Therapeutic Potential of Bacteroides fragilis SNBF-1 as a Next-Generation Probiotic: In Vitro Efficacy in Lipid and Carbohydrate Metabolism and Antioxidant Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism Isolation
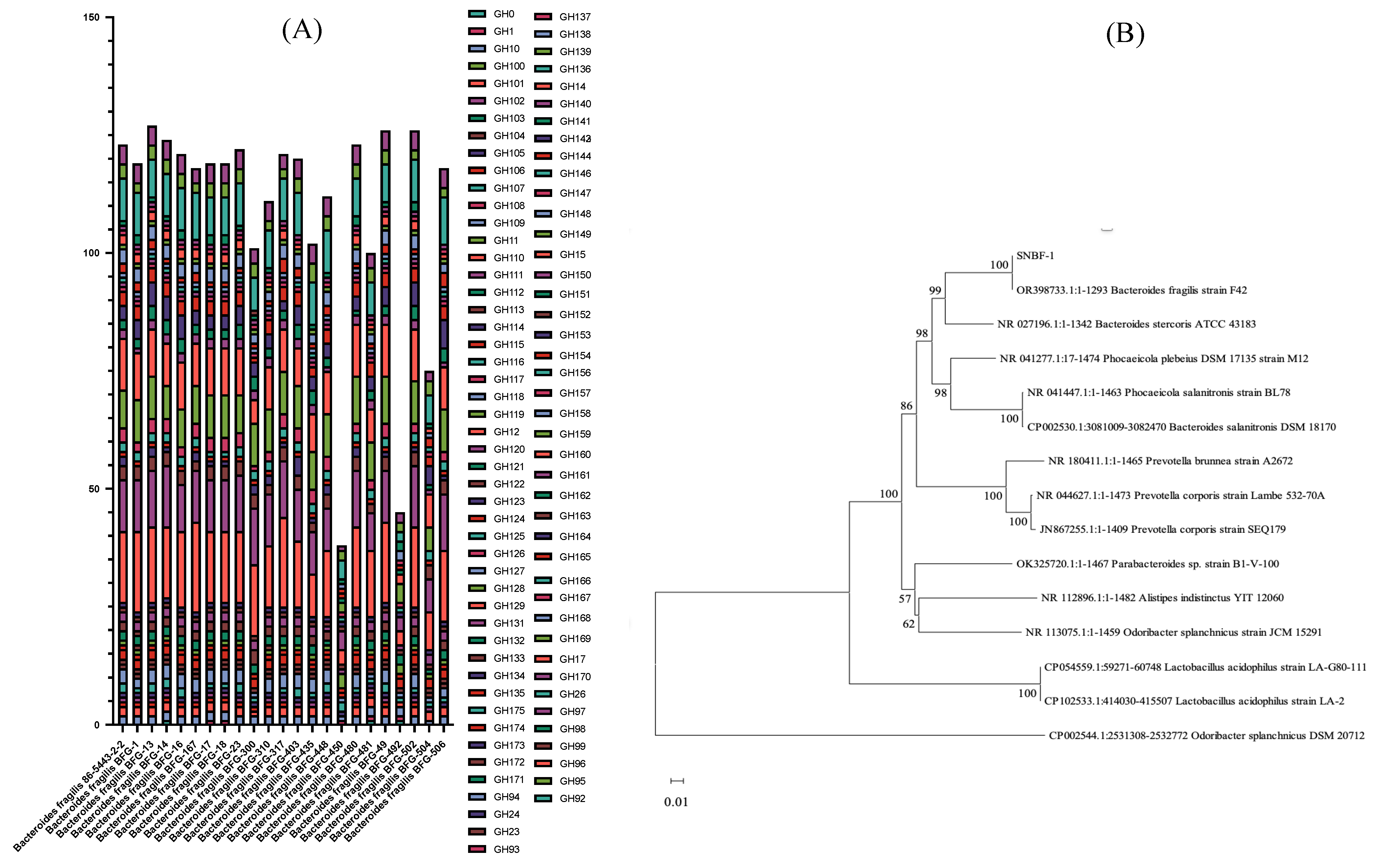
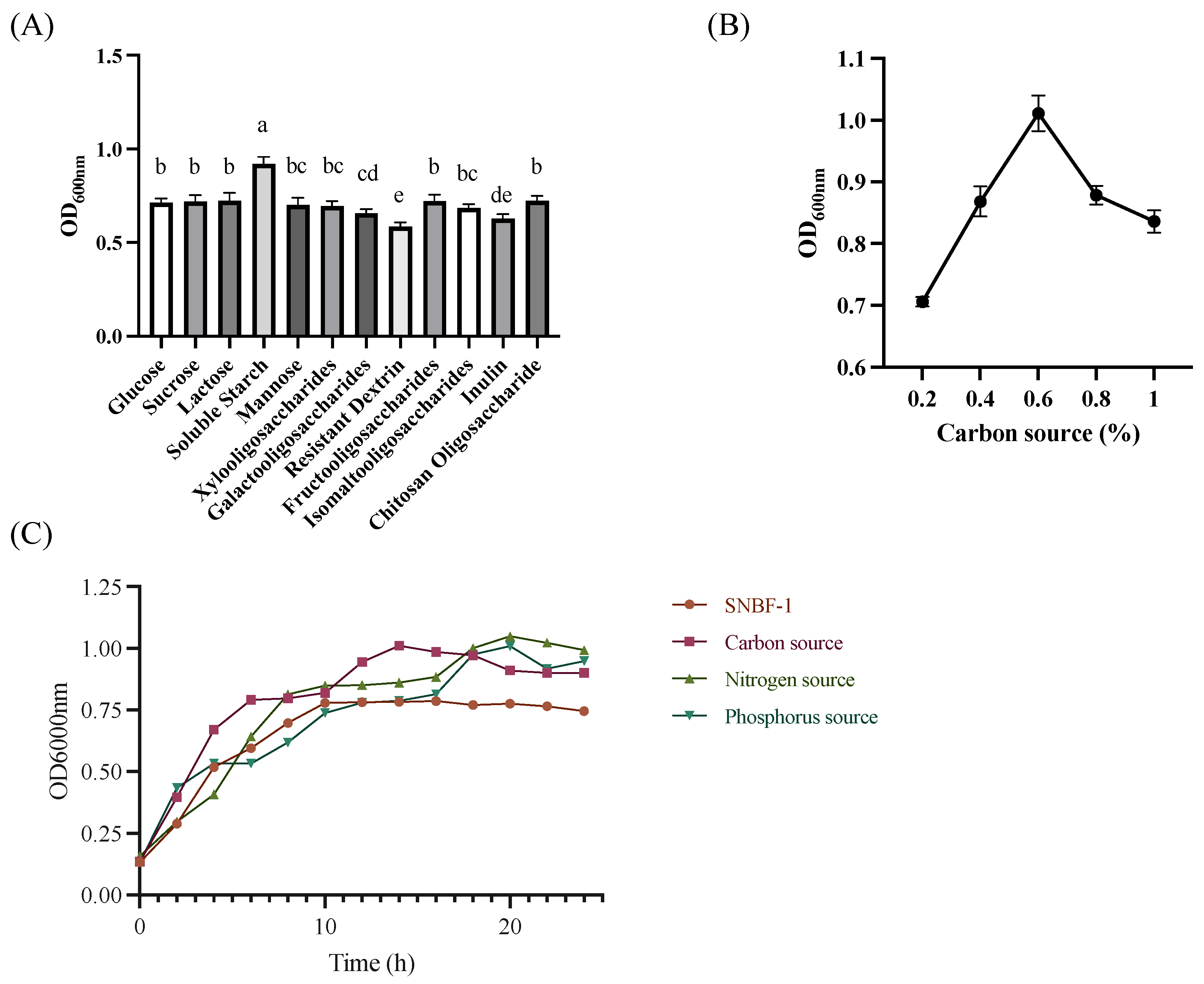
2.1.1. Genetic Analysis of Carbohydrate Metabolism for Enhanced Sugar Utilization and Antibiotic Resistance in Optimized Culture Medium
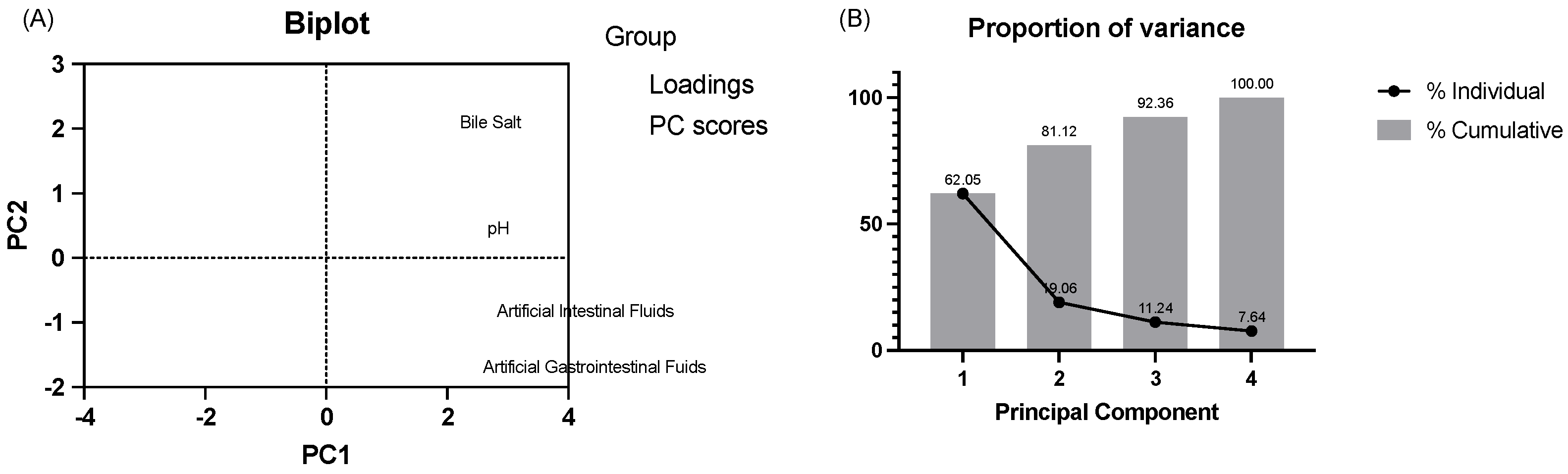
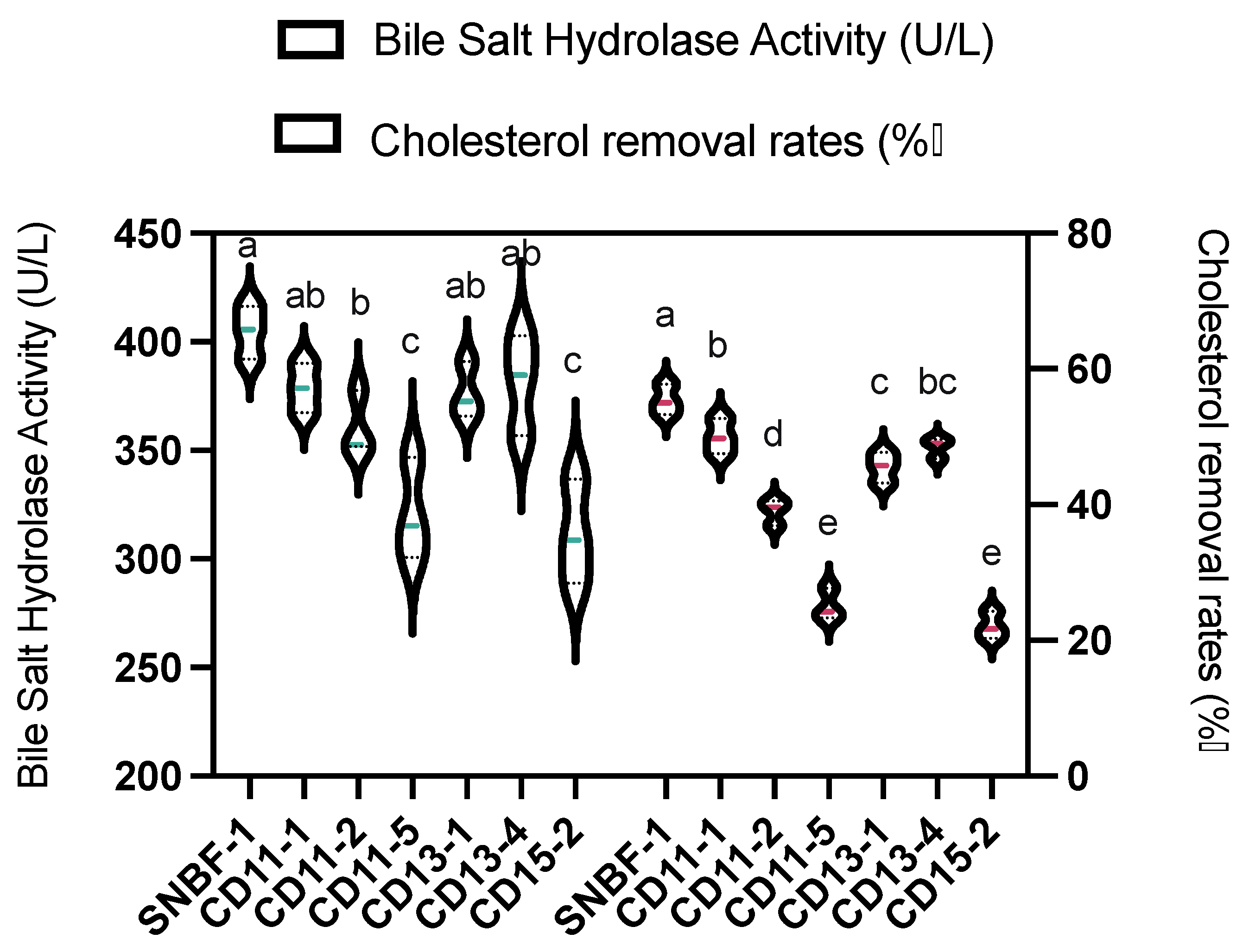
2.1.2. Bile Salt Hydrolase Activity and Survival in Simulated Gastrointestinal Conditions
2.1.3. Cell Surface Hydrophobicity
2.1.4. Auto-Aggregation Assay
2.1.5. Enterotoxin PCR and Hemolysis Tests
2.2. Characterization of B. fragilis
2.2.1. Growth Characteristics
2.2.2. Preparation of Samples
2.2.3. Cholesterol Removal Rate
2.2.4. Antibiotic Resistance Evaluation
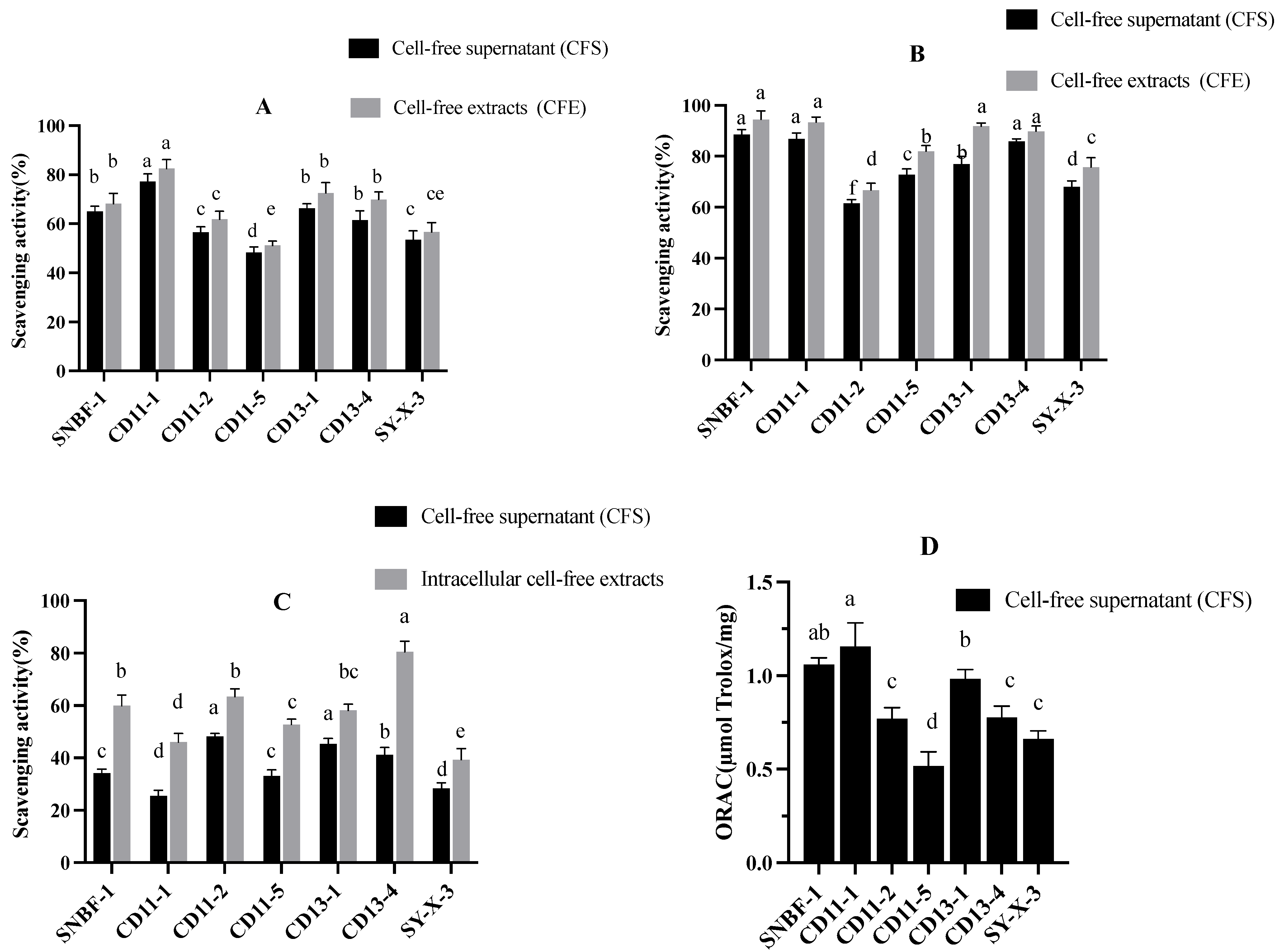
2.2.5. Antioxidant Assay:
DPPH Radical (DPPH•) Scavenging Effect
Hydroxyl Radical (•OH) Scavenging Activity
- Scavenging activity (%) %
- As: OD value of the sample with CPS.
- A0: OD value of the blank (distilled water instead of CPS).
- Ac: OD value of the control (no H2O2).
ABTS Radical (ABTS•+) Scavenging Activity
Oxygen Radical (O2 •) Absorbance Capacity Assay
2.3. In Vitro Investigation of Lipid Accumulation Reduction by B. fragilis Strain SNBF-1
2.3.1. Culture and Treatment of HepG2 Cells
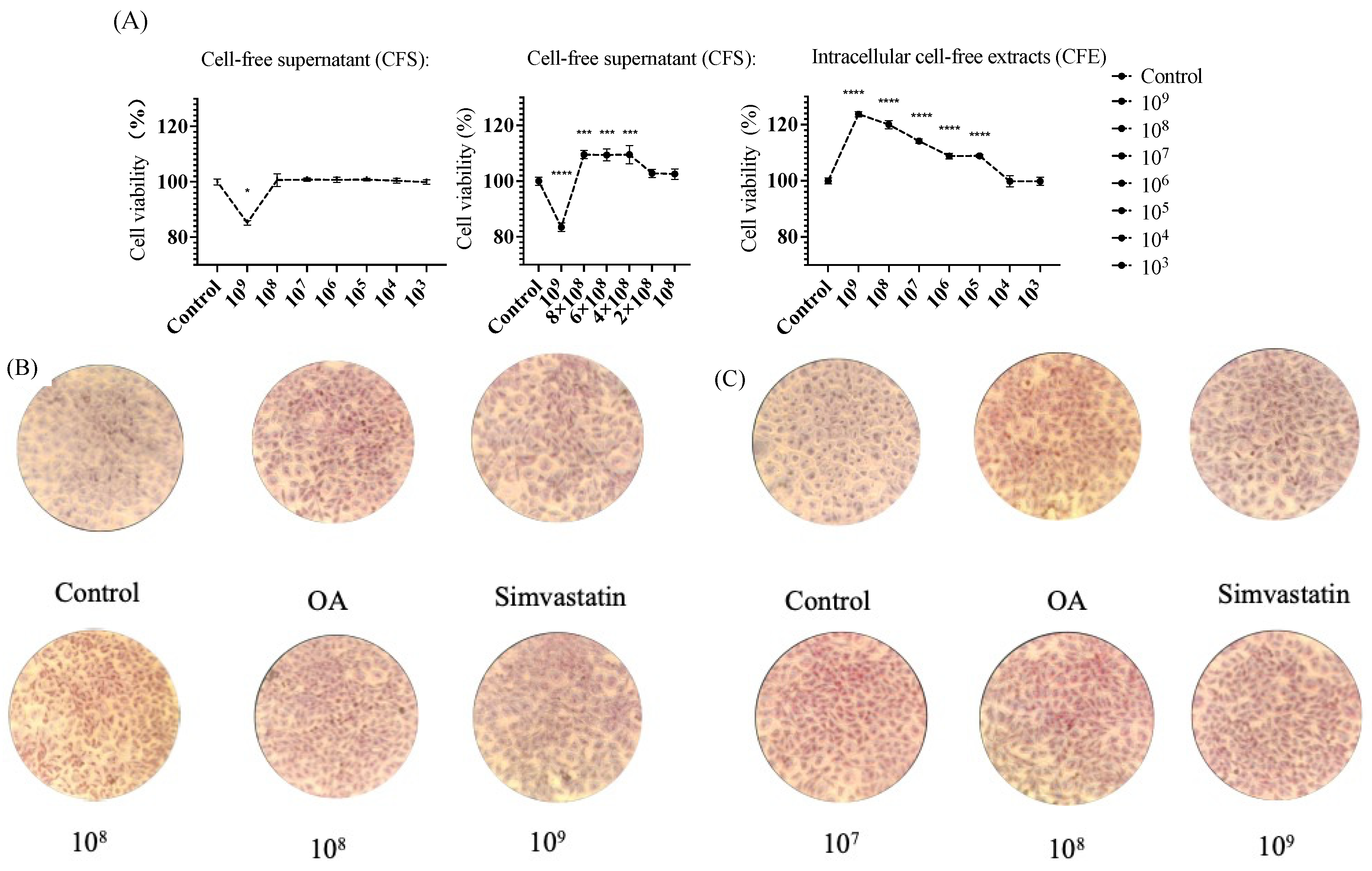
2.3.2. MTT Viability Assay
2.3.3. Oil Red O Stains Intracellular Lipids
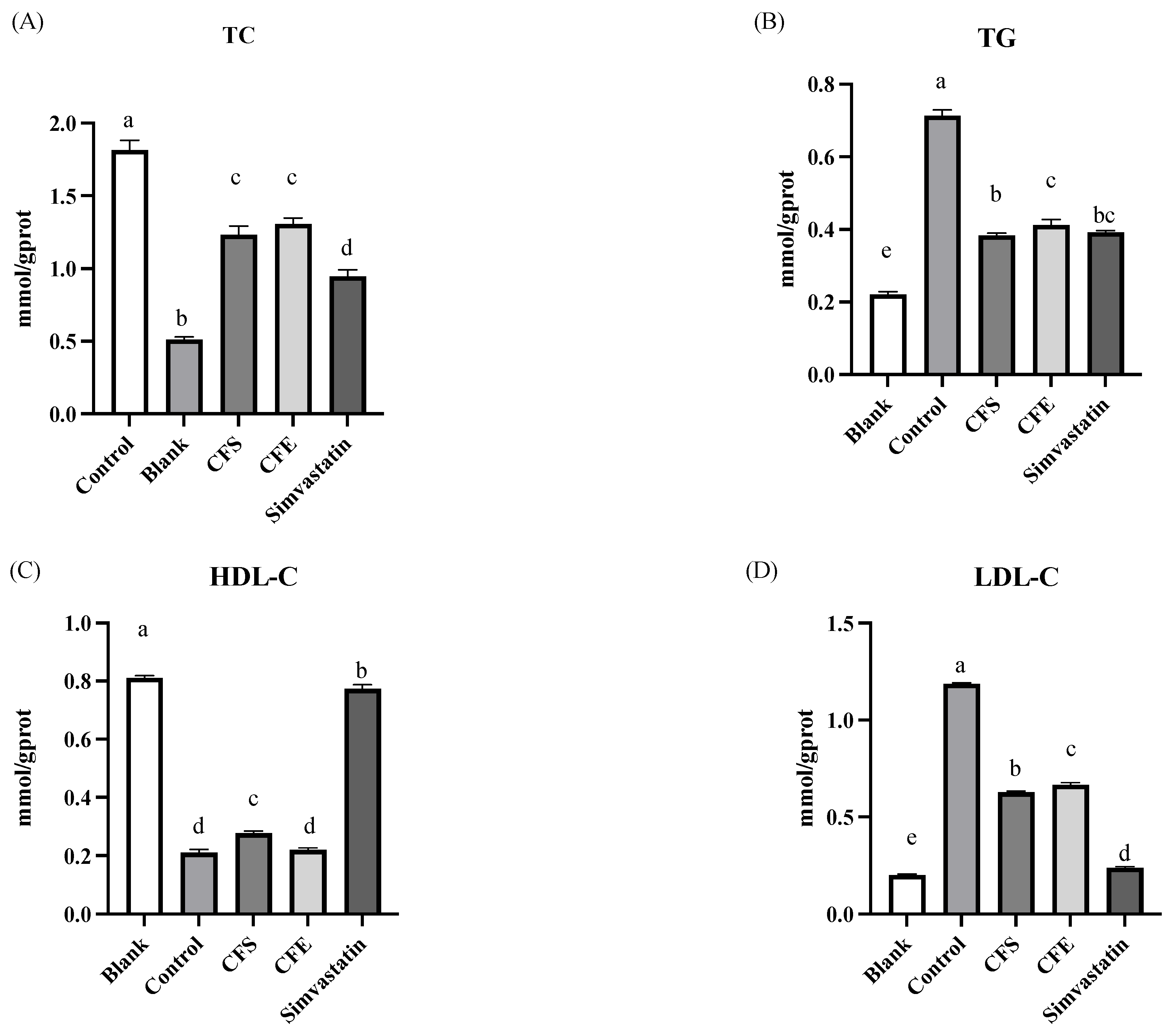
2.3.4. Lipid Accumulation, TC, TG, HDL-C, and LDL-C Assays
2.4. In Vitro Investigation of Glucose Metabolism by B. fragilis Strain SNBF-1
2.4.1. Inhibition of α-Amylase and α-Glucosidase Activity
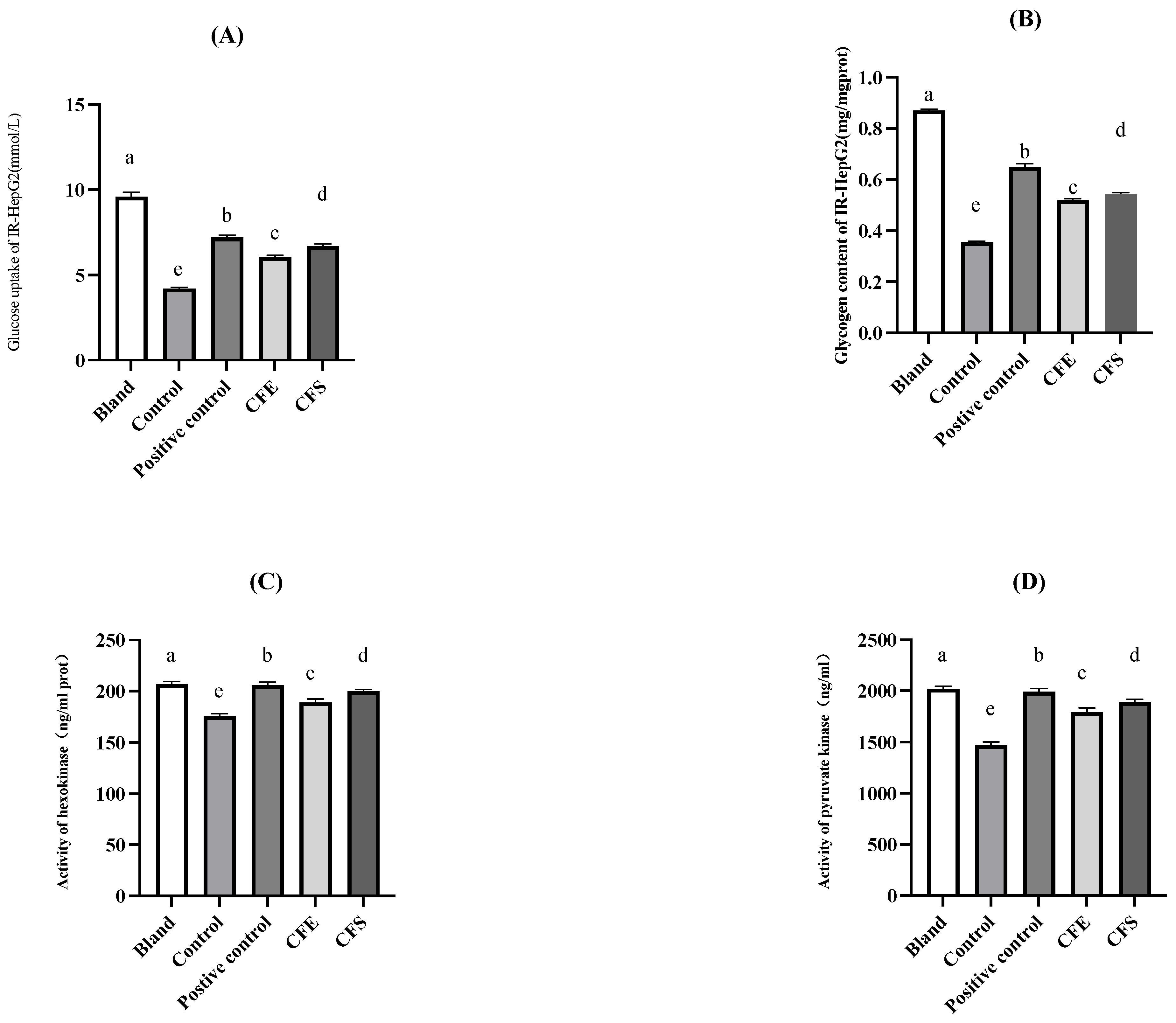
2.4.2. The IR-HepG2 Cell Model for Antidiabetic Test
2.5. Statistical Analysis
3. Results
3.1. Identification and Characterization of B. fragilis Isolated from Fecal Samples
3.1.1. Genetic Analysis of Carbohydrate Metabolism for Enhanced Sugar Utilization and Antibiotic Resistance in Optimized Culture Medium
3.1.2. Acid and Bile Tolerance Tests and Artificial Gastrointestinal and Intestinal Fluids
3.1.3. Cell Surface Hydrophobicity and Auto-Aggregation Assay
3.2. Characterization of B. fragilis
3.2.1. Cholesterol Removal Rate and Bile Salt Hydrolase Activity
3.2.2. Antibiotic Resistance Evaluation
3.2.3. Antioxidant Assay
3.2.4. MTT Viability Assay
3.2.5. Oil Red O Stains Intracellular Lipids
3.3. Lipid Accumulation, TG, and LDL-C Assays
3.4. In Vitro Investigation of Glucose Metabolism by B. fragilis Strain SNBF-1
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGF | Artificial gastric fluid |
| AIF | Artificial intestinal fluid |
| BAP | Blood agar plates |
| BBE | Bacteroides Bile Esculin |
| BHI | Brain Heart Infusion |
| CARD | Comprehensive Antibiotic Research Database |
| CAZy | Carbohydrate Active Enzyme Database |
| CFE | Cell-free extract |
| CFS | Cell-free supernatant |
| CGMCC | China General Microbiological Culture Collection Center |
| CFU | Colony-Forming Units |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DMSO | Dimethyl Sulfoxide |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| GH | Glycoside Hydrolase |
| HDL-C | High-density lipoprotein cholesterol |
| HK | Hexokinase |
| IR | Insulin-resistant |
| KEGG | Kyoto Encyclopedia of Genes and Genomes Database |
| LKV | Laked Vancomycin |
| LDL-C | Low-density lipoprotein cholesterol |
| MTT | Methylthiazolyldiphenyl-tetrazolium bromide |
| NGPs | Next-generation probiotics |
| ORAC | Oxygen radical absorbance capacity |
| PCR | Polymerase Chain Reaction |
| PK | Pyruvate kinase |
| TC | Total cholesterol |
References
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Cang, W.H.; Wu, J.R.; Ding, R.X.; Wang, W.M.; Li, N.; Shi, H.S.; Shi, L.; Lee, Y.K.; Wu, R.N. Potential of Probiotics as an Adjunct for Patients with Major Depressive Disorder. Mol. Nutr. Food Res. 2022, 66, 14. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Ghaderi, F.; Sotoodehnejadnematalahi, F.; Hajebrahimi, Z.; Fateh, A.; Siadat, S.D. Effects of active, inactive, and derivatives of Akkermansia muciniphila on the expression of the endocannabinoid system and PPARs genes. Sci. Rep. 2022, 12, 10031. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Singh, P.; Nataraj, B.H.; Kokkiligadda, A.; Naithani, H.; Azmal Ali, S.; Behare, P.V.; Nagpal, R. Fostering next-generation probiotics in human gut by targeted dietary modulation: An emerging perspective. Food Res. Int. 2021, 150, 110716. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.A.; Hall, L.J.; Walsh, S.K.; Leventhal, G.E.; Slack, E.; de Wouters, T.; Walter, J.; Clavel, T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. 2022, 15, 1095–1113. [Google Scholar] [CrossRef]
- Chang, C.J.; Lin, T.L.; Tsai, Y.L.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef]
- Ragan, M.V.; Wala, S.J.; Goodman, S.D.; Bailey, M.T.; Besner, G.E. Next-Generation Probiotic Therapy to Protect the Intestines From Injury. Front. Cell Infect. Microbiol. 2022, 12, 863949. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Xiao, Y.; Wang, Z.H.; Liu, X.; Alam, A.M.; Haran, J.P.; McCormick, B.A.; Shu, X.; Wang, X.; Ye, K. Bacteroides Fragilis in the gut microbiomes of Alzheimer’s disease activates microglia and triggers pathogenesis in neuronal C/EBPbeta transgenic mice. Nat. Commun. 2023, 14, 5471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, Q.; Liu, H.; Xu, J.; Huang, R.; Shen, B.; Guo, Y.; Ai, X.; Xu, J.; Zhao, X.; et al. Bacteroides fragilis strain ZY-312 facilitates colonic mucosa regeneration in colitis via motivating STAT3 signaling pathway induced by IL-22 from ILC3 secretion. Front. Immunol. 2023, 14, 1156762. [Google Scholar] [CrossRef]
- Chan, J.L.; Wu, S.; Geis, A.L.; Chan, G.V.; Gomes, T.A.M.; Beck, S.E.; Wu, X.; Fan, H.; Tam, A.J.; Chung, L.; et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019, 12, 164–177. [Google Scholar] [CrossRef]
- Kaźmierczak-Siedlecka, K.; Skonieczna-Żydecka, K.; Hupp, T.; Duchnowska, R.; Marek-Trzonkowska, N.; Połom, K. Next-generation probiotics—Do they open new therapeutic strategies for cancer patients? Gut Microbes 2022, 14, 2035659. [Google Scholar] [CrossRef]
- Oh, S.F.; Praveena, T.; Song, H.; Yoo, J.-S.; Jung, D.-J.; Erturk-Hasdemir, D.; Hwang, Y.S.; Lee, C.C.; Le Nours, J.; Kim, H.; et al. Host immunomodulatory lipids created by symbionts from dietary amino acids. Nature 2021, 600, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Li, W.; Tong, M.; Xu, J.; Chen, Z.; Yang, Y.; Zang, Y.; Jiao, X.; Liu, C.; Lim, B.; et al. Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome. Nat. Microbiol. 2023, 9, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Malek, A.; Ahmadi Badi, S.; Karimi, G.; Bizouarn, T.; Irian, S.; Siadat, S.D. The effect of Bacteroides fragilis and its postbiotics on the expression of genes involved in the endocannabinoid system and intestinal epithelial integrity in Caco-2 cells. J. Diabetes Metab. Disord. 2023, 22, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Livingston, S.J.; Kominos, S.D.; Yee, R.B. New medium for selection and presumptive identification of the Bacteroides fragilis group. J. Clin. Microbiol. 1978, 7, 448–453. [Google Scholar] [CrossRef]
- Kotzamanidis, C.; Kourelis, A.; Litopoulou-Tzanetaki, E.; Tzanetakis, N.; Yiangou, M. Evaluation of adhesion capacity, cell surface traits and immunomodulatory activity of presumptive probiotic Lactobacillus strains. Int. J. Food Microbiol. 2010, 140, 154–163. [Google Scholar] [CrossRef]
- Topçu, K.C.; Kaya, M.; Kaban, G. Probiotic properties of lactic acid bacteria strains isolated from pastırma. LWT 2020, 134, 110216. [Google Scholar] [CrossRef]
- Lin, M.Y.; Chang, F.J. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig. Dis. Sci. 2000, 45, 1617–1622. [Google Scholar] [CrossRef]
- Siddhuraju, P.; Manian, S. The antioxidant activity and free radical-scavenging capacity of dietary phenolic extracts from horse gram (Macrotyloma uniflorum (Lam.) Verdc.) seeds. Food Chem. 2007, 105, 950–958. [Google Scholar] [CrossRef]
- Cao, C.; Li, Y.; Wang, C.; Zhang, N.; Zhu, X.; Wu, R.; Wu, J. Purification, characterization and antitumor activity of an exopolysaccharide produced by Bacillus velezensis SN-1. Int. J. Biol. Macromol. 2020, 156, 354–361. [Google Scholar] [CrossRef]
- Ankolekar, C.; Pinto, M.; Greene, D.; Shetty, K. In vitro bioassay based screening of antihyperglycemia and antihypertensive activities of Lactobacillus acidophilus fermented pear juice. Innov. Food Sci. Emerg. Technol. 2012, 13, 221–230. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef]
- Alaaeldin, R.; Abdel-Rahman, I.A.M.; Hassan, H.A.; Youssef, N.; Allam, A.E.; Abdelwahab, S.F.; Zhao, Q.L.; Fathy, M. Carpachromene Ameliorates Insulin Resistance in HepG2 Cells via Modulating IR/IRS1/PI3k/Akt/GSK3/FoxO1 Pathway. Molecules 2021, 26, 7629. [Google Scholar] [CrossRef] [PubMed]
- Csepregi, R.; Temesfői, V.; Sali, N.; Poór, M.; Needs, P.W.; Kroon, P.A.; Kőszegi, T. A One-Step Extraction and Luminescence Assay for Quantifying Glucose and ATP Levels in Cultured HepG2 Cells. Int. J. Mol. Sci. 2018, 19, 2670. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. An optimized culture medium to isolate Lactobacillus fermentum strains from the human intestinal tract. Food Funct. 2021, 12, 6740–6754. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Yang, S.; Zhang, Y.; Qian, K.; Zhang, Z.; Liu, Y.; Wang, Y.; Bai, Y.; Fan, H.; Zhao, X.; et al. Corrigendum: Bacteroides fragilis Prevents Clostridium difficile Infection in a Mouse Model by Restoring Gut Barrier and Microbiome Regulation. Front. Microbiol. 2019, 10, 601. [Google Scholar] [CrossRef]
- Vernay, T.; Cannie, I.; Gaboriau, F.; Gall, S.D.; Tamanai-Shacoori, Z.; Burel, A.; Jolivet-Gougeon, A.; Loréal, O.; Bousarghin, L. Bacteroides fragilis prevents Salmonella Heidelberg translocation in co-culture model mimicking intestinal epithelium. Benef. Microbes 2020, 11, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Chatzidaki-Livanis, M.; Geva-Zatorsky, N.; Comstock, L.E. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. USA 2016, 113, 3627–3632. [Google Scholar] [CrossRef]
- Ramakrishna, C.; Kujawski, M.; Chu, H.; Li, L.; Mazmanian, S.K.; Cantin, E.M. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat. Commun. 2019, 10, 2153. [Google Scholar] [CrossRef]
- He, Q.; Niu, M.; Bi, J.; Du, N.; Liu, S.; Yang, K.; Li, H.; Yao, J.; Du, Y.; Duan, Y. Protective effects of a new generation of probiotic Bacteroides fragilis against colitis in vivo and in vitro. Sci. Rep. 2023, 13, 15842. [Google Scholar] [CrossRef]
- Antonyuk, S.V.; Sieminska, K.; Smiga, M.; Strange, R.W.; Wagner, M.; Barnett, K.J.; Olczak, T. Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes. FASEB J. 2023, 37, e22981. [Google Scholar] [CrossRef] [PubMed]
- Patrick, S. A tale of two habitats: Bacteroides fragilis, a lethal pathogen and resident in the human gastrointestinal microbiome. Microbiology 2022, 168, 001156. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.; Edgar, J.D.M.; Blakely, G.; Patrick, S. Antigenic mimicry of ubiquitin by the gut bacterium Bacteroides fragilis: A potential link with autoimmune disease. Clin. Exp. Immunol. 2018, 194, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Valdés-Mas, R.; Batard, E.; Zmora, N.; Dori-Bachash, M.; Suez, J.; Elinav, E. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat. Microbiol. 2021, 6, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, E.E.; Mollet, B.; deVos, W.M. Functionality of probiotics and intestinal lactobacilli: Light in the intestinal tract tunnel. Curr. Opin. Biotechnol. 1999, 10, 505–510. [Google Scholar] [CrossRef]
- Maldonado Galdeano, C.; Cazorla, S.I.; Lemme Dumit, J.M.; Vélez, E.; Perdigón, G. Beneficial Effects of Probiotic Consumption on the Immune System. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef]
- Sams, L.; Paume, J.; Giallo, J.; Carrière, F. Relevant pH and lipase for in vitro models of gastric digestion. Food Funct. 2016, 7, 30–45. [Google Scholar] [CrossRef]
- Yang, M.; Gu, Y.; Li, L.; Liu, T.; Song, X.; Sun, Y.; Cao, X.; Wang, B.; Jiang, K.; Cao, H. Bile Acid-Gut Microbiota Axis in Inflammatory Bowel Disease: From Bench to Bedside. Nutrients 2021, 13, 3143. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging issues in probiotic safety: 2023 perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Kierzkowska, M.; Majewska, A.; Szymanek-Majchrzak, K.; Sawicka-Grzelak, A.; Mlynarczyk, A.; Mlynarczyk, G. The presence of antibiotic resistance genes and bft genes as well as antibiotic susceptibility testing of Bacteroides fragilis strains isolated from inpatients of the Infant Jesus Teaching Hospital, Warsaw during 2007–2012. Anaerobe 2019, 56, 109–115. [Google Scholar] [CrossRef]
- Marr, L.; Biswas, D.; Daly, L.A.; Browning, C.; Vial, S.C.M.; Maskell, D.P.; Hudson, C.; Bertrand, J.A.; Pollard, J.; Ranson, N.A.; et al. Mechanism of glycogen synthase inactivation and interaction with glycogenin. Nat. Commun. 2022, 13, 3372. [Google Scholar] [CrossRef]
- Oboh, G.; Isaac, A.T.; Akinyemi, A.J.; Ajani, R.A. Inhibition of key enzymes linked to type 2 diabetes and sodium nitroprusside induced lipid peroxidation in rats’ pancreas by phenolic extracts of avocado pear leaves and fruit. Int. J. Biomed. Sci. 2014, 10, 208–216. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, A.; Keyhani-Nejad, F.; Pusec, C.M.; Goodman, L.; Geier, J.A.; Stoolman, J.S.; Stanczyk, P.J.; Nguyen, T.; Xu, K.; Suresh, K.V.; et al. Hexokinase 1 cellular localization regulates the metabolic fate of glucose. Mol. Cell 2022, 82, 1261–1277.e1269. [Google Scholar] [CrossRef] [PubMed]
| Drug Sensitive Tablets | Drug Content (mg/Piece) | B. fragilis Stains | ||||||
|---|---|---|---|---|---|---|---|---|
| SNBF-1 | CD11-1 | CD11-2 | CD11-5 | CD13-1 | CD13-4 | SY-X-3 | ||
| Ampicillin | 250 | R | R | R | R | R | R | R |
| Amoxicillin | 250 | I | I | R | R | R | I | R |
| Gentamicin | 250 | S | S | R | R | R | R | R |
| Polymyxin E | 250 | R | R | R | R | R | R | R |
| Tetracycline | 100 | S | I | R | R | R | I | R |
| Cephalosporin | 100 | S | S | S | S | S | S | S |
| Chloramphenicol | 50 | S | S | S | I | S | S | S |
| Akamycin | 50 | I | S | I | I | I | I | R |
| Ciprofloxacin | 100 | R | R | R | R | R | R | R |
| Sulfadiazine | 250 | R | R | R | R | R | R | R |
| Antibiotic Resistance Genes | Drug_Class | Resistance_Mechanism | Antibiotic |
|---|---|---|---|
| vanT gene in vanG cluster | Glycopeptide antibiotic | Antibiotic target alteration | Vancomycin |
| CepA-44 | Cephalosporin | Antibiotic inactivation | - |
| adeF | Fluoroquinolone antibiotic; tetracycline antibiotic | Antibiotic efflux | Tetracycline |
| adeF | Fluoroquinolone antibiotic; tetracycline antibiotic | Antibiotic efflux | Tetracycline |
| Strains (%) | SNBF-1 | CD11-1 | CD11-2 | CD11-5 | CD13-1 | CD13-4 | SY-X-3 | |
|---|---|---|---|---|---|---|---|---|
| Acid Tolerance (pH) | 2.0 | 90.86 ± 1.23 a | 94.32 ± 2.76 ab | 90.69 ± 0.89 a | 100.04 ± 3.12 bc | 97.57 ± 1.47 b | 94.32 ± 2.05 ab | 88.32 ± 0.92 a |
| 3.0 | 92.94 ± 1.56 a | 103.01 ± 2.03 b | 91.28 ± 1.10 a | 108.79 ± 3.45 c | 98.20 ± 1.88 ab | 103.01 ± 2.21 b | 93.01 ± 1.34 a | |
| 4.0 | 93.46 ± 1.78 a | 103.28 ± 2.58 b | 95.00 ± 1.25 a | 114.78 ± 3.67 c | 103.32 ± 2.09 b | 103.28 ± 2.32 b | 98.28 ± 1.53 a | |
| Bile tolerance | 0.1% | 98.54 ± 1.34 Ca | 96.41 ± 2.78 Ba | 13.15 ± 0.43 Aa | 119.77 ± 2.99 Fa | 100.84 ± 1.15 Da | 129.67 ± 0.32 Gd | 108.14 ± 3.04 Ea |
| 0.2% | 107.14 ± 1.62 Cb | 104.72 ± 2.03 Bb | 116.94 ± 1.57 Eb | 122.52 ± 1.31 Fb | 104.19 ± 1.06 Ab | 112.27 ± 2.98 Da | 10,814.75 ± 72.07 Gc | |
| 0.3% | 111.83 ± 3.33 Cc | 109.23 ± 1.07 Ac | 123.68 ± 2.81 Ec | 125.21 ± 2.94 Fc | 110.86 ± 1.34 Bc | 112.27 ± 1.34 Db | 12,667.91 ± 241.22 Gd | |
| 0.4% | 124.39 ± 1.40 Bd | 117.94 ± 1.73 Ad | 140.39 ± 3.49 Fd | 127.77 ± 2.60 Cd | 129.67 ± 1.33 Dd | 135.36 ± 1.83 Ee | 13,438.85 ± 223.58 Ge | |
| 0.5% | 130.27 ± 1.48 Ce | 125.52 ± 1.66 Be | 142.22 ± 3.12 Fe | 130.75 ± 1.94 De | 140.81 ± 3.22 Ee | 121.52 ± 2.77 Ac | 1132.22 ± 3.22 Gb | |
| Artificial gastrointestinal fluids | 94.21 ± 1.12 a | 97.16 ± 0.24 a | 94.23 ± 0.23 ab | 91.74 ± 1.06 bc | 91.90 ± 0.74 bc | 93.98 ± 0.35 a | 94 ± 1.67 ab | |
| Artificial intestinal fluids | 155.9 ± 63.70 a | 158.51 ± 1.14 a | 140.90 ± 4.17 b | 138.32 ± 1.53 bc | 148.42 ± 1.10 a | 94.17 ± 1.44 ghi | 94.17 ± 1.44 ghi | |
| Bile salt hydrolase activity | 410.04 ± 12.29 a | 384.09 ± 11.40 ab | 366.02 ± 14.77 b | 326.31 ± 13.51 c | 381.85 ± 12.97 ab | 386.81 ± 13.17 ab | 316.79 ± 9.11 c | |
| Strains | Cell Surface Hydrophobicity (%) | Auto Aggregation (%) | ||
|---|---|---|---|---|
| 2 h | 8 h | 24 h | ||
| HC-LX-1 | 36.41 ± 1.30 d | 20.67 ± 4.33 | 40.60 ± 0.12 | 41.59 ± 1.51 |
| SNBF-1 | 47.23 ± 1.21 b | 23.44 ± 0.27 | 46.91 ± 2.54 | 52.88 ± 4.09 |
| CD4-1 | 39.87 ± 1.30 c | 26.94 ± 4.10 | 49.27 ± 1.02 | 65.84 ± 3.19 |
| CD11-1 | 45.32 ± 1.50 b | 31.32 ± 1.45 | 53.43 ± 2.76 | 67.80 ± 0.65 |
| CD11-2 | 40.34 ± 0.63 c | 19.12 ± 1.07 | 50.54 ± 3.56 | 66.52 ± 6.25 |
| CD13-4 | 51.48 ± 0.95 a | 38.33 ± 4.62 | 61.89 ± 1.45 | 77.79 ± 7.11 |
| SY-XB-1 | 40.01 ± 1.29 c | 24.37949 ± 3.04 | 49.2702 ± 2.11 | 70.54 ± 2.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cang, W.; Li, X.; Tang, J.; Wang, Y.; Mu, D.; Wu, C.; Shi, H.; Shi, L.; Wu, J.; Wu, R. Therapeutic Potential of Bacteroides fragilis SNBF-1 as a Next-Generation Probiotic: In Vitro Efficacy in Lipid and Carbohydrate Metabolism and Antioxidant Activity. Foods 2024, 13, 735. https://doi.org/10.3390/foods13050735
Cang W, Li X, Tang J, Wang Y, Mu D, Wu C, Shi H, Shi L, Wu J, Wu R. Therapeutic Potential of Bacteroides fragilis SNBF-1 as a Next-Generation Probiotic: In Vitro Efficacy in Lipid and Carbohydrate Metabolism and Antioxidant Activity. Foods. 2024; 13(5):735. https://doi.org/10.3390/foods13050735
Chicago/Turabian StyleCang, Weihe, Xuan Li, Jiayi Tang, Ying Wang, Delun Mu, Chunting Wu, Haisu Shi, Lin Shi, Junrui Wu, and Rina Wu. 2024. "Therapeutic Potential of Bacteroides fragilis SNBF-1 as a Next-Generation Probiotic: In Vitro Efficacy in Lipid and Carbohydrate Metabolism and Antioxidant Activity" Foods 13, no. 5: 735. https://doi.org/10.3390/foods13050735
APA StyleCang, W., Li, X., Tang, J., Wang, Y., Mu, D., Wu, C., Shi, H., Shi, L., Wu, J., & Wu, R. (2024). Therapeutic Potential of Bacteroides fragilis SNBF-1 as a Next-Generation Probiotic: In Vitro Efficacy in Lipid and Carbohydrate Metabolism and Antioxidant Activity. Foods, 13(5), 735. https://doi.org/10.3390/foods13050735






