A Magnetic Beads-Based Sandwich Chemiluminescence Enzyme Immunoassay for the Rapid and Automatic Detection of Lactoferrin in Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
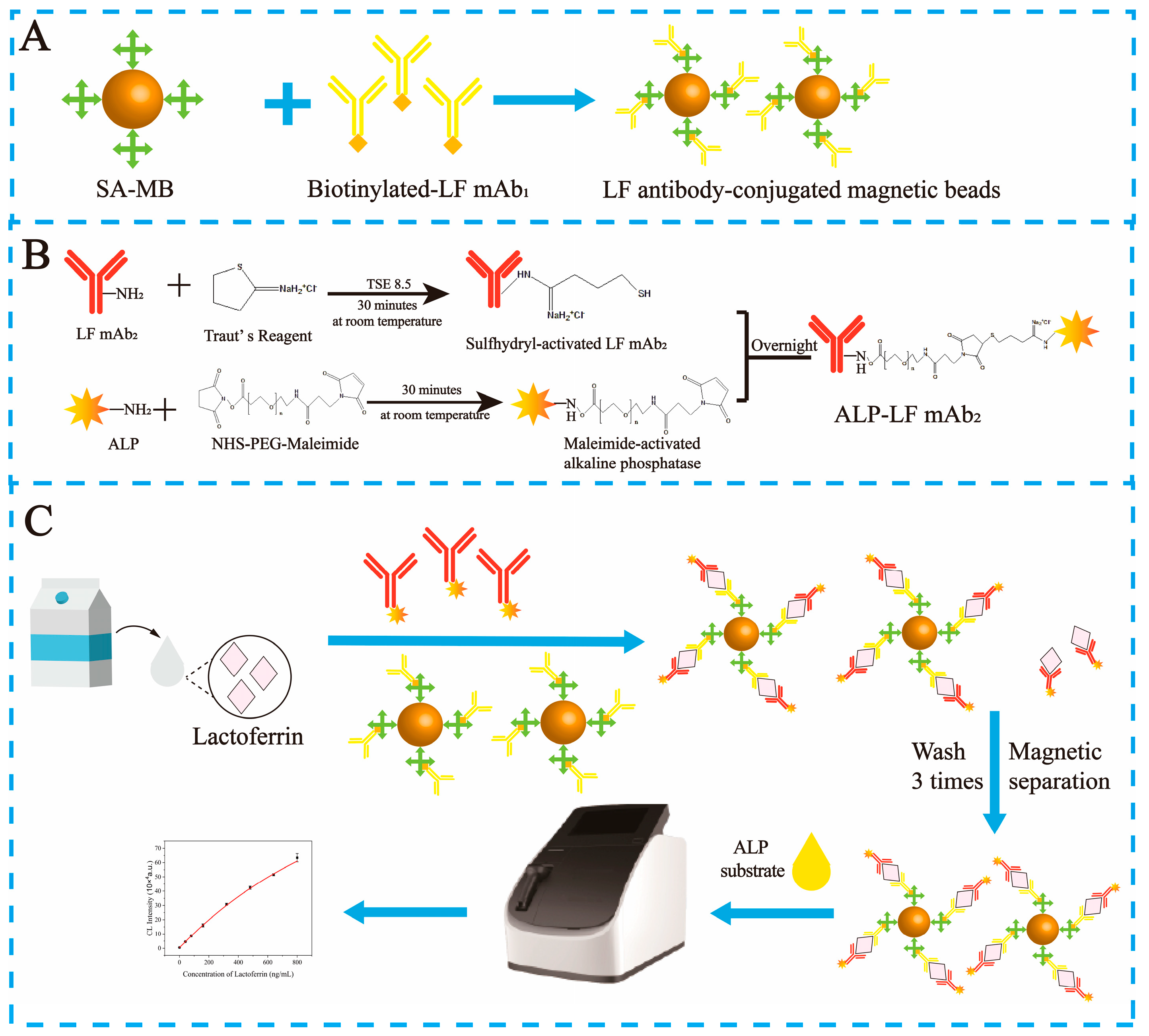
2.2. Preparation of mAb1-Labeled Biotin
2.3. Preparation of mAb2-Labeled Alkaline Phosphatase
2.4. Immobilization of the Biotin- Conjugated mAb1 on the Streptavidin Magnetic Beads
2.5. Establishment of MBs-sCLEIA for LF Detection
2.6. Data Analysis
2.7. Real Sample Detection
3. Results
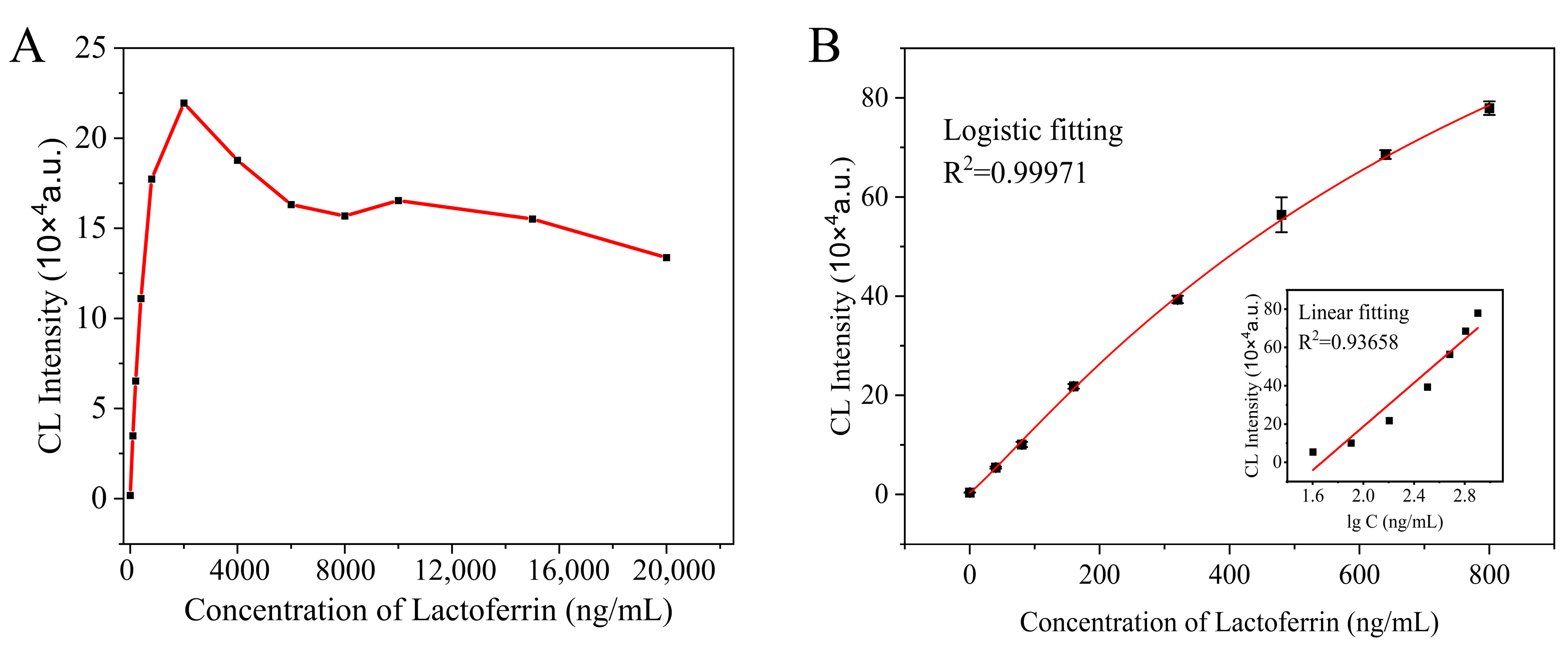
3.1. MBs−sCLEIA for Lactoferrin
3.2. Optimization of Experimental Conditions
3.3. Performance of MBs−sCLEIA
3.3.1. Accuracy and Precision
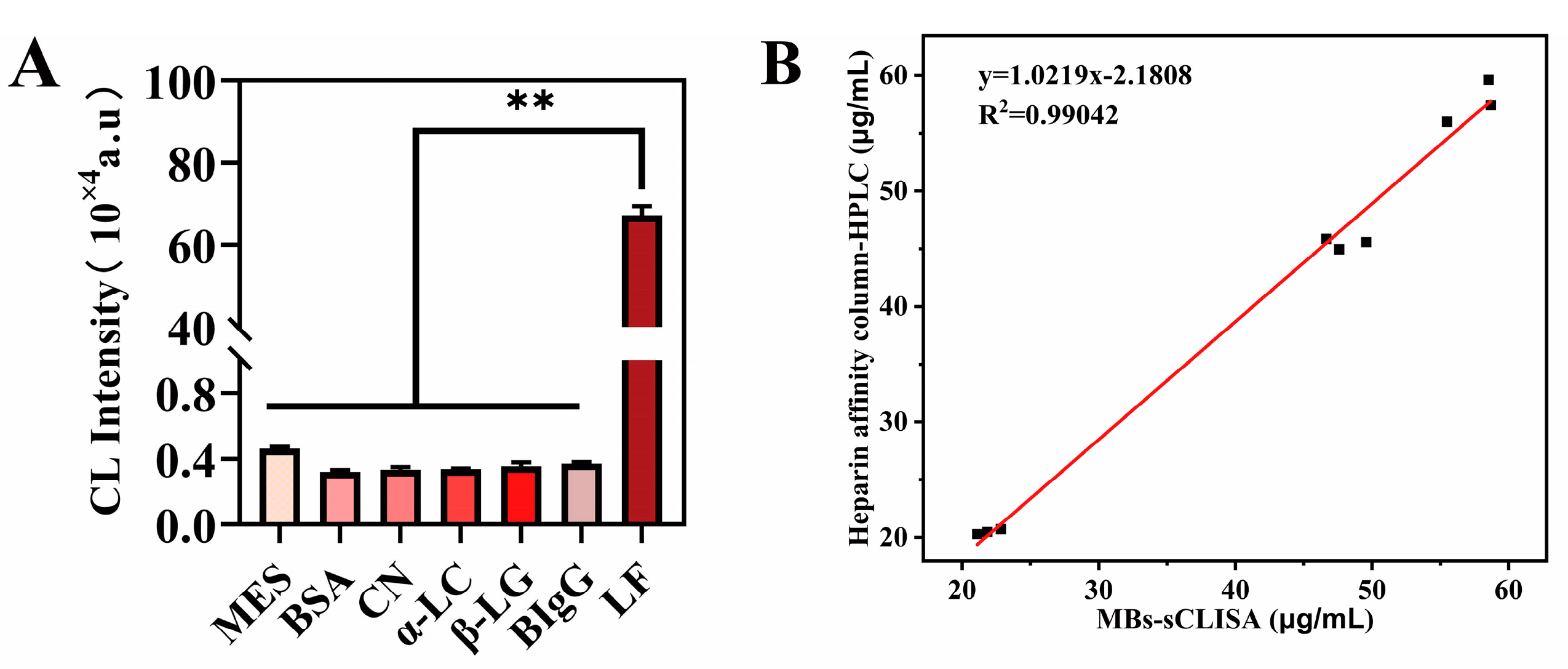
3.3.2. Specificity
3.4. Real Sample Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Caccavo, D.; Pellegrino, N.M.; Altamura, M.; Rigon, A.; Amati, L.; Amoroso, A.; Jirillo, E. Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. J. Endotoxin Res. 2002, 8, 403–417. [Google Scholar] [CrossRef]
- Adlerova, L.; Bartoskova, A.; Faldyna, M. Lactoferrin: A review. Veterinární Medicína 2008, 53, 457–468. [Google Scholar] [CrossRef]
- Cheng, J.B.; Wang, J.Q.; Bu, D.P.; Liu, G.L.; Zhang, C.G.; Wei, H.Y.; Zhou, L.Y.; Wang, J.Z. Factors affecting the lactoferrin concentration in bovine milk. J. Dairy Sci. 2008, 91, 970–976. [Google Scholar] [CrossRef]
- Safaeian, L.; Zabolian, H. Antioxidant effects of bovine lactoferrin on dexamethasone-induced hypertension in rat. ISRN Pharmacol. 2014, 2014, 943523. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, H.J.; Huang, L.Y.; Song, C.L.; Li, H.Q.; Zhao, X.H. Low-level Cu-fortification of bovine lactoferrin: Focus on its effect on in vitro anti-inflammatory activity in LPS-stimulated macrophages. Curr. Res. Food Sci. 2023, 6, 100520. [Google Scholar] [CrossRef]
- Jugert, C.S.; Didier, A.; Plotz, M.; Jessberger, N. Strain-specific antimicrobial activity of lactoferrin-based food supplements. J. Food Prot. 2023, 86, 100153. [Google Scholar] [CrossRef]
- Wotring, J.W.; Fursmidt, R.; Ward, L.; Sexton, J.Z. Evaluating the in vitro efficacy of bovine lactoferrin products against SARS-CoV-2 variants of concern. J. Dairy Sci. 2022, 105, 2791–2802. [Google Scholar] [CrossRef]
- Hu, P.; Zong, Q.; Zhao, Y.; Gu, H.; Liu, Y.; Gu, F.; Liu, H.Y.; Ahmed, A.A.; Bao, W.; Cai, D. Lactoferrin Attenuates Intestinal Barrier Dysfunction and Inflammation by Modulating the MAPK Pathway and Gut Microbes in Mice. J. Nutr. 2022, 152, 2451–2460. [Google Scholar] [CrossRef]
- Hwang, H.H.; Kim, H.S.; Lee, D.Y. Gastrointestinally absorbable lactoferrin-heparin conjugate with anti-angiogenic activity for treatment of brain tumor. J. Control Release 2023, 355, 730–744. [Google Scholar] [CrossRef]
- Ke, C.; Lan, Z.; Hua, L.; Ying, Z.; Humina, X.; Jia, S.; Weizheng, T.; Ping, Y.; Lingying, C.; Meng, M. Iron metabolism in infants: Influence of bovine lactoferrin from iron-fortified formula. Nutrition 2015, 31, 304–309. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, R.; Liu, J.; Lonnerdal, B. The bovine Lactoferrin-Osteopontin complex increases proliferation of human intestinal epithelial cells by activating the PI3K/Akt signaling pathway. Food Chem. 2020, 310, 125919. [Google Scholar] [CrossRef]
- Colombo, J.; Harris, C.L.; Wampler, J.L.; Zhuang, W.; Shaddy, D.J.; Liu, B.Y.; Wu, S.S. Improved Neurodevelopmental Outcomes at 5.5 Years of Age in Children Who Received Bovine Milk Fat Globule Membrane and Lactoferrin in Infant Formula through 12 Months: A Randomized Controlled Trial. J. Pediatr. 2023, 261, 113483. [Google Scholar] [CrossRef]
- El-Hawy, M.A.; Abd Al-Salam, S.A.; Bahbah, W.A. Comparing oral iron bisglycinate chelate, lactoferrin, lactoferrin with iron and iron polymaltose complex in the treatment of children with iron deficiency anemia. Clin. Nutr. ESPEN 2021, 46, 367–371. [Google Scholar] [CrossRef]
- Wagoner, T.B.; Foegeding, E.A. Whey protein–pectin soluble complexes for beverage applications. Food Hydrocoll. 2017, 63, 130–138. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Yamauchi, K.; Abe, F. Quality control of commercial bovine lactoferrin. Biometals 2018, 31, 313–319. [Google Scholar] [CrossRef]
- GB 14880-2012; National Food Safety Standard—Standard for the Use of Nutritional Fortification Substances in Foods. Ministry of Health of the People’s Republic of China, Standards Press of China: Beijing, China, 2012.
- Wang, M.; Zhang, F.; Tang, Y.; Mujahid Ali, M.; Shen, Z.; Atta Debrah, A.; Yuan, Q.; Du, Z. Boron-doped titania for separation and purification of lactoferrin in dairy products. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2022, 1212, 123501. [Google Scholar] [CrossRef]
- Chen, Y.; Qu, S.; Huang, Z.; Ren, Y.; Wang, L.; Rankin, S.A. Analysis and comparison of key proteins in Maiwa yak and bovine milk using high-performance liquid chromatography mass spectrometry. J. Dairy Sci. 2021, 104, 8661–8672. [Google Scholar] [CrossRef]
- Hurley, W.L.; Grieve, R.C.J.; Magura, C.E.; Hegarty, H.M.; Zou, S. Electrophoretic Comparisons of Lactoferrin from Bovine Mammary Secretions, Milk Neutrophils, and Human Milk. J. Dairy Sci. 1993, 76, 377–387. [Google Scholar] [CrossRef]
- Li, J.; Ding, X.; Chen, Y.; Song, B.; Zhao, S.; Wang, Z. Determination of bovine lactoferrin in infant formula by capillary electrophoresis with ultraviolet detection. J. Chromatogr. A 2012, 1244, 178–183. [Google Scholar] [CrossRef]
- Majka, G.; Spiewak, K.; Kurpiewska, K.; Heczko, P.; Stochel, G.; Strus, M.; Brindell, M. A high-throughput method for the quantification of iron saturation in lactoferrin preparations. Anal. Bioanal. Chem. 2013, 405, 5191–5200. [Google Scholar] [CrossRef]
- Fabian, O.; Daria, S.; Sonja, B.; Jörg, H. Development and validation of an enzyme-linked immunosorbent assay for the determination of bovine lactoferrin in various milk products. Int. Dairy J. 2022, 125, 105246. [Google Scholar] [CrossRef]
- Cooke, A.S.; Watt, K.A.; Albery, G.F.; Morgan, E.R.; Dungait, J.A.J. Lactoferrin quantification in cattle faeces by ELISA. PeerJ 2020, 8, e8631. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, C.; Zhang, J. Lactoferrin and Its Detection Methods: A Review. Nutrients 2021, 13, 2492. [Google Scholar] [CrossRef]
- Liu, L.; Kong, D.; Xing, C.; Zhang, X.; Kuang, H.; Xu, C. Sandwich immunoassay for lactoferrin detection in milk powder. Anal. Methods 2014, 6, 4742–4745. [Google Scholar] [CrossRef]
- Huang, J.; He, Z.; Cao, J.; Hong, J.; Wu, Z.; Gao, H.; Liao, X. Electrochemical immunosensor detection for lactoferrin in milk powder. Int. J. Electrochem. Sci. 2018, 13, 7816–7826. [Google Scholar] [CrossRef]
- Fateme, E.; Sala, A.H.; Ahmad, M.S. Impedimetric and single-frequency capacitance spectroscopy strategy in label-free rapid screening of lactoferrin. Sens. Actuators B Chem. 2022, 354, 131107. [Google Scholar] [CrossRef]
- Mauro, T.; Elisabetta, M.; Luigi, C.; Gabriele, F.; Gabriella, S.; Franco, M. Lactoferrin determination using flow or batch immunosensor surface plasmon resonance: Comparison with amperometric and screen-printed immunosensor methods. Sens. Actuators B Chem. 2013, 179, 215–225. [Google Scholar] [CrossRef]
- Fathi, N.; Saadati, A.; Alimohammadi, M.; Abolhassani, H.; Sharifi, S.; Rezaei, N.; Hasanzadeh, M. Biosensors for the detection of protein kinases: Recent progress and challenges. Microchem. J. 2022, 182, 107961. [Google Scholar] [CrossRef]
- Tan, H.; Chen, S.; Qiu, Y.; Ye, M.; Wu, Y.; Gao, J.; Meng, X.; Li, X.; Chen, H. Construction of sandwich ELISA method for quantitative detection of α-lactalbumin in hypoallergenic formula based on a novel monoclonal antibody prepared by the recombinant protein. Food Control 2023, 151, 109800. [Google Scholar] [CrossRef]
- Zhu, L.; Li, S.; Sun, L.; Zhao, J.; Huang, J.; Jiang, Y.; Wan, S.; Pavase, T.R.; Li, Z. Development and validation of a specific sandwich ELISA for determination of soybean allergens and its application in processed foods. Process Biochem. 2022, 117, 134–141. [Google Scholar] [CrossRef]
- He, Y.; Jiao, B. Detection of biotin-streptavidin interactions based on poly(thymine)-templated copper nanoparticles coupled with Exo III-aided DNA recycling amplification. Sens. Actuators B Chem. 2018, 265, 387–393. [Google Scholar] [CrossRef]
- Oberbichler, E.; Wiesauer, M.; Schlogl, E.; Stangl, J.; Faschinger, F.; Knor, G.; Gruber, H.J.; Hytonen, V.P. Competitive binding assay for biotin and biotin derivatives, based on avidin and biotin-4-fluorescein. Methods Enzym. 2020, 633, 1–20. [Google Scholar] [CrossRef]
- Chen, B.; Wang, C.; Wang, Y.M.; Liu, J.X. Effect of biotin on milk performance of dairy cattle: A meta-analysis. J. Dairy Sci. 2011, 94, 3537–3546. [Google Scholar] [CrossRef]
- Wang, N.; Jiang, X.; Xu, X.; Liu, Y.; Liu, L.; Lu, A.; Lu, J.; Luan, Y. An aptamer affinity column for purification and enrichment of lactoferrin in milk. J. Chromatogr. B 2021, 1178, 122724. [Google Scholar] [CrossRef]
- Pochet, S.; Arnould, C.; Debournoux, P.; Flament, J.; Rolet-Repecaud, O.; Beuvier, E. A simple micro-batch ion-exchange resin extraction method coupled with reverse-phase HPLC (MBRE-HPLC) to quantify lactoferrin in raw and heat-treated bovine milk. Food Chem. 2018, 259, 36–45. [Google Scholar] [CrossRef]
- Chen, M.; Wen, F.; Zhang, Y.; Li, P.; Zheng, N.; Wang, J. Determination of native lactoferrin in milk by HPLC on HiTrapTM Heparin HP column. Food Anal. Methods 2019, 12, 2518–2526. [Google Scholar] [CrossRef]
- Zhang, J.; Lai, S.; Cai, Z.; Chen, Q.; Huang, B.; Ren, Y. Determination of bovine lactoferrin in dairy products by ultra-high performance liquid chromatography-tandem mass spectrometry based on tryptic signature peptides employing an isotope-labeled winged peptide as internal standard. Anal. Chim. Acta 2014, 829, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Feng, C.; Wang, S.; Zhang, W.; Chen, M.; Jiang, H.; Feng, X. Selection of possible signature peptides for the detection of bovine lactoferrin in infant formulas by LC-MS/MS. PLoS ONE 2017, 12, e0184152. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Du, H.; Bai, L.; Zhang, Y.; Zhu, H.; Wang, Y. Poly (2-methyl-2-oxazoline) coating by thermally induced immobilization for determination of bovine lactoferrin in infant formula with capillary electrophoresis. Talanta 2017, 168, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, L.; Yang, G.; Irfan, M.; Wang, Z.; Fang, S.; Qu, F. High-efficiency selection of aptamers for bovine lactoferrin by capillary electrophoresis and its aptasensor application in milk powder. Talanta 2019, 205, 120088. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, J.; Liu, H.; Gao, Y.; Zhao, Q.; Ling, S.; Wang, S. Sensitive immunoassays based on specific monoclonal IgG for determination of bovine lactoferrin in cow milk samples. Food Chem. 2021, 338, 127820. [Google Scholar] [CrossRef] [PubMed]
- Shalini Devi, K.S.; Mahalakshmi, V.T.; Ghosh, A.R.; Kumar, A.S. Unexpected co-immobilization of lactoferrin and methylene blue from milk solution on a Nafion/MWCNT modified electrode and application to hydrogen peroxide and lactoferrin biosensing. Electrochim. Acta 2017, 244, 26–37. [Google Scholar] [CrossRef]
- AOAC Peer-Verified Methods Program, Manual on Policies and Procedures; AOAC International: Rockville, MD, USA, 1998.
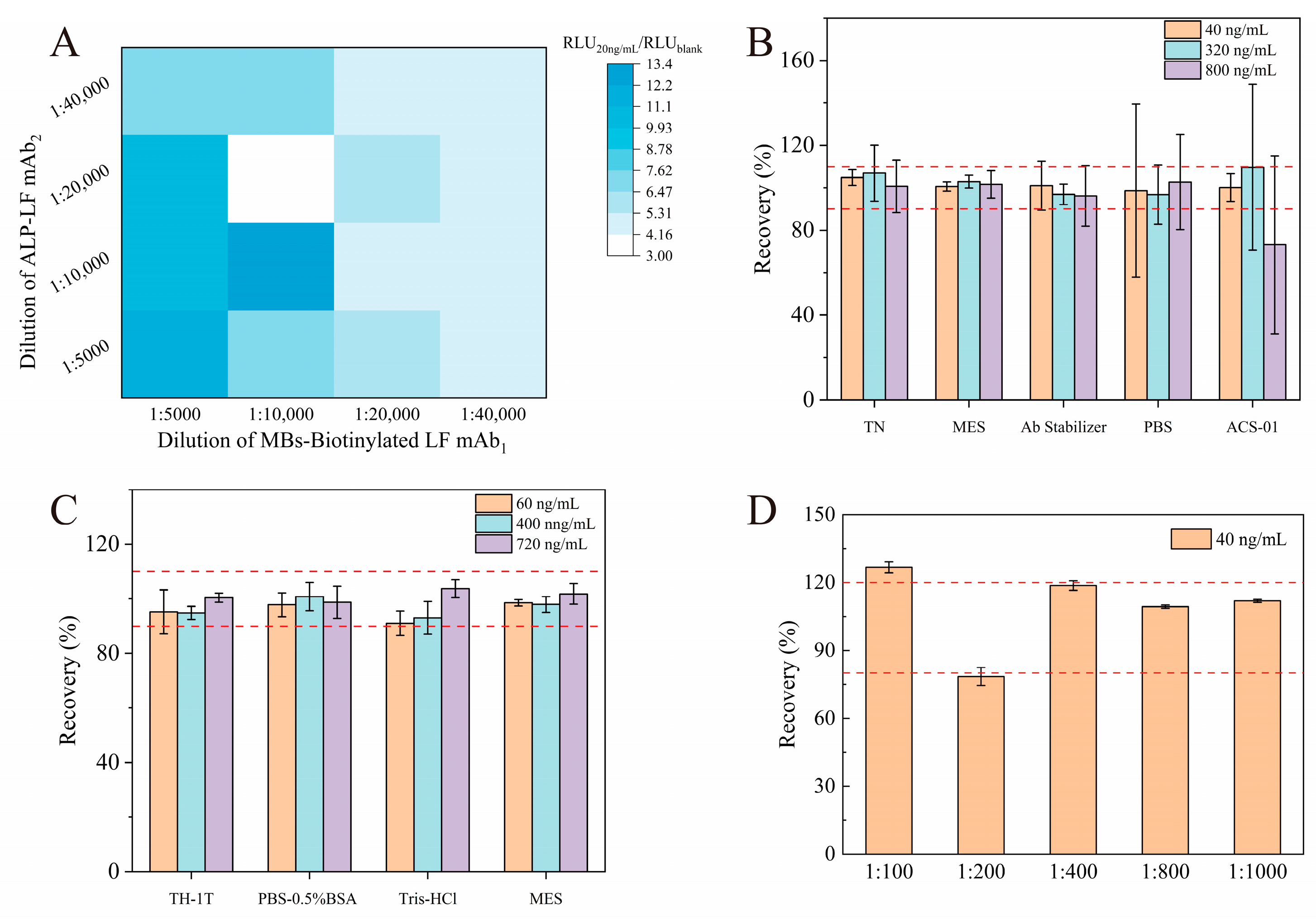
| Spiked (ng/mL) | Recovery (Mean ± SD) | |
|---|---|---|
| Logistic fitting | 60 | 98.5 ± 1.2% |
| 400 | 97.9 ± 2.9% | |
| 720 | 101.8 ± 3.9% | |
| Liner fitting | 60 | 101.8 ± 0.1% |
| 400 | 96.3 ± 0.8% | |
| 720 | 104.1 ± 1.1% |
| Materials or Strategy | LOD | Working Range | Test Time | Reference | |
|---|---|---|---|---|---|
| HPLC | Ion-exchange resin extraction | 2 µg /mL | 25–514 µg/mL | - | [36] |
| HiTrapTM Heparin HP column | 2 mg/L | 2–100 mg/L | - | [37] | |
| HPLC-MS/MS | Tryptic signature peptides as internal standard | 0.3 mg/100 g | 10–1000 nmol/L | - | [38] |
| 0.16 mg/100 g | 52.60–150.56 mg/100 g | - | [39] | ||
| Capillary electrophoresis | Thermally induced immobilization of poly (2-methyl-2-oxazoline) | 5.0 μg/mL | 10~500 μg/mL | <10 min | [40] |
| CE-Aptasensor | 1 nM | 4–128 nM | <15 min | [41] | |
| ELISA | Colloidal gold-based strip | 9.7 ng/mL | 9.76 ~ 625 ng/mL | <15 min | [42] |
| AuNFs-based strip | 2.4 ng/mL | ||||
| Competitive ELISA | 3.9 ng/mL | 100–520 ng/mL | >24 h | [22] | |
| Electrochemical detection | Impedance-derived single-frequency capacitive analysis | 65.2 nM | 125 nM–3.250 μM | <20 min | [27] |
| GCE/Nf-MWCNT@MB-LAF involved in the electron-shuttling process to facilitate the H2O2 reduction reaction | 3.2 μM | 25 μM–500 μM | 30 min | [43] | |
| Surface plasmon resonance transduction | SPR operating in batch mode method | 2.8 × 10−7 M | 0.5–3.50 × 10−6 M | >20 min | [28] |
| SPR operating in flow mode method | 5.0 × 10−8 M | 0.1–10.0 × 10−6 M | |||
| This work | MBs−sCLEIA | 2.85 ng/mL | 7.24–800 ng/mL | 17 min |
| Spiked (ng/mL) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|
| Mean ± SD | Recovery (%) | CV (%) | Mean ± SD | Recovery (%) | CV (%) | |
| 60 | 59.1 ± 0.7 | 98.5% | 1.2% | 60.4 ± 1.7 | 100.6% | 2.7% |
| 400 | 391.8 ± 11.8 | 97.9% | 2.9% | 400.7 ± 9.5 | 100.2% | 2.4% |
| 720 | 732.7 ± 28.1 | 101.8% | 3.9% | 735.9 ± 32.0 | 102.2% | 4.3% |
| MES | BSA | CN | α-LC | β-LG | BIgG | |
|---|---|---|---|---|---|---|
| BSA | 0.847 | |||||
| CN | 0.860 | 0.986 | ||||
| α-LC | 0.864 | 0.982 | 0.996 | |||
| β-LG | 0.884 | 0.962 | 0.976 | 0.980 | ||
| BIgG | 0.901 | 0.945 | 0.959 | 0.963 | 0.983 | |
| LF | 0.000 ** | 0.000 ** | 0.000 ** | 0.000 ** | 0.000 ** | 0.000 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, W.; Xuan, Z.; Liu, H.; Huang, K.; Guan, X.; Guo, B. A Magnetic Beads-Based Sandwich Chemiluminescence Enzyme Immunoassay for the Rapid and Automatic Detection of Lactoferrin in Milk. Foods 2024, 13, 953. https://doi.org/10.3390/foods13060953
Shen W, Xuan Z, Liu H, Huang K, Guan X, Guo B. A Magnetic Beads-Based Sandwich Chemiluminescence Enzyme Immunoassay for the Rapid and Automatic Detection of Lactoferrin in Milk. Foods. 2024; 13(6):953. https://doi.org/10.3390/foods13060953
Chicago/Turabian StyleShen, Wenjie, Zhihong Xuan, Hongmei Liu, Kai Huang, Xiao Guan, and Baoyuan Guo. 2024. "A Magnetic Beads-Based Sandwich Chemiluminescence Enzyme Immunoassay for the Rapid and Automatic Detection of Lactoferrin in Milk" Foods 13, no. 6: 953. https://doi.org/10.3390/foods13060953
APA StyleShen, W., Xuan, Z., Liu, H., Huang, K., Guan, X., & Guo, B. (2024). A Magnetic Beads-Based Sandwich Chemiluminescence Enzyme Immunoassay for the Rapid and Automatic Detection of Lactoferrin in Milk. Foods, 13(6), 953. https://doi.org/10.3390/foods13060953




