Abstract
Ligilactobacillus salivarius (basonym: Lactobacillus salivarius, L. salivarius) is a type of lactic acid bacteria (LAB) commonly found in the oropharyngeal-gastrointestinal tract (OGT). It has gained significant attention due to its probiotic and functional properties as well as its various health-promoting roles. L. salivarius strains exhibit strong resistance and adhesion in the OGT along with outstanding antioxidant and antimicrobial properties. Additionally, numerous L. salivarius strains have the ability to produce bacteriocins with antagonistic activity. These probiotic characteristics of L. salivarius indicate its remarkable potential in promoting favorable effects on human health. It has also been observed that L. salivarius has a positive effect on the composition of intestinal microbiota, thereby improving the metabolic profiling of intestinal microbiota, promoting a healthy and balanced internal environment. In recent years, multi-omics technologies such as genomics, transcriptomics, proteomics and metabolomics have been employed to gain a deeper understanding of the roles and mechanisms of L. salivarius associated with its functional properties. This review aims to provide an overview of the probiotic characteristics of L. salivarius, containing its specific interactions with the host microflora, as well as insights from omics studies.
1. Introduction
Ligilactobacillus salivarius, formerly named Lactobacillus salivarius, has been referred to as this species name for almost 70 years. It was initially described by Rogosa et al. in 1953 as an obligatory homo-fermentative lactic acid bacteria [1,2]. Over time, L. salivarius has undergone several reclassifications and is now recognized as a Gram-positive bacterium capable of both homotypic and parthenogenic heterotypic fermentation [3]. In the past three years, L. salivarius strains have been reclassified and moved from the Lactobacillus genus to a new genus called Ligilactobacillus, which consists of 16 species. The name Ligilactobacillus implies a combination, with a host-adapted lifestyle, specifically referring to the vertebrate host of L. salivarius [4].
L. salivarius is a non-motile, non-sporulating, oxidase- and catalase-negative, rod-shaped microorganism with a general size of 0.6–1.9 μm × 1.5–5 μm. L. salivarius exists in different niches and currently isolates are primarily derived from the intestines or feces of birds and mammals [5], e.g., geese, chickens, turkeys, pigeons, ducks, pigs and cattle. Moreover, it has also been shown to exist in the oral cavity, vagina and breast milk of humans, and in honeybee guts, and in foods such as grape wine, meat and St. Ivel cheese [5]. Recently, L. salivarius has received increasing attention as a potential probiotic, and various applications of L. salivarius strains have been explored.
Based on available data as of April 2023, the average genome size of L. salivarius strains that have been sequenced is 1.99 ± 0.14 Mbp, containing 1946.70 ± 382.15 genes with a GC content of 32.79 ± 0.16%. In addition to chromosomes, the genome typically includes a repA-type megaplasmid ranging from 100 to 380 kbp and small plasmids. Studies have shown significant differences in both chromosomal and plasmid sequences among L. salivarius strains, particularly in genes encoding glycoside hydrolases (GH), bacteriocins, proteases and EPS synthesis.
In terms of functional characteristics, most L. salivarius strains have a strong tolerance to acidic pH, resistance to the OGT conditions and adhesion to the intestinal mucosa, enabling them to effectively colonize in the host. In addition, excellent antioxidant and antibacterial properties have also demonstrated that this ligilactobacillus can have favorable effects on the host health. L. salivarius strains possess antagonistic properties against bacterial pathogens such as Salmonella enterica (S. enterica), Clostridium perfringens (C. perfringens), Staphylococcus aureus (S. aureus), Klebsiella pneumoniae (K. pneumoniae), Pasteurella multocida (P. multocida), Riemerella anatipestifer (R. anatipestifer) and Campylobacter sp. This ability is attributed to the production of lactic acid, H2O2 and bacteriocins, as well as the capacity to colonize the gut for an extended period, leading to the exclusion of unfavorable microflora [6]. Moreover, previous studies have shown that most L. salivarius strains can improve the composition of the intestinal microflora. Indeed, the correlation between L. salivarius strains and indigenous gut microbiota has emerged as a popular area in scientific research. On the other hand, studies have shown that L. salivarius strains possess several functional properties that are beneficial to the food industry. These properties include improving nutritional quality, enhancing flavor properties, exhibiting antioxidant and antimicrobial activities, increasing the shelf-life of foods, and reducing undesirable compounds [7]. Based on these favorable functional characteristics mentioned above, L. salivarius has been awarded the “generally recognized as safe” (GRAS) status by the United States Food and Drug Administration (FDA), and listed as a recommended biological agent intentionally added to food by the European Food Safety Authority (EFSA) and the Ministry of Public Health of China [8]. While studying the functional properties of L. salivarius, the multi-omics technologies including genomics, transcriptomics, proteomics and metabolomics have achieved rapid development. Among them, genomics and transcriptomics provide a strong guarantee for genetic information analysis and the functional gene identification of probiotics. Proteomics and metabolomics are very effective methods to study the adaptive mechanism of probiotics to physiological and environmental changes. In a recent study, Lugli et al. proposed a novel concept of “probiotic genomics”, which undoubtedly provides a proprietary identity card for each probiotic [9]. Therefore, omics technologies are growing in significance as their ability to elucidate the molecular mechanisms underlying the functional and probiotic properties of LAB. This review summarized the probiotics properties of L. salivarius as well as the application of omics approaches in L. salivarius strains. It provides a reference for subsequent research applications of L. salivarius strains.
2. Probiotic Properties and Roles of Ligilactobacillus salivarius
L. salivarius is an important member of the LAB family and it has been widely used as a probiotic due to its excellent characteristics (Table 1). Although early studies focused on the isolation of probiotic strains of L. salivarius and their bioactive metabolites (mainly bacteriocins), more research is now available to better understand the role of L. salivarius strains and their metabolites in various fields and their adaptability to environmental stresses.

Table 1.
Probiotic properties and applications of Ligilactobacillus salivarius.
2.1. Resistance to Oropharyngeal-Gastrointestinal Conditions
A requisite condition for any microorganism to be a probiotic is the ability to survive or pass the harsh conditions of the GT. The stomach is a very unfriendly environment, with its internal gastric juice consisting mainly of pepsin and hydrochloric acid (HCl). This directly contributes to the low pH (1.5–4.5) of the gastric environment. Therefore, acidic pH tolerance is considered to be the main criterion for probiotic screening. In this unfavorable environment, L. salivarius strains are potential probiotics by maintaining intracellular pH balancing, ATR signaling pathway and macromolecule protection and repair, among other strategies to achieve pH and bile tolerance [42]. In addition, the acid resistance of some LAB can also be improved by exposing themselves to non-lethal acidic conditions through acid resistance reactions [43]. It is known that different strains of the same LAB species have highly variable acid resistance and this is also true for L. salivarius strains. A study shows that L. salivarius IBB3154 showed good resistance and survived at low pH (value is 3.5) conditions [44]. Similarly, Sajedinejad et al. demonstrated that L. salivarius NK02 displayed significant tolerance to low pH conditions. Specifically, this strain exhibited survival rates (7 logs CFU/mL) when exposed to simulated gastric juice (pH 2.2). Furthermore, L. salivarius NK02 was also found to be significantly tolerant to bile, lysozyme, and 1–5% NaCl [37].
In addition to gastric acid, probiotics must also tolerate exposure to bile salts in the small intestine. Bile salt is a sodium or potassium salt formed by the combination of bile acids secreted by hepatocytes with glycine or taurine. It is the main component of bile involved in fat digestion and absorption. Cell membrane disruption, DNA damage, intracellular acidification, oxidative stress and metabolic changes triggered by bile salts, pose a serious threat to the colonization and survival of probiotics [45]. Therefore, bile tolerance is another major criterion for screening potential probiotic strains. L. salivarius strains, an OGT natural flora, have been exposed to bile salt stress for a long time and have evolved various mechanisms to cope with bile salt toxicity, mainly including the production of bile salt hydrolases (BSHs), alteration of cell membrane composition and structure, and the use of a transport system to transfer bile salts [46]. BSH is considered to be the main component of bile tolerance in LAB. It is responsible for catalyzing the depolymerization of glycine and taurine residues in cholesterol. Although the exact nature of how bile salt depolymerization limits negative effects on cellular homeostasis is unknown, many literatures have reported that bile salt hydrolase expression is associated with bile resistance in many lactobacilli species [47]. However, Fang et al. demonstrated that BSH is not the primary determinant of bile resistance in L. salivarius strains, and may have additional biological importance because of its varying effects upon bile as a signaling molecule in the host [48]. Pan et al. also showed that there was no association between bile salt hydrolase and bile salt tolerance in L. salivarius strains and indicated that bile salt tolerance in L. salivarius strains was associated with processes such as peptidoglycan synthesis, the phosphotransferase system (PTS) and DNA damage repair [49]. This is consistent with the results of Wang et al. [50] and Lv et al. [51].
In addition to gastrointestinal microbiota, oral microbiota is also part of the human microbiome. In the daily diet process, it is essential to add salt, vinegar and other food ingredients to various diets, and these high osmotic pressure foods will inevitably affect the survival and growth of lactic acid bacteria in the oral tract. Therefore, probiotics must also tolerate a variety of complex conditions. L. salivarius AR809 was isolated from a healthy adult oral cavity with good resistance to acidic pH, bile, lysozyme and H2O2 [35,42].
It can be seen that most of the L. salivarius have the characteristics of acid and bile salt tolerance. Certainly, tolerance to various conditions could impact the probiotic potential of different strains of L. salivarius. Nevertheless, on the other hand, it is essential to recognize that resistance to low pH and bile is crucial for probiotics, but it does not guarantee that acid pH-tolerant and bile-tolerant strains will exhibit probiotic properties.
2.2. Adherence to the Intestinal Mucosa and Extracellular Matrix Components
Adhesion is an important prerequisite for the colonization and function of LAB strains in the OGT, which is directly related to cell wall components such as adhesins, polysaccharides and proteins (Figure 1). It includes two steps, non-specific adhesion and specific adhesion. Through adhesion, LAB can be permanently colonized in the IEC membrane, enhance the signal exchange between cells, promote the stability of intestinal flora, regulate the body’s immunity, form a biological barrier, and resist pathogenic bacteria [52]. Therefore, adhesion is also one of the important criteria for screening probiotic lactic acid bacteria in vitro. In this case, many researchers have begun to study the adhesion ability of L. salivarius strains. A study conducted by Jia et al. [35] demonstrated that in a cell adhesion assay simulating the human oral environment, the extent of adhesion to FaDu cells of L. salivarius AR809 (31.1%) was significantly higher than that of L. plantarum AR113 (4.44%) and L. plantarum AR195 (8.28%). In addition, L. salivarius AR809 also significantly reduced the adhesion effect of S. aureus to FaDu cells through exclusion, competition and displacement. In a similar way, Dash et al. demonstrated that the L. salivarius F14 strain had good adhesion to the Caco-2 cells through the co-culture model in vitro, and it also could significantly inhibit the adhesion process of S. typhimurium ST-Xen 33 on Caco-2 cells [10]. For their part, Zhang et al. [53] and Ren et al. [54] showed that L. salivarius strains had good adhesion to the Caco-2 cells which could be associated with the presence of genes that encode different proteins attributed to adhesion to different extracellular matrices and intestinal mucus [46]. Of course, we should also list more in vivo results to confirm that L. salivarius strains can adhere to the intestinal mucosa/extracellular matrix components and persist.
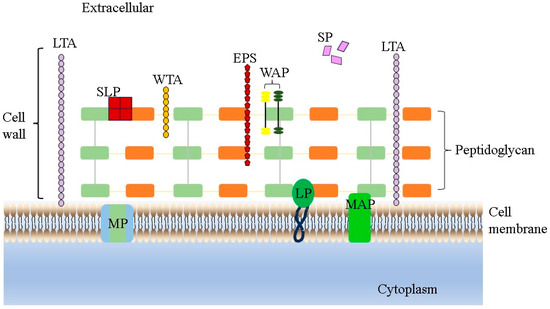
Figure 1.
LAB cell wall structure. LTA represents lipoteichoic acids, WTA represents wall teichoic acids, EPS represents exopolysaccharides, WAP represents wall-associated proteins, SLP represents surface layer proteins, SP represents secreted proteins, MP represents membrane proteins, LP represents lipoproteins and MAP represents membrane-associated proteins.
2.3. Antioxidant Activity
Oxidative stress is when the balance between anti-oxidants and prooxidants in the cell is disturbed, resulting in DNA hydroxylation, protein denaturation, lipid peroxidation and ultimately cell apoptosis. It is the fundamental reason for aging and aging-related diseases, which can induce diabetes, atherosclerosis, arthritis, hyperlipidemia, cardiovascular diseases and many other diseases [55]. In recent years, a large number of studies have shown that lactic acid bacteria can remove the active oxygen molecules in the intestine, so that the active oxygen molecules in the body remain relatively stable, thereby reducing the body damage caused by the oxidation reaction. These lactobacilli exert antioxidant effects mainly through scavenging reactive oxygen radicals in and around cells, chelating metal ions, alleviating lipid peroxidation, modulating their antioxidant defense system, regulating the host cell antioxidant defense system, and modulating host cell antioxidant-related signaling pathways [56]. With the gradual increase in reports on the antioxidant effect of lactobacillus, the understanding of the antioxidant function of lactobacillus is also increasing. Zhang et al. identified a strain of Limosilactobacillus fermentum YLF016, which contained various antioxidant enzyme encoding genes, as well as having a strong ability to scavenge 2,2-diphenyl-1-picrylhydrazyl (DPPH) and hydroxyl radicals (-OH) scavenging ability [57]. Zhai et al. evaluated the antioxidant activity of 10 lactobacillus strains and found that L. plantarum CCFM 8661 exhibited the strongest lipid peroxidation inhibitory activity [58].
Currently, there have been studies on the in vivo/vitro antioxidant function evaluation of L. salivarius strains at home and abroad. Zhang et al. studied the protective mechanism of L. salivarius by establishing an alcohol injury model [59]. Specifically, L. salivarius M18-6 protected mouse hepatocytes from alcohol-induced oxidative stress damage by downregulating serum alanine transaminase (ALT) levels, upregulating superoxide dismutase (SOD) levels and activating the keap1-Nrf2 signaling pathway. This research provided a scientific basis for the clinical application and product development of the L. salivarius strain for alcohol injury. Wang et al. revealed the molecular mechanism of L. salivarius Ren in response to bile-induced oxidative stress by transcriptomics and proteomics [60]. L. salivarius MG242 isolated from the human vagina by Kang et al. exhibited good antioxidant properties with DPPH radical scavenging rate and 2, 2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) of 56.9% and 97.1%, respectively [61]. Chooruk et al. investigated the antioxidant activities of 201 LAB strains in vitro [62]. Among them, the antioxidant capacity of L. salivarius differed significantly from other lactobacillus, which also indicates the specificity of antioxidant properties of different species and strains. The supplementation of L. salivarius AP-32 in Parkinsonian rats modulates short-chain fatty acid (SCFA) production, increases antioxidant enzyme activity and protects mitochondria from reactive oxygen species (ROS)-induced damage [63]. Therefore, the antioxidant potential of L. salivarius can provide ideas for anti-aging related research.
2.4. Active Metabolites and Its Antimicrobial Activity
Although the traditional view is that only live probiotics can exert their probiotic effects, more and more studies have shown that some of the probiotic properties of probiotics are closely related to the active products produced by their metabolism. At present, the active metabolites of probiotics mainly include EPS, bacteriocins, organic acids, SCFAs, vitamins and some bioactive enzymes and small molecules (Figure 2). These active metabolites have been proven to have anti-inflammatory, antimicrobial, antioxidation, immune regulating properties useful for the prevention or treatment of a variety of metabolic diseases. Antimicrobial properties are one of the most notable characteristics of probiotics. This can be achieved through competition for nutrients and adhesion space, inducing environments that are harmful to pathogens, and that produce antimicrobial metabolites and regulate immune responses [46]. This subsection is devoted to the antimicrobial activity of L. salivarius strains.
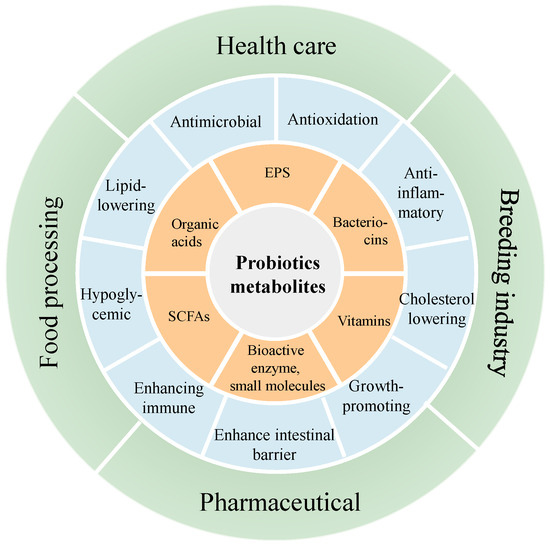
Figure 2.
Active metabolites of probiotics, their functions and applications.
L. salivarius strains can produce several antimicrobial active compounds that exhibit antagonistic activity against pathogenic organisms. First, many L. salivarius strains are good producers of many small proteins such as antimicrobial peptides and bacteriocins. This allows them to compete with other bacteria in the environment and disrupt the cell walls and cell membranes of pathogenic bacteria, causing the intracellular material to leach out, thus producing an antibacterial effect [64,65,66,67]. L. salivarius PS7 showed strong antagonistic activity against ten acute otitis media (AOM) related pathogens (Streptococcus pneumoniae MP07, Streptococcus pyogenes MP03, S. aureus MP29, Staphylococcus epidermidis MP33, Alloiococcus otitidis MP02, Enterococcus faecalis MP64, Haemophilus influenzae MP04, Moraxella catarrhalis MP08, P. aeruginosa MP24 and E. coli MP69) [28]. Similarly, Martín et al. found that L. salivarius CECT5713 inhibited the growth of Gram-negative bacteria (including, E. coli CECT4076, K. pneumoniae CECT 142, K. oxytoca CECT 860T and Proteus vulgaris CECT484), Gram-positive bacteria (E. faecium P21, E. faecalis TAB28, Listeria monocytogenes ScottA, L. monocytogenes Ohio, L. innocua RdC, S. aureus CECT5191, S. epidermidis CECT 231, Lactococcus lactis MG1614 and Latilactobacillus sakei NCFB2714) and the yeasts Rhodotorula mucilaginosa CECT10359, this effect being greater in L. monocytogenes Ohio and K. oxytoca CECT 860T [68]. The antimicrobial effect of both strains of L. salivarius was attributed not only to the presence of bacteriocins but it was also related to the formation of organic acids (such as common lactic acid and SCFAs) and H2O2 that leads to a change in the medium, which can alter the development of indicator organisms [69]. Furthermore, L. salivarius strains can also produce EPS with natural antibacterial activity, which is usually attributed to its anti-biofilm effect [70]. Bikric et al. found that EPSBIS312 and EPSBIS722 derived from L. salivarius BIS312 and L. salivarius BIS722, respectively, could significantly inhibit the biofilm formation of Enter. faecalis 29212, Staph. aureus EB1 and E. coli ATCC 11229, and were significantly higher than commercial inulin [71]. This indicates that these two EPS may become substitutes for plant prebiotics (such as inulin) in poultry.
Moreover, many L. salivarius strains also possesses antiviral activity. L. salivarius YM33 isolated from the feces of nursing piglets has good activity against pig epidemic diarrhea virus (PEDV) and can significantly down-regulate proinflammatory cytokine levels [72]. This research was the first demonstration of the antiviral activity of L. salivarius against PEDV. Similarly, Shojadoost et al. reported that L. salivarius JTBo9 enhanced the antiviral activity of macrophages of chicken against avian influenza virus infection via virus titer reduction and increased the expression of IL-1β and IFN-γ virus titer in an in vitro cell model [73]. It is also because of the antimicrobial activity of L. salivarius strains that it is widely used in the breeding industry. In conclusion, the antimicrobial activity of L. salivarius provides favorable conditions for its application in various fields.
2.5. Host OGT Micro-Ecosystem Modulation
The host gut is a complex micro-ecosystem consisting of communities of bacteria, viruses, archaea, fungi and protozoa that live in the GT [74], known as the intestinal flora. Gut microbiota performs useful functions including fermenting unused energy substrates, training the immune system with metabolic end products, maintaining the intestinal epithelium, synthesizing host vitamins [75], producing hormones that induce host fat accumulation, metabolizing dietary and pharmaceutical compounds and controlling immune function, and even affects behavior via the gut-brain-axis [76].
Given these facts, intestinal microflora plays a key role in the maintenance of host health and the pathogenesis of many diseases [77]. Therefore, gut microbiota eubiosis is essential for the prevention of infectious and non-infectious diseases and for preventing disturbances (also known as dysbiosis) in the balance of the microbial community equilibrium [78]. However, the gut microbiota is an open micro-ecosystem whose composition and/or activity can be influenced by many elements, including the mode of birth, sex, host inheritance, immune system and host health or disease status, geographic location, social economic factors, diet, the use of therapeutic drugs, etc. [79]. In point of fact, the intestinal microbiota is constantly exposed to transient exogenous microorganisms transmitted through food, as demonstrated by Veiga et al. [80]. In this regard, probiotics can regulate the composition of gut microbiota and correct the abnormal response of the immune system [81], thereby exerting different favorable effects on host health (Figure 3). In summary, probiotics may be a therapeutic strategy to regulate intestinal flora and improve human disease [60,82]. In this sense, several studies have found that different strains of L. salivarius have a significant regulatory effect on intestinal flora (Table 2).
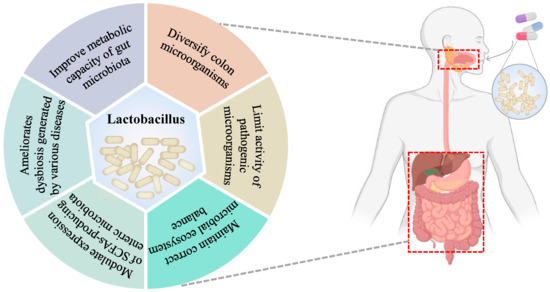
Figure 3.
Effects on the gut microbiota following administration of lactobacillus.

Table 2.
Effect of different Ligilactobacillus salivarius strains on the intestinal microbiota.
For instance, Moturi et al. explored the fecal microflora composition in piglets fed two different L. salivarius strains during the lactation period [94]. Specifically, significantly fewer OTUs and a lower phylogenetic diversity index and Chao index of bacteria were observed after supplementation with L. salivarius from normal piglets compared to the control group, suggesting that probiotics may inhibit bacterial growth. Similarly to these results, Riboulet-Bisso et al. stated that the application of L. salivarius UCC118 wild type reduced the number of Gram-negative bacteria present in the intestine of pigs [95]. In addition, L. salivarius 144 (from fast-growing pigs)-treated piglets showed a significant increase in the abundance of beneficial bacteria and a decrease in the abundance of C. perfringens, which may be related to their antimicrobial activity, and these changes in microbial communities may reduce the susceptibility of weaned piglets to pathogenic infections at weaning. Furthermore, L. salivarius (LS144 and LS160) supplementation could promote the growth of villus in all of the intestinal segments. On the contrary, in a recent study, Wei et al. found that the ingestion of L. salivarius WZ1 corrected the reduced species abundance and species diversity of the intestinal flora caused by E. coli K88 infection and increased the abundance of beneficial bacteria (Lactobacillus and Bifidobacterium), as well as decreased the abundance of harmful bacteria (Ralstonia and Helicobacter) [41]. Similar results were obtained by Xin et al. who found that the Shannon, Chao1 and Ace indices of the Sinocyclocheilus grahami gut microbiome were all found to increase after feeding supplemented with L. salivarius S01, suggesting that L. salivarius S01 can promote gut microbial diversity and abundance in the host [83]. These results show that the probiotic function of different L. salivarius strains also varies greatly.
Xu et al. found that the supplementation of L. salivarius CML352 in late phase improved the gut microflora composition of laying hens [88]. Specifically, this strain significantly reduced the relative abundance of the phylum Firmicutes and increased the relative abundance of the phylum Bacteroidetes, thus resulting in a significant decrease in F/B ratio. Many human studies have consistently shown that the F/B ratio is positively correlated with the degree of obesity [96]. Moreover, the gene expression levels related to methanogenesis from acetate in the L. salivarius CML352 group were significantly lower than in the control group, which has been presumed to reduce fat deposition and obesity [97]. Thus, L. salivarius strains may have the potential to reduce obesity. Additionally, the increased abundance of Bacteroidetes may promote gut health in chickens. Simply speaking, the majority of the species in the Bacteroidetes phylum are producers of SCFAs [98], which play a significant role in innate immunity, are an important source of energy for IEC, and maintain epithelial barrier function and inhibit pro-inflammatory cytokines. In addition, eggshell strength and thickness increased significantly (p < 0.05) after feeding L. salivarius CML352. This may be due to the fact that feeding L. salivarius CML352 increases the vitamin content of the gut and promotes the absorption of mineral elements [99].
For their part, Lv et al. investigated the influence of five LAB on dysbiosis produced in a rat model with acute liver injury induced by D-galactosamine [19]. They discovered that L. salivarius LI01 and Pediococcus pentosaceus LI05 were beneficial in preventing acute liver failure. Specifically, L. salivarius LI01 significantly decreased alanine aminotransferase and aspartate aminotransferase levels, inhibited total bilirubin accumulation, reduced the histological abnormalities of both the liver and the terminal ileum, prevented bacterial translocation, increased the serum interleukin 10 (IL-10) and/or interferon-γ (IFN-γ) levels, and resulted in a cecal microbiome that differed from that of the liver injury control. In addition, L. salivarius LI01 also attenuated liver fibrosis by increasing microbial abundance, improving the integrity of the intestinal barrier, and reducing plasma endotoxin levels and regulating Toll-like receptors (TLR) gene expression, among other mechanisms [90]. Furthermore, it also synergized with Bifidobacterium to significantly improve the symptoms of D-galactosamine-induced liver failure in rats [91]. It can be seen that in the complex OGT environment, the regulation of intestinal flora may be attributed to the synergistic effect of multiple probiotics. In conclusion, the above studies provide strong evidence for the prevention and treatment of liver injury and demonstrate that gut microbiome homeostasis contributes to the improvement of liver disease.
3. Multi-Omics Approach to Understanding the Role of Ligilactobacillus salivarius
Omics technology belongs to the concept of systems biology, mainly including genomics, transcriptomics, proteomics and metabolomics, etc. With the continuous improvement and evolution of omics technology research, combined with chemometric tools, people have a comprehensive and in-depth understanding of the mechanism behind the functional and specific interactions between probiotics and hosts (Figure 4). Researchers have successfully revealed the genetic information of different microorganisms with various phenotypes using these methods. Therefore, these are very useful tools to bridge the gap between genetic information and cell-specific metabolites. Among these, genomics allows for the identification of functional genes contained in the target strain and transcriptomics, proteomics and metabolomics allow for the quantification of mRNA, proteins and metabolites (<1500 Daltons), respectively, under specific physiological conditions. Therefore, it is worth noting that all of these methods are completely different from traditional characterization methods. Considering the complexity and uniqueness of organisms, a single method is not enough to characterize organisms. Most lactobacillus has been widely used in food and pharmaceutical fields. It is only recently, however, that advances in technology and methods have revealed the mechanisms that explain the beneficial effects of these bacteria on the host [100]. To date, many scholars have integrated various “omics” approaches to understanding the functional role of LAB, including L. salivarius [51].
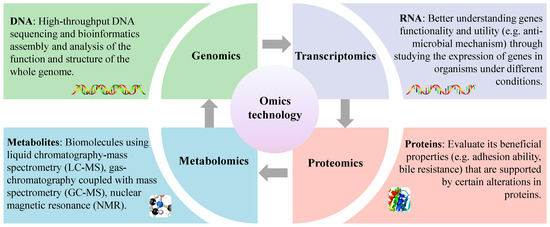
Figure 4.
Understanding probiotic functional properties using an omics approach.
3.1. Genomics Approach to Comprehend the Role of Ligilactobacillus salivarius
With the rise of genomics, LAB have important research value for human production and life, and gradually become the focus of research. It is expected that this will reveal the diversity and evolution of LAB at the molecular level, enable analysis of their physiological and metabolic mechanisms, and uncover functional genes related to important traits in order to accelerate the breeding and transformation of excellent strains, and provide a basis for the efficient utilization of LAB and improvement in the industrial-level control of fermentation.
Based on the genome database of NCBI (https://www.ncbi.nlm.nih.gov/genome, accessed on 11 May 2023), United States, 22 complete genome maps of L. salivarius have been constructed as of May 2023. Yang et al. determined the whole genome of L. salivarius AR809 and it contains 1967 genes, of which a total of 1593 genes encoding proteins, 79 tRNA genes and 22 rRNA genes were found on the circular chromosome, and 240, 28, 3 and 2 genes encoding proteins were found on plasmid pA-pD, respectively [42]. In addition, genomic analysis also screened a series of genes related to beneficial properties, such as carbohydrate metabolism, environmental stress and adhesion [42], which provide valuable guidance for the oral colonization of L. salivarius AR809. Sun et al. reported the whole genome sequence of L. salivarius Ren, which contained a 1,751,565 bp circular chromosome and two plasmids [101]. Bioinformatics analysis identified several genes important for gastrointestinal tolerance and adhesion, such as genes involved in acid and bile salt tolerance responses, genes encoding S-adenosylmethionine synthase, and fibronectin-binding protein genes [101]. In addition, it found that the absence of an α-glycerophosphate oxidase coding gene in L. salivarius ATCC11741, which fails to degrade 4-hydroxyaminoquinoline 1-oxide (4-HAQO), suggested it may be one of the reasons for the significant differences between L. salivarius Ren and ATCC11741 in the secretion of H2O2 and degradation of 4-HAQO [101].
3.2. Transcriptomics Approach to Comprehend the Role of Ligilactobacillus salivarius
Microarray and RNA sequencing (RNA-Seq) are two main techniques currently used for transcriptomic research [102]. In the probiotics field, the transcriptome has been used to study the molecular mechanisms involved in environmental stress responses, e.g., tolerance to the stress of acid and bile salts in the GIT [51]. In addition, transcriptomics help to reveal molecular strategies for probiotic interactions with the host or other microbiota, including changes in metabolic profiles, signaling pathways regulation, cell growth and communication (intercellular signaling) [103]. Song et al. used RNA sequencing analysis to identify the first well-characterized endogenous constitutive promoter library from L. salivarius, providing a useful toolbox for the subsequent metabolic engineering and synthetic biology of L. salivarius and other prokaryotes [104]. Xia et al. fused a multi-omics strategy of transcriptomics, metabolomics and cytokine arrays to explore the effects of colonization with L. salivarius LI01 on growth, immunity and metabolism in germ-free rats [105]. Similarly, Lv et al. used transcriptome sequencing combined with proteome, and proposed the first model of a bile stress response mechanism for L. salivarius LI01, which provides a reference for the subsequent bile salt resistance mechanism of L. salivarius [51]. The combination of transcriptomics and other omics is one of the most common methods to solve experimental problems. Considering the specific environment, the integrated study of transcriptome combined with other technologies can be used in many fields, e.g., studying the role of LAB in food spoilage, potential probiotic properties of LAB strains, etc.
3.3. Proteomics Approach to Comprehend the Role of Ligilactobacillus salivarius
Proteomics mainly studies the dynamic changes in proteins during development and their responses to internal or external stimuli [106]. Based on the complexity of protein structure, researchers have developed a comprehensive proteomics technique to deeply analyze all of the proteins present in a sample [107]. In recent years, the proteomics research on L. salivarius has been increasing (Table 3). Kang et al. used 2D gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-ToF/ToF MS) analysis and identified potential secreted proteins that may be responsible for the antimicrobial activity [108]. A total of 21 secreted proteins were identified, of which five were produced by L. salivarius. The LysM domain protein was a peptidoglycan binding protein that may cause the lysis of S. aureus upon binding to the cell wall but does not affect lactobacillus. Kelly et al. identified three proteins, DnaK, Ef-Ts and pyruvate kinase, in the cell wall of L. salivarius UCC118 by combining proteomics analysis with enzymatic techniques [109]. These proteins may play an important role in adhesion and promoting host immune perception. In addition, the proteome can also be used for strain identification. Hamza et al. used MALDI-TOF spectroscopy and 16S rDNA sequencing to identify 67 isolates [110]. All identified isolates were L. salivarius and L. plantarum. Proteomic analysis can help to understand the different characteristics of L. salivarius and identify the proteomic profile of individual characteristics, which can be used as biomarkers for the initial selection of potential probiotic strains.

Table 3.
Potential functional properties of Ligilactobacillus salivarius revealed using omics techniques.
3.4. Metabolomics Approach to Comprehend the Role of Ligilactobacillus salivarius
The metabolome is believed to be the result of the genome, transcriptome and proteome, and directly influences the molecular phenotype of microbial cells. Its application in probiotics research has been developed rapidly in the past few years. Metabolomics has been used to map metabolic pathways and reveal microbial metabolic networks, e.g., studying types and changes of metabolites in food, evaluating the effect of probiotics on the metabolic activity of resident microflora and characterizing microbial molecules secreted during the industrial production of probiotics. Zhu et al. used genomics and metabolomics to explore the alleviation mechanism of the L. salivarius strain on nonalcoholic fatty liver disease (NAFLD) [112]. Specifically, most of the 250 metabolites from L. salivarius SNK-6 are involved in multiple metabolic pathways, including amino acid and lipid metabolism. Studies have reported that the L. salivarius strain can effectively alleviate liver injury by regulating liver lipid metabolism. In addition, a further analysis of the metabolomics data showed that butyric acid, acetic acid and propionic acid were the main SCFAs produced by L. salivarius strain, while cholic acid (CA), ursodeoxycholic acid (UDCA), chenodeoxycholic acid (CDCA) and tauroursodeoxycholic acid (TCA) were the four most abundant bile acids in the metabolites of L. salivarius SNK-6. Studies have shown that they are important signaling molecules that regulate lipid metabolism genes and can alleviate lipid accumulation and inflammation in NAFLD rats. Bile acids play an important role in regulating lipid, glucose and energy metabolism, and are essential for protecting hepatocytes from cholesterol and bile acid toxicity. CA shows a specific affinity for bile acid receptors, and as a signal molecule, it has a unique role in regulating liver lipid metabolism. UDCA is used to treat primary biliary cirrhosis, activate PKC and MAPK signaling and anti-inflammatory hepatocyte pathways, and promote bile HCO3− secretion to reduce cholestasis and liver damage. CDCA can reduce body weight and improve glucose tolerance. TCA can reduce hepatic steatosis, intestinal inflammation and insulin resistance in mice.
In summary, we found that genomics and transcriptomics can analyze the genetic information and functional genes of L. salivarius, and proteomics and metabolomics can delve into the adaptive mechanism of L. salivarius in response to physiological and environmental changes. Therefore, when we study L. salivarius, the combined use of the above four histological approaches can comprehensively reveal the complex mechanisms behind the functions and beneficial properties of L. salivarius from multiple perspectives, including genetics, expression and metabolism.
4. Conclusions
In recent years, L. salivarius has gained increasing attention from researchers due to its tolerance to the oral cavity and gastrointestinal environment, as well as its ability to adhere to human cells. Studies have shown that L. salivarius strains have various physiological functions, such as anti-oxidation, immune regulation, and maintaining the balance of intestinal flora, providing a theoretical and experimental basis for the development of functional lactic acid bacteria products and healthy foods with the potential to lower blood fat and prevent cardiovascular diseases. Despite the recognized functional properties of different L. salivarius strains, further research is needed to fully understand the role this microbe plays in host health. Omics approaches are becoming increasingly important in this field of research as they enhance the understanding of the functions and mechanisms behind the probiotic properties of beneficial bacteria such as L. salivarius. Genomics and transcriptomics can reveal the genetic information of L. salivarius strains, analyze their physiological and metabolic mechanisms, and discover important functional genes. Proteomics and metabolomics can study the effect of L. salivarius strains on the mechanisms of adaptation to physiological and environmental changes, such as adhesion, biofilm formation, antagonistic capacity, and tolerance. However, there are still many questions that need to be clarified, emphasizing the importance of further research in the field of multi-omics techniques to increase our understanding of how L. salivarius exerts its probiotic activity.
Author Contributions
Y.Y. wrote the draft manuscript. X.S., Z.X., Y.X. and G.W. conceived and edited the manuscript. L.A. supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32101928), National Science Foundation for Distinguished Young Scholars (No. 32025029), Shanghai Municipal Education Commission Scientific Research and Innovation Project (2101070007800120), and Shanghai 460 Food Microbiology Engineering Research Center (grant No. 19DZ2281100).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Rogosa, M.; Sharpe, M.E. An Approach to the Classification of the Lactobacilli. J. Appl. Bacteriol. 1960, 22, 329–340. [Google Scholar]
- Rogosa, M.; Wiseman, R.F.; Mitchell Joyce, A.; Disraely, M.N.; Beaman, A.J. Species Differentiation of Oral Lactobacilli from Man Including Descriptions of Lactobacillus salivarius Nov Spec and Lactobacillus cellobiosus Nov Spec. J. Bacteriol. 1953, 65, 681–699. [Google Scholar] [CrossRef]
- Neville, B.A.; O’Toole, P.W. Probiotic Properties Of Lactobacillus salivarius and Closely Related Lactobacillus Species. Future Microbiol. 2010, 5, 759–774. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Dec, M.; Stępień-Pyśniak, D.; Puchalski, A.; Hauschild, T.; Pietras-Ożga, D.; Ignaciuk, S.; Urban-Chmiel, R. Biodiversity of Ligilactobacillus salivarius Strains from Poultry and Domestic Pigeons. Animals 2021, 11, 972. [Google Scholar] [CrossRef]
- Puchalski, A.; Nowaczek, A.; Wernicki, A.; Dec, M. Antimicrobial Activity of Lactobacillus Strains of Chicken Origin against Bacterial Pathogens. Int. Microbiol. 2016, 19, 57–67. [Google Scholar]
- de los Reyes-Gavilán, C.G.; Fernández, M.; Hudson, J.A.; Korpela, R. Role of Microorganisms Present in Dairy Fermented Products in Health and Disease. Biomed. Res. Int. 2015, 2015, 204173. [Google Scholar] [CrossRef]
- Zhai, Q.; Shen, X.; Cen, S.; Zhang, C.; Tian, F.; Zhao, J.; Zhang, H.; Xue, Y.; Chen, W. Screening of Lactobacillus salivarius Strains from the Feces of Chinese Populations and the Evaluation of their Effects against Intestinal Inflammation in Mice. Food Funct. 2020, 11, 221–235. [Google Scholar] [CrossRef]
- Lugli, G.A.; Longhi, G.; Alessandri, G.; Mancabelli, L.; Tarracchini, C.; Fontana, F.; Turroni, F.; Milani, C.; Di Pierro, F.; van Sinderen, D.; et al. The Probiotic Identity Card: A Novel “Probiogenomics” Approach to Investigate Probiotic Supplements. Front. Microbiol. 2022, 12, 790881. [Google Scholar] [CrossRef]
- Dash, J.; Sethi, M.; Deb, S.; Parida, D.; Kar, S.; Mahapatra, S.; Minz, A.P.; Pradhan, B.; Prasad, P.; Senapati, S. Biochemical, Functional and Genomic Characterization of a New Probiotic Ligilactobacillus salivarius F14 from the Gut of Tribes of Odisha. World. J. Microb. Biot. 2023, 39, 171. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Yang, R.S.; Lin, Y.C.; Xin, W.G.; Zhou, H.Y.; Wang, F.; Zhang, Q.L.; Lin, L.B. Assessment of the Safety and Probiotic Characteristics of Lactobacillus salivarius CGMCC20700 Based on Whole-Genome Sequencing and Phenotypic Analysis. Front. Microbiol. 2023, 14, 1120263. [Google Scholar] [CrossRef]
- Chouraddi, R.; Kumar, S.; Kumar, B.; Bhatia, M.; Varada, V.V.; Tyagi, N.; Mallapa, R.H. Techno-Functional Characterization of Fecal Lactobacilli Isolates of Bos indicus Calves for Probiotic Properties. Vet. Res. Commun. 2023, 47, 1285–1302. [Google Scholar] [CrossRef]
- Fernández, L.; Castro, I.; Arroyo, R.; Alba, C.; Beltrán, D.; Rodríguez, J.M. Immunomodulation of the Vaginal Ecosystem by Ligilactobacillus salivarius CECT 30632 Improves Pregnancy Rates among Women with Infertility of Unknown Origin or Habitual Abortions. Nutrients 2023, 15, 362. [Google Scholar] [CrossRef]
- Cuevas-Gómez, I.; de Andrés, J.; Cardenas, N.; Espinosa-Martos, I.; Jiménez, E. Safety Assessment and Characterisation of Ligilactobacillus salivarius PS21603 as Potential Feed Additive for Swine. Benef. Microbes 2022, 13, 397–406. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro Selection Criteria for Probiotic Bacteria of Human Origin: Correlation with in vivo Findings1,2,3,4. Am. J. Clin. Nutr. 2001, 73, 386s–392s. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, J.; Zhang, M.; Ge, S.; Zhao, L. Probiotic Lactobacillus salivarius Ren Prevent Dimethylhydrazine-Induced Colorectal Cancer through Protein Kinase B Inhibition. Appl. Microbiol. Biot. 2020, 104, 7377–7389. [Google Scholar] [CrossRef]
- Wang, R.; Jiang, L.; Zhang, M.; Zhao, L.; Hao, Y.; Guo, H.; Sang, Y.; Zhang, H.; Ren, F. The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-Layer Proteins. Sci. Rep. 2017, 7, 44029. [Google Scholar] [CrossRef]
- Zhang, M.; Qiao, X.; Zhao, L.; Jiang, L.; Ren, F. Lactobacillus salivarius REN Counteracted Unfavorable 4-Nitroquinoline-1-Oxide-Induced Changes in Colonic Microflora of Rats. J. Microbiol. 2011, 49, 877–883. [Google Scholar] [CrossRef]
- Lv, L.X.; Hu, X.J.; Qian, G.R.; Zhang, H.; Lu, H.F.; Zheng, B.W.; Jiang, L.; Li, L.J. Administration of Lactobacillus salivarius Li01 or Pediococcus pentosaceus Li05 Improves Acute Liver Injury Induced by D-Galactosamine in Rats. Appl. Microbiol. Biot. 2014, 98, 5619–5632. [Google Scholar] [CrossRef]
- Lara-Villoslada, F.; Sierra, S.; Díaz-Ropero, M.P.; Olivares, M.; Xaus, J. Safety Assessment of the Human Milk-Isolated Probiotic Lactobacillus salivarius CECT5713. J. Dairy Sci. 2007, 90, 3583–3589. [Google Scholar] [CrossRef]
- Pérez-Cano, F.J.; Dong, H.; Yaqoob, P. In vitro Immunomodulatory Activity of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713: Two Probiotic Strains Isolated from Human Breast Milk. Immunobiology 2010, 215, 996–1004. [Google Scholar] [CrossRef]
- Lee, T.H.; Park, D.; Kim, Y.J.; Lee, I.; Kim, S.; Oh, C.T.; Kim, J.Y.; Yang, J.; Jo, S.K. Lactobacillus salivarius BP121 Prevents Cisplatin-Induced Acute Kidney Injury by Inhibition of Uremic Toxins such as Indoxyl Sulfate and P-Cresol Sulfate via Alleviating Dysbiosis. Int. J. Mol. Med. 2020, 45, 1130–1140. [Google Scholar] [CrossRef]
- Oh, H.W.; Jeun, G.H.; Jin, L.; Chun, T.H.; Kim, S.H. Probiotics Inhibit Lipopolysaccharide-Induced Interleukin-8 Secretion from Intestinal Epithelial Cells. Food Sci. Animal. Resour. 2012, 32, 434–440. [Google Scholar] [CrossRef][Green Version]
- Zhou, B.; Albarracin, L.; Indo, Y.; Arce, L.; Masumizu, Y.; Tomokiyo, M.; Islam, M.A.; Garciacastillo, V.; Ikedaohtsubo, W.; Nochi, T. Selection of Immunobiotic Ligilactobacillus salivarius Strains from the Intestinal Tract of Wakame-Fed Pigs: Functional and Genomic Studies. Microorganisms 2020, 8, 1659. [Google Scholar] [CrossRef]
- Daniel, C.; Poiret, S.; Goudercourt, D.; Dennin, V.; Leyer, G.; Pot, B. Selecting Lactic Acid Bacteria for their Safety and Functionality by Use of a Mouse Colitis Model. Appl. Environ. Microb. 2006, 72, 5799–5805. [Google Scholar] [CrossRef]
- Jiménez, E.; Manzano, S.; Schlembach, D.; Arciszewski, K.; Martin, R.; Ben Amor, K.; Roelofs, M.; Knol, J.; Rodríguez, J.M.; Abou-Dakn, M.; et al. Ligilactobacillus salivarius PS2 Supplementation during Pregnancy and Lactation Prevents Mastitis: A Randomised Controlled Trial. Microorganisms 2021, 9, 1933. [Google Scholar] [CrossRef]
- Quilodrán-Vega, S.; Albarracin, L.; Mansilla, F.; Arce, L.; Zhou, B.; Islam, M.A.; Tomokiyo, M.; Al Kassaa, I.; Suda, Y.; Kitazawa, H.; et al. Functional and Genomic Characterization of Ligilactobacillus salivarius TUCO-L2 Isolated from Lama Glama Milk: A Promising Immunobiotic Strain to Combat Infections. Front. Microbiol. 2020, 11, 608752. [Google Scholar] [CrossRef]
- Cárdenas, N.; Martín, V.; Arroyo, R.; López, M.; Carrera, M.; Badiola, C.; Jiménez, E.; Rodríguez, J.M. Prevention of Recurrent Acute Otitis Media in Children through the Use of Lactobacillus salivarius PS7, a Target-Specific Probiotic Strain. Nutrients 2019, 11, 376. [Google Scholar] [CrossRef]
- Rejish Kumar, V.J.; Seo, B.J.; Mun, M.R.; Kim, C.J.; Lee, I.; Kim, H.; Park, Y.H. Putative Probiotic Lactobacillus spp. from Porcine Gastrointestinal Tract Inhibit Transmissible Gastroenteritis Coronavirus and Enteric Bacterial Pathogens. Trop. Anim. Health Prod. 2010, 42, 1855–1860. [Google Scholar]
- Pascual, M.; Hugas, M.; Badiola Jose, I.; Monfort Josep, M.; Garriga, M. Lactobacillus salivarius CTC2197 Prevents Salmonella Enteritidis Colonization in Chickens. Appl. Environ. Microb. 1999, 65, 4981–4986. [Google Scholar] [CrossRef]
- Lone, A.; Mottawea, W.; Ait Chait, Y.; Hammami, R. Dual Inhibition of Salmonella enterica and Clostridium perfringens by New Probiotic Candidates Isolated from Chicken Intestinal Mucosa. Microorganisms 2021, 9, 166. [Google Scholar] [CrossRef]
- Thirabunyanon, M.; Hongwittayakorn, P. Potential Probiotic Lactic Acid Bacteria of Human Origin Induce Antiproliferation of Colon Cancer Cells via Synergic Actions in Adhesion to Cancer Cells and Short-Chain Fatty Acid Bioproduction. Appl. Biochem. Biotech. 2013, 169, 511–525. [Google Scholar] [CrossRef]
- Tsai, C.C.; Lin, P.P.; Hsieh, Y.M. Three Lactobacillus Strains from Healthy Infant Stool Inhibit Enterotoxigenic Escherichia coli Grown in vitro. Anaerobe 2008, 14, 61–67. [Google Scholar] [CrossRef]
- Jia, G.; Liu, X.; Che, N.; Xia, Y.; Wang, G.; Xiong, Z.; Zhang, H.; Ai, L. Human-Origin Lactobacillus salivarius AR809 Protects against Immunosuppression in S. aureus-Induced Pharyngitis via Akt-Mediated NF-κB and Autophagy Signaling Pathways. Food Funct. 2020, 11, 270–284. [Google Scholar] [CrossRef]
- Jia, G.C.; Che, N.; Xia, Y.J.; Lai, P.F.; Xiong, Z.Q.; Wang, G.Q.; Zhang, H.; Ai, L.Z. Adhesion to Pharyngeal Epithelium and Modulation of Immune Response: Lactobacillus salivarius AR809, a Potential Probiotic Strain Isolated from the Human Oral Cavity. J. Dairy Sci. 2019, 102, 6738–6749. [Google Scholar] [CrossRef]
- Jung, J.I.; Baek, S.M.; Nguyen, T.H.; Kim, J.W.; Kang, C.H.; Kim, S.; Imm, J.Y. Effects of Probiotic Culture Supernatant on Cariogenic Biofilm Formation and RANKL-Induced Osteoclastogenesis in RAW 264.7 Macrophages. Molecules 2021, 26, 733. [Google Scholar] [CrossRef]
- Sajedinejad, N.; Paknejad, M.; Houshmand, B.; Sharafi, H.; Jelodar, R.; Shahbani Zahiri, H.; Noghabi, K.A. Lactobacillus salivarius NK02: A Potent Probiotic for Clinical Application in Mouthwash. Probiotics Antimicrob. 2018, 10, 485–495. [Google Scholar] [CrossRef]
- Kang, C.H.; Han, S.H.; Kim, Y.; Paek, N.S.; So, J.S. In Vitro Probiotic Properties of Lactobacillus salivarius MG242 Isolated from Human Vagina. Probiotics Antimicrob. 2018, 10, 343–349. [Google Scholar] [CrossRef]
- Zárate, G.; Nader-Macias, M.E. Influence of Probiotic Vaginal Lactobacilli on in Vitro Adhesion of Urogenital Pathogens to Vaginal Epithelial Cells. Lett. Appl. Microbiol. 2006, 43, 174–180. [Google Scholar] [CrossRef]
- de Llano, D.G.; Arroyo, A.; Cárdenas, N.; Rodríguez, J.M.; Moreno-Arribas, M.V.; Bartolomé, B. Strain-Specific Inhibition of the Adherence of Uropathogenic Bacteria to Bladder Cells by Probiotic Lactobacillus spp. Pathog. Dis. 2017, 75, ftx043. [Google Scholar] [CrossRef]
- Wei, Z.; He, Z.; Wang, T.; Wang, X.; Wang, T.; Long, M. Lactobacillus salivarius WZ1 Inhibits the Inflammatory Injury of Mouse Jejunum Caused by Enterotoxigenic Escherichia coli K88 by Regulating the TLR4/NF-kB/MyD88 Inflammatory Pathway and Gut Microbiota. Microorganisms 2023, 11, 657. [Google Scholar] [CrossRef]
- Yang, Y.; Song, X.; Xiong, Z.; Xia, Y.; Wang, G.; Ai, L. Complete Genome Sequence of Lactobacillus salivarius AR809, a Probiotic Strain with Oropharyngeal Tract Resistance and Adhesion to the Oral Epithelial Cells. Curr. Microbiol. 2022, 79, 280. [Google Scholar] [CrossRef]
- Foster, J.; Hall, H. Inducible Ph Homeostasis and the Acid Tolerance Response of Salmonella Typhimurium. J. Bacteriol. 1991, 173, 5129–5135. [Google Scholar] [CrossRef]
- Aleksandrzak-Piekarczyk, T.; Puzia, W.; Żylińska, J.; Cieśla, J.; Górecki, R.K. Potential of Lactobacillus plantarum IBB3036 and Lactobacillus salivarius IBB3154 to Persistence in Chicken after in Ovo Delivery. MicrobiologyOpen 2018, 8, e00620. [Google Scholar] [CrossRef]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile Resistance Mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef]
- Fidanza, M.; Panigrahi, P.; Kollmann, T.R. Lactiplantibacillus Plantarum–Nomad and Ideal Probiotic. Front. Microbiol. 2021, 12, 712236. [Google Scholar] [CrossRef]
- Pfeiler, E.A.; Klaenhammer, T.R. Role of Transporter Proteins in Bile Tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 2009, 75, 6013–6016. [Google Scholar] [CrossRef]
- Fang, F.; Li, Y.; Bumann, M.; Raftis Emma, J.; Casey Pat, G.; Cooney Jakki, C.; Walsh Martin, A.; O’Toole Paul, W. Allelic Variation of Bile Salt Hydrolase Genes in Lactobacillus salivarius Does not Determine Bile Resistance Levels. J. Bacteriol. 2009, 191, 5743–5757. [Google Scholar] [CrossRef]
- Pan, Q.; Shen, X.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Comparative Genomic Analysis Determines the Functional Genes Related to Bile Salt Resistance in Lactobacillus salivarius. Microorganisms 2021, 9, 2038. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Y.; Luo, A.; Heng, X.; Liu, J.; Wang, H.; Chu, W. Lactobacillus salivarius CPU-01 Ameliorates Temozolomide-Induced Intestinal Mucositis by Modulating Gut Microbiota, Maintaining Intestinal Barrier, and Blocking Pro-Inflammatory Cytokines. Probiotics Antimicrob. 2022, 15, 1079–1091. [Google Scholar] [CrossRef]
- Lv, L.X.; Yan, R.; Shi, H.Y.; Shi, D.; Fang, D.Q.; Jiang, H.Y.; Wu, W.R.; Guo, F.F.; Jiang, X.W.; Gu, S.L.; et al. Integrated Transcriptomic and Proteomic Analysis of the Bile Stress Response in Probiotic Lactobacillus salivarius LI01. J. Proteomics 2017, 150, 216–229. [Google Scholar] [CrossRef]
- Yadav, A.K.; Tyagi, A.; Kumar, A.; Saklani, A.C.; Grover, S.; Batish, V.K. Adhesion of Indigenous Lactobacillus plantarum to Gut Extracellular Matrix and Its Physicochemical Characterization. Arch. Microbiol. 2015, 197, 155–164. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, F.; Jiang, L.; Liu, R.; Zhang, L.; Lei, X.; Li, J.; Jiang, J.; Guo, H.; Fang, B. Lactobacillus salivarius REN Inhibits Rat Oral Cancer Induced by 4-Nitroquioline 1-Oxide. Cancer Prev. Res. 2013, 6, 686–694. [Google Scholar] [CrossRef]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Li, X.; Tian, M.; Du, S.; Guo, H.; Liu, C.; Zhu, N.; et al. Inhibition of Staphylococcus aureus Adherence to Caco-2 Cells by Lactobacilli and Cell Surface Properties that Influence Attachment. Anaerobe 2012, 18, 508–515. [Google Scholar] [CrossRef]
- Liu, S. Research on the Antioxidant Ability and Mechanism of Different Lactic Acid Bacteria. Master’s thesis, Northeast Agricultural University, Harbin, China, 2015. (In Chinese). [Google Scholar]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Kulyar, M.F.e.A.; Pan, H.; Li, K.; Li, A.; Mo, Q.; Wang, Y.; Dong, H.; Bao, Y.; et al. Complete Genome Analysis of Lactobacillus fermentum YLF016 and Its Probiotic Characteristics. Microbial. Pathogenesis 2022, 162, 105212. [Google Scholar] [CrossRef]
- Zhai, Q.; Yin, R.; Yu, L.; Wang, G.; Tian, F.; Yu, R.; Zhao, J.; Liu, X.; Chen, Y.Q.; Zhang, H.; et al. Screening of Lactic Acid Bacteria with Potential Protective Effects against Cadmium Toxicity. Food Control 2015, 54, 23–30. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Jing, N.; Zhang, H.; Xie, Y.; Liu, H.; Shan, X.; Ren, J.; Jin, J. Bifidobacterium animalis A12 and Lactobacillus salivarius M18-6 Alleviate Alcohol Injury by Keap1-Nrf2 Pathway and Thioredoxin System. Foods 2023, 12, 439. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Zhao, L.; Zhao, Y.; Yang, G.; Wang, C.; Gao, L.; Niu, C.; Li, S. Lactobacillus plantarum DP189 Reduces A-SYN Aggravation in MPTP-Induced Parkinson’s Disease Mice via Regulating Oxidative Damage, Inflammation, and Gut Microbiota Disorder. J. Agric. Food Chem. 2022, 70, 1163–1173. [Google Scholar] [CrossRef]
- Kang, C.H.; Kim, J.S.; Park, H.M.; Kim, S.; Paek, N.-S. Antioxidant Activity and Short-Chain Fatty Acid Production of Lactic Acid Bacteria Isolated from Korean Individuals and Fermented Foods. 3 Biotech. 2021, 11, 217. [Google Scholar] [CrossRef]
- Chooruk, A.; Piwat, S.; Teanpaisan, R. Antioxidant Activity of Various Oral Lactobacillus Strains. J. Appl. Microbiol. 2017, 123, 271–279. [Google Scholar] [CrossRef]
- Nurrahma, B.A.; Tsao, S.P.; Wu, C.H.; Yeh, T.H.; Hsieh, P.S.; Panunggal, B.; Huang, H.Y. Probiotic Supplementation Facilitates Recovery of 6-OHDA-Induced Motor Deficit via Improving Mitochondrial Function and Energy Metabolism. Front. Aging Neurosci. 2021, 13, 668775. [Google Scholar] [CrossRef]
- Domalaon, R.; Zhanel, G.G.; Schweizer, F. Short Antimicrobial Peptides and Peptide Scaffolds as Promising Antibacterial Agents. Curr. Top. Med. Chem. 2016, 16, 1217–1230. [Google Scholar] [CrossRef]
- Langa, S.; Arqués, J.; Medina, M.; Landete, J. Coproduction of Colicin V and Lactic Acid Bacteria Bacteriocins in Lactococci and Enterococci Strains of Biotechnological Interest. J. Appl. Microbiol. 2017, 122, 1159–1167. [Google Scholar] [CrossRef]
- Ołdak, A.; Zielińska, D. Bacteriocins from Lactic Acid Bacteria as an Alternative to Antibiotics. Postep. Hig. Med. Dosw. 2017, 71, 328–338. [Google Scholar] [CrossRef]
- Pei, J.; Li, X.; Han, H.; Tao, Y. Purification and Characterization of Plantaricin SLG1, a Novel Bacteriocin Produced by Lb. Plantarum Isolated from Yak Cheese. Food Control 2018, 84, 111–117. [Google Scholar] [CrossRef]
- Martín, R.; Jiménez, E.; Olivares, M.; Marín, M.L.; Fernández, L.; Xaus, J.; Rodríguez, J.M. Lactobacillus salivarius CECT 5713, a Potential Probiotic Strain Isolated from Infant Feces and Breast Milk of a Mother–Child Pair. Int. J. Food Microbiol. 2006, 112, 35–43. [Google Scholar] [CrossRef]
- Wasfi, R.; Abd El-Rahman, O.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. Inhibit Growth, Biofilm Formation and Gene Expression of Caries-Inducing Streptococcus Mutans. J. Cell. Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef]
- Wang, K.; Niu, M.; Song, D.; Song, X.; Zhao, J.; Wu, Y.; Lu, B.; Niu, G. Preparation, Partial Characterization and Biological Activity of Exopolysaccharides Produced from Lactobacillus fermentum S1. J. Biosci. Bioeng. 2020, 129, 206–214. [Google Scholar] [CrossRef]
- Bikric, S.; Aslim, B.; Dincer, İ.; Yuksekdag, Z.; Ulusoy, S.; Yavuz, S. Characterization of Exopolysaccharides (EPSs) Obtained from Ligilactobacillus salivarius Strains and Investigation at the Prebiotic Potential as an Alternative to Plant Prebiotics at Poultry. Probiotics Antimicrob. 2022, 14, 49–59. [Google Scholar] [CrossRef]
- Chen, Y.M.; Limaye, A.; Chang, H.W.; Liu, J.R. Screening of Lactic Acid Bacterial Strains with Antiviral Activity against Porcine Epidemic Diarrhea. Probiotics Antimicrob. 2022, 14, 546–559. [Google Scholar] [CrossRef]
- Shojadoost, B.; Kulkarni, R.R.; Brisbin, J.T.; Quinteiro-Filho, W.; Alkie, T.N.; Sharif, S. Interactions between Lactobacilli and Chicken Macrophages Induce Antiviral Responses against Avian Influenza Virus. Res. Vet. Sci. 2019, 125, 441–450. [Google Scholar] [CrossRef]
- Enaud, R.; Vandenborght, L.E.; Coron, N.; Bazin, T.; Prevel, R.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Lamireau, T.; Delhaes, L. The Mycobiome: A Neglected Component in the Microbiota-Gut-Brain Axis. Microorganisms 2018, 6, 22. [Google Scholar] [CrossRef]
- Lavelle, A.; Hill, C. Gut Microbiome in Health and Disease: Emerging Diagnostic Opportunities. Gastroenterol. Clin. 2019, 48, 221–235. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Chen, P.; Chen, X.; Pan, L.; Han, L.; Zhu, T. Alteration of the Gut Microbiota in Missed Abortion. Indian J. Microbiol. 2023, 63, 106–119. [Google Scholar] [CrossRef]
- Pham, V.T.; Dold, S.; Rehman, A.; Bird, J.K.; Steinert, R.E. Vitamins, the Gut Microbiome and Gastrointestinal Health in Humans. Nutr. Res. 2021, 95, 35–53. [Google Scholar] [CrossRef]
- Al-Rashidi, H.E. Gut Microbiota and Immunity Relevance in Eubiosis and Dysbiosis. Saudi J. Biol. Sci. 2022, 29, 1628–1643. [Google Scholar] [CrossRef]
- Surono, I.S.; Simatupang, A.; Kusumo, P.D.; Waspodo, P.; Verbruggen, S.; Verhoeven, J.; Venema, K. Effect of Different Functional Food Supplements on the Gut Microbiota of Prediabetic Indonesian Individuals during Weight Loss. Nutrients 2022, 14, 781. [Google Scholar] [CrossRef]
- Veiga, P.; Pons, N.; Agrawal, A.; Oozeer, R.; Guyonnet, D.; Brazeilles, R.; Faurie, J.-M.; van Hylckama Vlieg, J.E.T.; Houghton, L.A.; Whorwell, P.J.; et al. Changes of the Human Gut Microbiome Induced by a Fermented Milk Product. Sci. Rep. 2014, 4, 6328. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Y.; Wang, X.; Wang, S.; Bi, D. The Impact of Lactobacillus plantarum on the Gut Microbiota of Mice with DSS-Induced Colitis. BioMed Res. Int. 2019, 2019, 3921315. [Google Scholar]
- Ng, S.C.; Hart, A.L.; Kamm, M.A.; Stagg, A.J.; Knight, S.C. Mechanisms of Action of Probiotics: Recent Advances. Inflamm. Bowel Dis. 2009, 15, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.G.; Li, X.D.; Lin, Y.C.; Jiang, Y.H.; Xu, M.Y.; Zhang, Q.L.; Wang, F.; Lin, L.B. Whole Genome Analysis of Host-Associated Lactobacillus salivarius and the Effects on Hepatic Antioxidant Enzymes and Gut Microorganisms of Sinocyclocheilus Grahami. Front. Microbiol. 2022, 13, 1014970. [Google Scholar] [CrossRef]
- Hong, Y.; Zhou, Z.; Yu, L.; Jiang, K.; Xia, J.; Mi, Y.; Zhang, C.; Li, J. Lactobacillus salivarius and Lactobacillus agilis Feeding Regulates Intestinal Stem Cells Activity by Modulating Crypt Niche in Hens. Appl. Microbiol. Biot. 2021, 105, 8823–8835. [Google Scholar] [CrossRef]
- Saint-Cyr, M.J.; Haddad, N.; Taminiau, B.; Poezevara, T.; Quesne, S.; Amelot, M.; Daube, G.; Chemaly, M.; Dousset, X.; Guyard-Nicodème, M. Use of the Potential Probiotic Strain Lactobacillus salivarius SMXD51 to Control Campylobacter jejuni in Broilers. Int. J. Food Microbiol. 2017, 247, 9–17. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Faseleh Jahromi, M.; Liang, J.B.; Ramasamy, K.; Sieo, C.C.; Ho, Y.W. Effects of a Lactobacillus salivarius Mixture on Performance, Intestinal Health and Serum Lipids of Broiler Chickens. PLoS ONE 2017, 12, e0175959. [Google Scholar] [CrossRef]
- Chen, F.; Zhu, L.; Qiu, H. Isolation and Probiotic Potential of Lactobacillus salivarius and Pediococcus pentosaceus in Speci C Pathogen Free Chickens. Rev. Bras. De Ciência Avícola 2017, 19, 325–332. [Google Scholar] [CrossRef]
- Xu, C.; Wei, F.; Yang, X.; Feng, Y.; Liu, D.; Hu, Y. Lactobacillus salivarius CML352 Isolated from Chinese Local Breed Chicken Modulates the Gut Microbiota and Improves Intestinal Health and Egg Quality in Late-Phase Laying Hens. Microorganisms 2022, 10, 726. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Yan, H.; Ning, Z.; Wang, Z. Lactobacillus salivarius SNK-6 Activates Intestinal Mucosal Immune System by Regulating Cecal Microbial Community Structure in Laying Hens. Microorganisms 2022, 10, 1469. [Google Scholar] [CrossRef]
- Shi, D.; Lv, L.; Fang, D.; Wu, W.; Hu, C.; Xu, L.; Chen, Y.; Guo, J.; Hu, X.; Li, A.; et al. Administration of Lactobacillus salivarius LI01 or Pediococcus pentosaceus LI05 Prevents CCl4-Induced Liver Cirrhosis by Protecting the Intestinal Barrier in Rats. Sci. Rep. 2017, 7, 6927. [Google Scholar] [CrossRef]
- Zhuge, A.; Li, S.; Yuan, Y.; Li, B.; Li, L. The Synergy of Dietary Supplements Lactobacillus salivarius LI01 and Bifidobacterium longum TC01 in Alleviating Liver Failure in Rats Treated with D-Galactosamine. Food Funct. 2021, 12, 10239–10252. [Google Scholar] [CrossRef] [PubMed]
- Sierra, S.; Lara-Villoslada, F.; Sempere, L.; Olivares, M.; Boza, J.; Xaus, J. Intestinal and Immunological Effects of Daily Oral Administration of Lactobacillus salivarius CECT5713 to Healthy Adults. Anaerobe 2010, 16, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Kyoung, H.; Park, K.I.; Oh, S.; Song, M.; Kim, Y. Postbiotic Heat-Killed Lactobacilli Modulates on Body Weight Associated with Gut Microbiota in a Pig Model. AMB Express 2022, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Moturi, J.; Kim, K.Y.; Hosseindoust, A.; Lee, J.H.; Xuan, B.; Park, J.; Kim, E.B.; Kim, J.S.; Chae, B.J. Effects of Lactobacillus salivarius Isolated from Feces of Fast-Growing Pigs on Intestinal Microbiota and Morphology of Suckling Piglets. Sci. Rep. 2021, 11, 6757. [Google Scholar] [CrossRef]
- Riboulet-Bisson, E.; Sturme, M.H.J.; Jeffery, I.B.; O’Donnell, M.M.; Neville, B.A.; Forde, B.M.; Claesson, M.J.; Harris, H.; Gardiner, G.E.; Casey, P.G.; et al. Effect of Lactobacillus salivarius Bacteriocin Abp118 on the Mouse and Pig Intestinal Microbiota. PLoS ONE 2012, 7, e31113. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Morton, J.M. The Human Gut Microbiome: A Review of the Effect of Obesity and Surgically Induced Weight Loss. JAMA Surg. 2013, 148, 563–569. [Google Scholar] [CrossRef]
- Samuel, B.S.; Gordon, J.I. A Humanized Gnotobiotic Mouse Model of Host–Archaeal–Bacterial Mutualism. Proc. Natl. Acad. Sci. USA 2006, 103, 10011–10016. [Google Scholar] [CrossRef]
- Li, N.; Yang, J.; Zhang, J.; Liang, C.; Wang, Y.; Chen, B.; Zhao, C.; Wang, J.; Zhang, G.; Zhao, D.; et al. Correlation of Gut Microbiome Between ASD Children and Mothers and Potential Biomarkers for Risk Assessment. Genom. Proteom. Bioinf. 2019, 17, 26–38. [Google Scholar] [CrossRef]
- Mohan, B.; Kadirvel, R.; Natarajan, A.; Bhaskaran, M. Effect of Probiotic Supplementation on Growth, Nitrogen Utilisation and Serum Cholesterol in Broilers. Brit. Poultry Sci. 1996, 37, 395–401. [Google Scholar] [CrossRef]
- Sharma, H.; Ozogul, F.; Bartkiene, E.; Rocha, J.M. Impact of Lactic Acid Bacteria and Their Metabolites on the Techno-Functional Properties and Health Benefits of Fermented Dairy Products. Crit. Rev. Food Sci. 2021, 63, 4819–4841. [Google Scholar] [CrossRef]
- Sun, E.; Ren, F.; Liu, S.; Ge, S.; Zhang, M.; Guo, H.; Jiang, L.; Zhang, H.; Zhao, L. Complete Genome Sequence of Lactobacillus salivarius Ren, a Probiotic Strain with Anti-Tumor Activity. J. Biotechnol. 2015, 210, 57–58. [Google Scholar] [CrossRef]
- Xu, Y.J. Foodomics: A Novel Approach for Food Microbiology. TrAC Trends Anal. Chem. 2017, 96, 14–21. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Binda, S.; Bron, P.A.; Gross, G.; Hill, C.; van Hylckama Vlieg, J.E.T.; Lebeer, S.; Satokari, R.; Ouwehand, A.C. Understanding Mode of Action can Drive the Translational Pipeline towards more Reliable Health Benefits for Probiotics. Curr. Opin. Biotech. 2019, 56, 55–60. [Google Scholar] [CrossRef]
- Song, X.; Zhang, X.; Xiong, Z.; Xia, Y.; Wu, Y.; Ai, L.; Xu, H.; Tian, Y.; Yang, Y.; Wang, G. Characterization of Endogenous Constitutive Promoters from Lactobacillus salivarius for Finely-Tuning Gene Expression. Food Biosci. 2022, 50, 101980. [Google Scholar] [CrossRef]
- Xia, J.; Jiang, S.; Lv, L.; Wu, W.; Wang, Q.; Xu, Q.; Ye, J.; Fang, D.; Li, Y.; Wu, J.; et al. Modulation of the Immune Response and Metabolism in Germ-Free Rats Colonized by the Probiotic Lactobacillus salivarius LI01. Appl. Microbiol. Biot. 2021, 105, 1629–1645. [Google Scholar] [CrossRef] [PubMed]
- Larance, M.; Lamond, A.I. Multidimensional Proteomics for Cell Biology. Nat. Rev. Mol. Cell Biol. 2015, 16, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Kang, M.S.; Lim, H.S.; Oh, J.S.; Lim, Y.j.; Wuertz-Kozak, K.; Harro, J.M.; Shirtliff, M.E.; Achermann, Y. Antimicrobial Activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog. Dis. 2017, 75, ftx009. [Google Scholar] [CrossRef]
- Kelly, P.; Maguire, P.B.; Bennett, M.; Fitzgerald, D.J.; Edwards, R.J.; Thiede, B.; Treumann, A.; Collins, J.K.; O’Sullivan, G.C.; Shanahan, F.; et al. Correlation of Probiotic Lactobacillus salivarius Growth Phase with Its Cell Wall-Associated Proteome. FEMS Microbiol. Lett. 2005, 252, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ait Seddik, H.; Bendali, F.; Cudennec, B.; Drider, D. Anti-Pathogenic and Probiotic Attributes of Lactobacillus salivarius and Lactobacillus plantarum Strains Isolated from Feces of Algerian Infants and Adults. Res. Microbiol. 2017, 168, 244–254. [Google Scholar] [CrossRef]
- de Andrés, J.; Jiménez, E.; Espinosa-Martos, I.; Rodríguez, J.M.; García-Conesa, M.T. An Exploratory Search for Potential Molecular Targets Responsive to the Probiotic Lactobacillus salivarius PS2 in Women with Mastitis: Gene Expression Profiling vs. Interindividual Variability. Front. Microbiol. 2018, 9, 2166. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, R.; Huang, J.; Xiao, C.; Yang, Y.; Wang, H.; He, D.; Yan, H.; Yang, C. Lactobacillus salivarius SNK-6 Regulates Liver Lipid Metabolism Partly via the Mir-130a-5p/MBOAT2 Pathway in a NAFLD Model of Laying Hens. Cells 2022, 11, 4133. [Google Scholar] [CrossRef]
- Vázquez-Fresno, R.; Llorach, R.; Marinic, J.; Tulipani, S.; Garcia-Aloy, M.; Espinosa-Martos, I.; Jiménez, E.; Rodríguez, J.M.; Andres-Lacueva, C. Urinary Metabolomic Fingerprinting after Consumption of a Probiotic Strain in Women with Mastitis. Pharmacol. Res. 2014, 87, 160–165. [Google Scholar] [CrossRef]
- Lu, H.; Chen, L.; Pan, X.; Yao, Y.; Zhang, H.; Zhu, X.; Lou, X.; Zhu, C.; Wang, J.; Li, L.; et al. Lactitol Supplementation Modulates Intestinal Microbiome in Liver Cirrhotic Patients. Front. Med. 2021, 8, 762930. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yu, L.; Cao, J.; Yu, J.; Lin, Z.; Hong, Y.; Jiang, S.; Chen, C.; Mi, Y.; Zhang, C.; et al. Lactobacillus salivarius Promotion of Intestinal Stem Cell Activity in Hens Is Associated with Succinate-Induced Mitochondrial Energy Metabolism. mSystems 2022, 7, e00903–e00922. [Google Scholar] [CrossRef]
- Qi, N.L.; Gong, X.; Yang, C.L.; Cheng, Z.H.; Li, J.H. 1H NMR-Based Metabolic Profile of Lactobacillus salivarius FDB89 under Osmotic Stress. Appl. Ecol. Environ. Res. 2018, 16, 3489–3500. [Google Scholar] [CrossRef]
- Jin, W.; Zhang, Z.; Zhu, K.; Xue, Y.; Xie, F.; Mao, S. Comprehensive Understanding of the Bacterial Populations and Metabolites Profile of Fermented Feed by 16S rRNA Gene Sequencing and Liquid Chromatography–Mass Spectrometry. Metabolites 2019, 9, 239. [Google Scholar] [CrossRef]
- Fuochi, V.; Coniglio, M.A.; Laghi, L.; Rescifina, A.; Caruso, M.; Stivala, A.; Furneri, P.M. Metabolic Characterization of Supernatants Produced by Lactobacillus spp. with in vitro Anti-Legionella Activity. Front. Microbiol. 2019, 10, 1403. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).