | 0.678 ± 0.687 ab | 2.612 ± 2.066 b |
| AH (n = 84) | 0.376 ± 0.257 b | 1.940 ± 1.113 c | 1.928 ± 1.831 c | 4.245 ± 2.786 c |
| Pretreatments | PLS-DA | SVM | ||
|---|---|---|---|---|
| Calibration Set (%) | Prediction Set (%) | Calibration Set (%) | Prediction Set (%) | |
| ORI | 98.45 | 95.88 | 98.73 | 80.95 |
| D1 | 99.69 | 98.97 | 99.37 | 91.43 |
| D2 | 98.45 | 97.94 | 99.05 | 95.24 |
| SG | 99.07 | 97.94 | 98.73 | 80.95 |
| MSC | 99.07 | 96.91 | 97.46 | 85.71 |
| SNV | 99.69 | 97.94 | 100.00 | 75.24 |
| Chemical Indexes | Models | Calibration Set | Prediction Set | Chemical Indexes | Models | Calibration Set | Prediction Set | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSEC | R2 | RMSEP | RPD | R2 | RMSEC | R2 | RMSEP | RPD | ||||
| Tan I | ORI-BPNN | 0.965 | 0.079 | 0.924 | 0.118 | 3.35 | Tan IIA | ORI-BPNN | 0.945 | 0.282 | 0.917 | 0.332 | 3.34 |
| D1-BPNN | 0.948 | 0.108 | 0.861 | 0.163 | 2.53 | D1-BPNN | 0.949 | 0.275 | 0.871 | 0.431 | 2.63 | ||
| D2-BPNN | 0.950 | 0.095 | 0.846 | 0.172 | 2.51 | D2-BPNN | 0.899 | 0.384 | 0.820 | 0.503 | 2.33 | ||
| SG-BPNN | 0.966 | 0.079 | 0.917 | 0.125 | 3.18 | SG-BPNN | 0.918 | 0.348 | 0.885 | 0.391 | 2.66 | ||
| MSC-BPNN | 0.971 | 0.071 | 0.919 | 0.121 | 3.41 | MSC-BPNN | 0.939 | 0.297 | 0.919 | 0.328 | 3.28 | ||
| SNV-BPNN | 0.961 | 0.083 | 0.930 | 0.114 | 3.44 | SNV-BPNN | 0.957 | 0.258 | 0.886 | 0.393 | 2.81 | ||
| ORI-PLSR | 0.960 | 0.081 | 0.934 | 0.117 | 3.90 | ORI-PLSR | 0.940 | 0.296 | 0.864 | 0.422 | 2.66 | ||
| D1-PLSR | 0.950 | 0.091 | 0.906 | 0.136 | 3.24 | D1-PLSR | 0.946 | 0.281 | 0.821 | 0.489 | 2.32 | ||
| D2-PLSR | 0.940 | 0.100 | 0.910 | 0.132 | 3.25 | D2-PLSR | 0.895 | 0.391 | 0.826 | 0.478 | 2.32 | ||
| SG-PLSR | 0.970 | 0.072 | 0.931 | 0.123 | 3.79 | SG-PLSR | 0.935 | 0.308 | 0.851 | 0.442 | 2.51 | ||
| MSC-PLSR | 0.955 | 0.087 | 0.938 | 0.110 | 4.03 | MSC-PLSR | 0.950 | 0.269 | 0.853 | 0.439 | 2.53 | ||
| SNV-PLSR | 0.976 | 0.063 | 0.932 | 0.120 | 3.83 | SNV-PLSR | 0.932 | 0.316 | 0.841 | 0.455 | 2.39 | ||
| ORI-RF | 0.9600 | 0.119 | 0.880 | 0.218 | 1.64 | ORI-RF | 0.946 | 0.400 | 0.860 | 0.578 | 1.78 | ||
| D1-RF | 0.9795 | 0.084 | 0.923 | 0.175 | 2.16 | D1-RF | 0.974 | 0.291 | 0.919 | 0.448 | 2.21 | ||
| D2-RF | 0.9761 | 0.099 | 0.928 | 0.173 | 2.06 | D2-RF | 0.955 | 0.390 | 0.915 | 0.472 | 1.93 | ||
| SG-RF | 0.9622 | 0.117 | 0.868 | 0.227 | 1.59 | SG-RF | 0.952 | 0.385 | 0.853 | 0.592 | 1.73 | ||
| MSC-RF | 0.9641 | 0.110 | 0.891 | 0.211 | 1.77 | MSC-RF | 0.952 | 0.373 | 0.867 | 0.572 | 1.91 | ||
| SNV-RF | 0.9690 | 0.102 | 0.902 | 0.196 | 2.03 | SNV-RF | 0.959 | 0.354 | 0.902 | 0.485 | 2.18 | ||
| CTS | ORI-BPNN | 0.966 | 0.327 | 0.911 | 0.534 | 3.03 | Total (Tan I + Tan IIA + CTS) | ORI-BPNN | 0.956 | 0.658 | 0.933 | 0.803 | 3.83 |
| D1-BPNN | 0.881 | 0.612 | 0.860 | 0.659 | 2.56 | D1-BPNN | 0.939 | 0.778 | 0.886 | 1.106 | 2.96 | ||
| D2-BPNN | 0.897 | 0.563 | 0.711 | 0.966 | 1.68 | D2-BPNN | 0.939 | 0.758 | 0.803 | 1.384 | 2.15 | ||
| SG-BPNN | 0.927 | 0.483 | 0.911 | 0.527 | 3.25 | SG-BPNN | 0.917 | 0.888 | 0.920 | 0.880 | 3.24 | ||
| MSC-BPNN | 0.881 | 0.607 | 0.913 | 0.529 | 3.01 | MSC-BPNN | 0.951 | 0.699 | 0.927 | 0.856 | 3.41 | ||
| SNV-BPNN | 0.926 | 0.480 | 0.907 | 0.556 | 2.79 | SNV-BPNN | 0.943 | 0.794 | 0.940 | 0.759 | 4.01 | ||
| ORI-PLSR | 0.927 | 0.455 | 0.830 | 0.783 | 2.25 | ORI-PLSR | 0.934 | 0.770 | 0.929 | 0.866 | 3.53 | ||
| D1-PLSR | 0.901 | 0.530 | 0.770 | 0.914 | 1.92 | D1-PLSR | 0.967 | 0.547 | 0.864 | 1.206 | 2.66 | ||
| D2-PLSR | 0.897 | 0.538 | 0.711 | 0.911 | 1.75 | D2-PLSR | 0.940 | 0.733 | 0.859 | 1.216 | 2.44 | ||
| SG-PLSR | 0.927 | 0.582 | 0.911 | 0.718 | 2.39 | SG-PLSR | 0.945 | 0.705 | 0.919 | 0.927 | 3.32 | ||
| MSC-PLSR | 0.907 | 0.513 | 0.817 | 0.811 | 2.14 | MSC-PLSR | 0.931 | 0.787 | 0.909 | 0.985 | 3.02 | ||
| SNV-PLSR | 0.903 | 0.525 | 0.823 | 0.798 | 2.17 | SNV-PLSR | 0.949 | 0.675 | 0.907 | 0.986 | 3.11 | ||
| ORI-RF | 0.950 | 0.550 | 0.878 | 0.956 | 1.50 | ORI-RF | 0.971 | 0.738 | 0.912 | 1.369 | 1.99 | ||
| D1-RF | 0.968 | 0.441 | 0.894 | 0.890 | 1.63 | D1-RF | 0.977 | 0.948 | 0.906 | 1.374 | 2.03 | ||
| D2-RF | 0.960 | 0.494 | 0.904 | 0.842 | 1.78 | D2-RF | 0.977 | 0.687 | 0.927 | 1.275 | 2.05 | ||
| SG-RF | 0.949 | 0.542 | 0.876 | 0.947 | 1.56 | SG-RF | 0.963 | 0.828 | 0.907 | 1.418 | 1.89 | ||
| MSC-RF | 0.953 | 0.519 | 0.888 | 0.879 | 1.83 | MSC-RF | 0.971 | 0.717 | 0.917 | 1.287 | 2.30 | ||
| SNV-RF | 0.958 | 0.495 | 0.872 | 0.930 | 1.73 | SNV-RF | 0.970 | 0.748 | 0.913 | 1.343 | 2.06 | ||
| Model | Methods | LV | Number | Calibration Set (%) | Prediction Set (%) |
|---|---|---|---|---|---|
| D1-PLS-DA | SPA | 5 | 51 | 100 | 100 |
| VISSA | 13 | 150 | 100 | 100 |
| Chemical | Model | Method | Wavelengths Number | Calibration Set | Prediction Set | |||
|---|---|---|---|---|---|---|---|---|
| R2 | RMSEC | R2 | RMSEP | RPD | ||||
| Tan I | MSC-PLSR | SPA | 21 | 0.931 | 0.107 | 0.937 | 0.109 | 3.94 |
| VISSA | 131 | 0.955 | 0.087 | 0.940 | 0.113 | 4.08 | ||
| Tan IIA | ORI-BPNN | SPA | 33 | 0.906 | 0.369 | 0.905 | 0.357 | 3.17 |
| VISSA | 104 | 0.952 | 0.263 | 0.902 | 0.362 | 3.12 | ||
| CTS | SG-BPNN | SPA | 36 | 0.962 | 0.345 | 0.910 | 0.528 | 3.24 |
| VISSA | 100 | 0.931 | 0.476 | 0.902 | 0.560 | 3.62 | ||
| Total | SNV-BPNN | SPA | 33 | 0.941 | 0.759 | 0.933 | 0.830 | 3.44 |
| VISSA | 111 | 0.959 | 0.626 | 0.931 | 0.830 | 3.58 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Y.; Yan, B.; Xiong, F.; Bai, R.; Wang, S.; Guo, L.; Yang, J. Tanshinone Content Prediction and Geographical Origin Classification of Salvia miltiorrhiza by Combining Hyperspectral Imaging with Chemometrics. Foods 2024, 13, 3673. https://doi.org/10.3390/foods13223673
Dai Y, Yan B, Xiong F, Bai R, Wang S, Guo L, Yang J. Tanshinone Content Prediction and Geographical Origin Classification of Salvia miltiorrhiza by Combining Hyperspectral Imaging with Chemometrics. Foods. 2024; 13(22):3673. https://doi.org/10.3390/foods13223673
Chicago/Turabian StyleDai, Yaoyao, Binbin Yan, Feng Xiong, Ruibin Bai, Siman Wang, Lanping Guo, and Jian Yang. 2024. "Tanshinone Content Prediction and Geographical Origin Classification of Salvia miltiorrhiza by Combining Hyperspectral Imaging with Chemometrics" Foods 13, no. 22: 3673. https://doi.org/10.3390/foods13223673
APA StyleDai, Y., Yan, B., Xiong, F., Bai, R., Wang, S., Guo, L., & Yang, J. (2024). Tanshinone Content Prediction and Geographical Origin Classification of Salvia miltiorrhiza by Combining Hyperspectral Imaging with Chemometrics. Foods, 13(22), 3673. https://doi.org/10.3390/foods13223673





