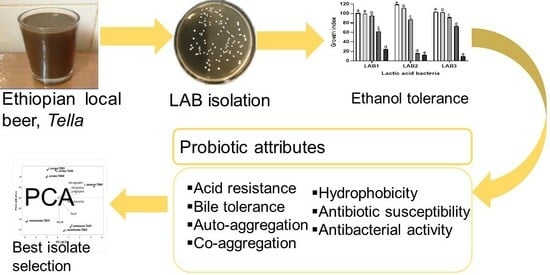
Characterization of Autochthonous Lactic Acid Bacteria Isolated from a Traditional Ethiopian Beverage, Tella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tella Samples
2.2. LAB Isolation
2.3. Phenotypic Identification of Purified Isolates
2.4. Genotypic Identification of Presumptive LAB Isolates
2.5. Alcohol Tolerance of LAB Isolates
2.6. Probiotic Characterization of LAB Isolates
2.6.1. Preparation of LAB and Bacterial Culture
2.6.2. Acid Resistance
2.6.3. Bile Salt Tolerance
2.6.4. Cell Auto-Aggregation
2.6.5. Co-Aggregation
2.6.6. Cell Surface Hydrophobicity
2.7. Antibiotic Susceptibility
2.8. Antibacterial Activity
2.9. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Phenotypic Characterization of Presumptive LAB Isolates
3.2. Genotypic Identification
3.3. Alcohol Tolerance Ability
3.4. Probiotic Properties
3.4.1. Resistance to Low pH and Bile Salts
3.4.2. Auto-Aggregation and Co-Aggregation Properties
3.4.3. Hydrophobicity
3.5. Antibiotic Susceptibility
3.6. Antibacterial Activity
3.7. Functional Starter Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fentie, E.G.; Emire, S.A.; Demsash, H.D.; Dadi, D.W.; Shin, J.-H. Cereal-and fruit-based Ethiopian traditional fermented alcoholic beverages. Foods 2020, 9, 1781. [Google Scholar] [CrossRef] [PubMed]
- Yohannes, T.; Melak, F.; Siraj, K. Preparation and physicochemical analysis of some Ethiopian traditional alcoholic beverages. Afr. J. Food Sci. 2013, 7, 399–403. [Google Scholar] [CrossRef]
- Samuel, S.; Berhanu, A. The microbiology of Tella fermentation. Sinet 1991, 14, 81–92. [Google Scholar]
- Shewakena, S.; Chandravanshi, B.; Debebe, A. Levels of total polyphenol, flavonoid, tannin and antioxidant activity of selected Ethiopian fermented traditional beverages. Int. Food Res. J. 2017, 24, 2033–2040. [Google Scholar] [CrossRef]
- Andualem, B.; Shiferaw, M.; Berhane, N. Isolation and characterization of Saccaromyces cervisiae yeasts isolates from Tella for beer production. Annu. Res. Rev. Biol. 2017, 15, 1–12. [Google Scholar] [CrossRef]
- Ashenafi, M. A review on the microbiology of indigenous fermented foods and beverages of Ethiopia. Ethiop. J. Biol. Sci. 2006, 5, 189–245. [Google Scholar] [CrossRef]
- Tekle, B.; Anuradha Jabasingh, S.; Fantaw, D.; Gebreslassie, T.; Ram Mohan Rao, S.; Baraki, H.; Gebregziabher, K. An insight into the Ethiopian traditional alcoholic beverage: Tella processing, fermentation kinetics, microbial profiling and nutrient analysis. LWT Food Sci. Technol. 2019, 107, 9–15. [Google Scholar] [CrossRef]
- Berhanu, A. Microbial profile of Tella and the role of “gesho” (Rhamnus prinoides) as bittering and antimicrobial agent in traditional Tella (beer) production. Int. Food Res. 2014, 21, 357–365. [Google Scholar]
- Gupta, S.; Mohanty, U.; Majumdar, R.K. Isolation and characterization of lactic acid bacteria from traditional fermented fish product Shidal of India with reference to their probiotic potential. LWT Food Sci. Technol. 2021, 146, 111641. [Google Scholar] [CrossRef]
- Holzapfel, W.H. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef]
- Hotessa, N.; Robe, J. Ethiopian indigenous traditional fermented beverage: The role of the microorganisms toward nutritional and safety value of fermented beverage. Int. J. Microbiol. 2020, 2020, 8891259. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Enujiugha, V.N.; Badejo, A.A. Probiotic potentials of cereal-based beverages. Crit. Rev. Food Sci. Nutr. 2017, 57, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa-Avila, C.R.; García-Gamboa, R.; Chedraui-Urrea, J.J.; García-Cayuela, T. Exploring the potential of probiotic-enriched beer: Microorganisms, fermentation strategies, sensory attributes, and health implications. Food Res. Int. 2023, 175, 113717. [Google Scholar] [CrossRef] [PubMed]
- Min, M.; Bunt, C.R.; Mason, S.L.; Hussain, M.A. Non-dairy probiotic food products: An emerging group of functional foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 2626–2641. [Google Scholar] [CrossRef] [PubMed]
- Alemu, F.; Amhaselassie, T.; Kelbessa, U.; Elias, S. Methanol, fusel oil, and ethanol contents of some Ethiopian traditional alcoholic beverages. Sinet 1991, 14, 19–27. [Google Scholar]
- Lacerda, I.C.; Miranda, R.L.; Borelli, B.M.; Nunes, Á.C.; Nardi, R.M.; Lachance, M.-A.; Rosa, C.A. Lactic acid bacteria and yeasts associated with spontaneous fermentations during the production of sour cassava starch in Brazil. Int. J. Food Microbiol. 2005, 105, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.C.; Hussey, M.A. Gram Stain Protocols, 3rd ed.; American Society for Microbiology: Washington, DC, USA, 2005; pp. 113–144. [Google Scholar]
- Reiner, K. Catalase Test Protocol; American Society for Microbiology: Washington, DC, USA, 2010; pp. 1–7. [Google Scholar]
- Speranza, B.; Racioppo, A.; Beneduce, L.; Bevilacqua, A.; Sinigaglia, M.; Corbo, M.R. Autochthonous lactic acid bacteria with probiotic aptitudes as starter cultures for fish-based products. Food Microbiol. 2017, 65, 244–253. [Google Scholar] [CrossRef]
- Singh, H.; Du, J.; Singh, P.; Yi, T.H. Extracellular synthesis of silver nanoparticles by Pseudomonas sp. THG-LS1. 4 and their antimicrobial application. J. Pharm. Anal. 2018, 8, 258–264. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, A.; Altieri, C.; Corbo, M.R.; Sinigaglia, M.; Ouoba, L.I.I. Characterization of lactic acid bacteria isolated from Italian Bella di Cerignola table olives: Selection of potential multifunctional starter cultures. J. Food Sci. 2010, 75, M536–M544. [Google Scholar] [CrossRef] [PubMed]
- Vijayalakshmi, S.; Adeyemi, D.E.; Choi, I.Y.; Sultan, G.; Madar, I.H.; Park, M.-K. Comprehensive in silico analysis of lactic acid bacteria for the selection of desirable probiotics. LWT Food Sci. Technol. 2020, 130, 109617. [Google Scholar] [CrossRef]
- Mallappa, R.H.; Singh, D.K.; Rokana, N.; Pradhan, D.; Batish, V.K.; Grover, S. Screening and selection of probiotic Lactobacillus strains of Indian gut origin based on assessment of desired probiotic attributes combined with principal component and heatmap analysis. LWT Food Sci. Technol. 2019, 105, 272–281. [Google Scholar] [CrossRef]
- Muñoz-Provencio, D.; Llopis, M.; Antolín, M.; De Torres, I.; Guarner, F.; Pérez-Martínez, G.; Monedero, V. Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch. Microbiol. 2009, 191, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Haghshenas, B.; Nami, Y.; Abdullah, N.; Radiah, D.; Rosli, R.; Khosroushahi, A.Y. Anti-proliferative effects of Enterococcus strains isolated from fermented dairy products on different cancer cell lines. J. Funct. Foods 2014, 11, 363–374. [Google Scholar] [CrossRef]
- Won, S.-M.; Chen, S.; Park, K.W.; Yoon, J.-H. Isolation of lactic acid bacteria from kimchi and screening of Lactobacillus sakei ADM14 with anti-adipogenic effect and potential probiotic properties. LWT Food Sci. Technol. 2020, 126, 109296. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2020; pp. 1–16. [Google Scholar]
- Sakoui, S.; Derdak, R.; Addoum, B.; Pop, O.L.; Vodnar, D.C.; Suharoschi, R.; Soukri, A.; El Khalfi, B. The first study of probiotic properties and biological activities of lactic acid bacteria isolated from Bat guano from Er-rachidia, Morocco. LWT Food Sci. Technol. 2022, 159, 113224. [Google Scholar] [CrossRef]
- Amelia, R.; Philip, K.; Pratama, Y.E.; Purwati, E. Characterization and probiotic potential of lactic acid bacteria isolated from dadiah sampled in West Sumatra. Food Sci. Technol. 2020, 41, 746–752. [Google Scholar] [CrossRef]
- Chen, X.; Wang, T.; Jin, M.; Tan, Y.; Liu, L.; Liu, L.; Li, C.; Yang, Y.; Du, P. Metabolomics analysis of growth inhibition of Lactobacillus plantarum under ethanol stress. Int. J. Food Sci. Technol. 2020, 55, 3441–3454. [Google Scholar] [CrossRef]
- Wang, J.; Lu, C.; Xu, Q.; Li, Z.; Song, Y.; Zhou, S.; Zhang, T.; Luo, X. Bacterial diversity and lactic acid bacteria with high alcohol tolerance in the fermented grains of soy sauce aroma type baijiu in North China. Foods 2022, 11, 1794. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Jiranek, V.; Hayes, A.M.; Grbin, P.R. Isolation and characterization of high-ethanol-tolerance lactic acid bacteria from Australian Wine. Foods 2022, 11, 1231. [Google Scholar] [CrossRef] [PubMed]
- Evaluation of Health and Nutritional Properties of Powder Milk and Live Lactic Acid Bacteria; FAO/WHO: Cordoba, Argentina, 2001; pp. 1–34.
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-Q.; Meng, X.-C.; Zhang, B.-R.; Wang, Y.; Shang, Y.-L. Influence of cell surface properties on adhesion ability of bifidobacteria. World J. Microbiol. Biotechnol. 2010, 26, 1999–2007. [Google Scholar] [CrossRef]
- Chandran, A.; Duary, R.K.; Grover, S.; Batish, V.K. Relative expression of bacterial and host specific genes associated with probiotic survival and viability in the mice gut fed with Lactobacillus plantarum Lp91. Microbiol. Res. 2013, 168, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, J.K.; Kumar, A.; Duary, R.K.; Mohanty, A.K.; Grover, S.; Batish, V.K. Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS ONE 2009, 4, e8099. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, N.C.; de Ruiz, C.S.; Otero, M.C.; Sesma, F.; Nader-Macías, M.E. Lactic acid bacteria isolated from young calves—Characterization and potential as probiotics. Res. Vet. Sci. 2012, 92, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.-X.; Yang, Z.-Q.; Li, Z.-H.; Chen, S.-L.; Luo, Z.-L. Probiotic properties of lactic acid bacteria isolated from stool samples of longevous people in regions of Hotan, Xinjiang and Bama, Guangxi, China. Anaerobe 2008, 14, 313–317. [Google Scholar] [CrossRef]
- Cesena, C.; Morelli, L.; Alander, M.; Siljander, T.; Tuomola, E.; Salminen, S.; Mattila-Sandholm, T.; Vilpponen-Salmela, T.; von Wright, A. Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. J. Dairy Sci. 2001, 84, 1001–1010. [Google Scholar] [CrossRef]
- Rokana, N.; Mallappa, R.H.; Batish, V.K.; Grover, S. Interaction between putative probiotic Lactobacillus strains of Indian gut origin and Salmonella: Impact on intestinal barrier function. LWT Food Sci. Technol. 2017, 84, 851–860. [Google Scholar] [CrossRef]
- Campana, R.; van Hemert, S.; Baffone, W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Vidhyasagar, V.; Jeevaratnam, K. Evaluation of Pediococcus pentosaceus strains isolated from Idly batter for probiotic properties in vitro. J. Funct. Food. 2013, 5, 235–243. [Google Scholar] [CrossRef]
- Ferreira, C.L.; Grześkowiak, L.; Collado, M.C.; Salminen, S. In vitro evaluation of Lactobacillus gasseri strains of infant origin on adhesion and aggregation of specific pathogens. J. Food Prot. 2011, 74, 1482–1487. [Google Scholar] [CrossRef] [PubMed]
- Vlková, E.; Rada, V.; Šmehilová, M.; Killer, J. Auto-aggregation and co-aggregation ability in Bifidobacteria and Clostridia. Folia Microbiol. 2008, 53, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Alcántara, A.M.; Wacher, C.; Llamas, M.G.; López, P.; Pérez-Chabela, M.L. Probiotic properties and stress response of thermotolerant lactic acid bacteria isolated from cooked meat products. LWT Food Sci. Technol. 2018, 91, 249–257. [Google Scholar] [CrossRef]
- Xu, H.; Jeong, H.; Lee, H.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Oh, A.; Daliri, E.B.-M.; Oh, D.H. Screening for potential probiotic bacteria from Korean fermented soybean paste: In vitro and Caenorhabditis elegans model testing. LWT Food Sci. Technol. 2018, 88, 132–138. [Google Scholar] [CrossRef]
- Schillinger, U.; Guigas, C.; Heinrich Holzapfel, W. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 2005, 15, 1289–1297. [Google Scholar] [CrossRef]
- Bhargava, K.; Zhang, Y. Multidrug-resistant coagulase-negative Staphylococci in food animals. J. Appl. Microbiol. 2012, 113, 1027–1036. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpińska, M.; Łaniewska-Trokenheim, Ł. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin–phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015, 46, 222–226. [Google Scholar] [CrossRef]
- Han, J.; Chen, D.; Li, S.; Li, X.; Zhou, W.-W.; Zhang, B.; Jia, Y. Antibiotic susceptibility of potentially probiotic Lactobacillus strains. Ital. J. Food Sci. 2015, 27, 282–289. [Google Scholar]
- Botta, C.; Langerholc, T.; Cencič, A.; Cocolin, L. In vitro selection and characterization of new probiotic candidates from table olive microbiota. PLoS ONE 2014, 9, e94457. [Google Scholar] [CrossRef] [PubMed]
- Elkins, C.A.; Mullis, L.B. Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 2004, 70, 7200–7209. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Pillidge, C.; Gopal, P.; Gill, H. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005, 98, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, M.; Simpson, P.; O’connor, E.; Ross, R.; Stanton, C. Susceptibility of Pediococcus spp. to antimicrobial agents. J. Appl. Microbiol. 2007, 102, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Borriello, S.; Hammes, W.; Holzapfel, W.; Marteau, P.; Schrezenmeir, J.; Vaara, M.; Valtonen, V. Safety of probiotics that contain Lactobacilli or Bifidobacteria. Clin. Infect. Dis. 2003, 36, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Wang, J.; Wu, C.; Shen, Z.; Fu, X.; Yan, Y.; Zhang, Q.; Schwarz, S.; Shen, J. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 2012, 56, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Cruxen, C.E.; Funck, G.D.; Haubert, L.; da Silva Dannenberg, G.; de Lima Marques, J.; Chaves, F.C.; da Silva, W.P.; Fiorentini, Â.M. Selection of native bacterial starter culture in the production of fermented meat sausages: Application potential, safety aspects, and emerging technologies. Food Res. Int. 2019, 122, 371–382. [Google Scholar] [CrossRef]
- Fei, Y.; Li, L.; Zheng, Y.; Liu, D.; Zhou, Q.; Fu, L. Characterization of Lactobacillus amylolyticus L6 as potential probiotics based on genome sequence and corresponding phenotypes. LWT Food Sci. Technol. 2018, 90, 460–468. [Google Scholar] [CrossRef]
- Holzapfel, W. Use of starter cultures in fermentation on a household scale. Food Control 1997, 8, 241–258. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef]
- Shah, N.P. Functional cultures, and health benefits. Int. Dairy J. 2007, 17, 1262–1277. [Google Scholar] [CrossRef]




| Collection Cites | No. of LAB -Positive Samples | No. of Single Isolates | Isolate Code 1 | Physiological Test | No. of Presumptive LAB Isolates 2 | |
|---|---|---|---|---|---|---|
| No. of Gram-Positive Isolates | No. of Catalase -Negative Isolates | |||||
| Addis Ababa (n = 5) | 5 | 16 | TAA01–TAA16 | 13 | 6 | 3 |
| Debre Birhan (n = 5) | 3 | 14 | TDB17–TDB30 | 9 | 6 | 6 |
| Debre Markos (n = 5) | 5 | 13 | TDM31–TDM43 | 11 | 7 | 6 |
| Isolate Code | Cell Morphology | Acidification (ΔpH > 1) 1 | Genotype Identification | |||
|---|---|---|---|---|---|---|
| The Closest Type Strain | Query Length (bp) | Identity (%) | Accession Number | |||
| TAA01 | Coccus | + | Pediococcus pentosaceus DSM 20336 T | 1441 | 100.0 | KX886792.1 |
| TAA04 | Bacillus | + | Lactiplantibacillus curvatus JCM 1096 T | 1440 | 100.0 | LC063167.1 |
| TAA14 | Bacillus | − | ND | ND | ND | ND |
| TDB18 | Streptococcus | − | ND | ND | ND | ND |
| TDB19 | Coccus | + | Leuconostoc mesenteroides JCM 9700 T | 1418 | 100.0 | LC063167.1 |
| TDB21 | Bacillus | + | Lactiplantibacillus curvatus JCM 1096 T | 1435 | 99.9 | LC063167.1 |
| TDB22 | Coccus | + | Leuconostoc mesenteroides JCM 9700 T | 1417 | 100.0 | LC096223.1 |
| TDB23 | Coccus | − | ND | ND | ND | ND |
| TDB24 | Bacillus | − | ND | ND | ND | ND |
| TDM32 | Bacillus | − | ND | ND | ND | ND |
| TDM34 | Streptobacillus | − | ND | ND | ND | ND |
| TDM35 | Bacillus | − | ND | ND | ND | ND |
| TDM38 | Bacillus | − | ND | ND | ND | ND |
| TDM40 | Bacillus | + | Lactiplantibacillus curvatus JCM 1096 T | 1440 | 100.0 | LC063167.1 |
| TDM41 | Bacillus | + | Lactiplantibacillus plantarum JCM 1149 T | 1438 | 100.0 | LC064896.1 |
| LAB Isolates | Auto- Aggregation (%) | Co-Aggregation (%) | Hydrophobicity (%) | |||
|---|---|---|---|---|---|---|
| E. coli ATCC 43895 | S. Enteritidis ATCC 13076 | S. aureus ATCC 25923 | Xylene | Chloroform | ||
| P. pentosaceus TAA01 | 31.7 ± 0.1 b | 23.3 ± 0.1 c | 29.4 ± 0.8 a | 32.0 ± 0.1 b | 32.2 ± 0.6 c | 35.3 ± 0.3 c |
| L. curvatus TAA04 | 33.4 ± 1.1 b | 26.5 ± 0.4 b | 26.6 ± 0.1 b | 27.0 ± 0.4 c | 31.7 ± 0.6 c | 39.8 ± 0.6 b |
| L. mesenteroides TDB19 | 24.5 ± 1.9 c | 19.5 ± 0.3 f | 20.7 ± 0.2 d | 21.6 ± 0.1 e | 17.0 ± 0.4 e | 18.0 ± 0.1 g |
| L. curvatus TDB21 | 33.8 ± 1.4 b | 23.8 ± 0.2 c | 21.2 ± 0.1 d | 24.2 ± 0.4 d | 28.1 ± 0.9 d | 25.9 ± 0.6 f |
| L. mesenteroides TDB22 | 32.3 ± 0.5 b | 20.5 ± 0.5 e | 29.9 ± 0.8 a | 20.2 ± 0.1 f | 34.5 ± 0.2 b | 28.1 ± 0.1 e |
| L. curvatus TDM40 | 34.5 ± 1.8 b | 21.9 ± 0.4 d | 23.2 ± 0.4 c | 24.0 ± 0.4 d | 29.2 ± 1.2 d | 31.6 ± 0.6 d |
| L. plantarum TDM41 | 44.9 ± 1.7 a | 34.0 ± 0.5 a | 23.5 ± 0.5 c | 41.4 ± 0.2 a | 45.4 ± 0.1 a | 52.1 ± 0.1 a |
| LAB Isolates | Antibiotics 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | KAN | STR | ERY | TET | CHL | PEN | SXT | CIP | AZM | |
| P. pentosaceus TAA01 | R | R | R | R | S | S | S | I | R | R | S |
| L. curvatus TAA04 | R | R | R | R | S | S | S | S | R | I | S |
| L. mesenteroides TDB19 | R | R | R | R | S | S | S | I | R | I | S |
| L. curvatus TDB21 | R | R | R | R | S | S | S | S | R | R | S |
| L. mesenteroides TDB22 | R | R | R | R | S | S | S | I | R | R | S |
| L. curvatus TDM40 | R | R | R | R | S | S | S | S | R | I | S |
| L. plantarum TDM41 | R | R | R | R | S | S | S | I | S | R | S |
| LAB Isolates | Inhibition Zone (mm) | ||
|---|---|---|---|
| E. coli ATCC 43895 | S. Enteritidis ATCC 13076 | S. aureus ATCC 25923 | |
| P. pentosaceus TAA01 | 16.0 ± 0.7 | 16.3 ± 0.8 | 12.7 ± 0.5 |
| L. curvatus TAA04 | 12.0 ± 0.7 | 13.0 ± 1.0 | ND |
| L. mesenteroides TDB19 | ND | ND | 9.7 ± 0.5 |
| L. curvatus TDB21 | 11.0 ± 1.4 | 9.7 ± 0.6 | 13.5 ± 0.7 |
| L. mesenteroides TDB22 | 14.0 ± 0.7 | 17.0 ± 1.0 | 14.0 ± 1.0 |
| L. curvatus TDM40 | 11.3 ± 1.2 | 9.0 ± 1.0 | ND |
| L. plantarum TDM41 | 14.7 ± 1.1 | 16.0 ± 1.0 | 14.0 ± 1.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yehuala, G.A.; Shibeshi, N.T.; Kim, S.-H.; Park, M.-K. Characterization of Autochthonous Lactic Acid Bacteria Isolated from a Traditional Ethiopian Beverage, Tella. Foods 2024, 13, 575. https://doi.org/10.3390/foods13040575
Yehuala GA, Shibeshi NT, Kim S-H, Park M-K. Characterization of Autochthonous Lactic Acid Bacteria Isolated from a Traditional Ethiopian Beverage, Tella. Foods. 2024; 13(4):575. https://doi.org/10.3390/foods13040575
Chicago/Turabian StyleYehuala, Gashaw Assefa, Nurelegne Tefera Shibeshi, Su-Hyeon Kim, and Mi-Kyung Park. 2024. "Characterization of Autochthonous Lactic Acid Bacteria Isolated from a Traditional Ethiopian Beverage, Tella" Foods 13, no. 4: 575. https://doi.org/10.3390/foods13040575
APA StyleYehuala, G. A., Shibeshi, N. T., Kim, S.-H., & Park, M.-K. (2024). Characterization of Autochthonous Lactic Acid Bacteria Isolated from a Traditional Ethiopian Beverage, Tella. Foods, 13(4), 575. https://doi.org/10.3390/foods13040575