The Probiotic Kluyveromyces lactis JSA 18 Alleviates Obesity and Hyperlipidemia in High-Fat Diet C57BL/6J Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Cells, and Animals
2.1.1. Strains
2.1.2. Cells
2.1.3. Animals
2.2. Test for Tolerance to Acid and Bile
2.3. Self-Cohesion, Hydrophobicity Experiments, and Adhesion of Kluyveromyces lactate to Cells
2.4. Insulin Resistance (ITT) Analysis
2.5. Blood, Liver, WAT, Cecum, and Feces Collection
2.6. Serum Biochemical Parameter Analysis
2.7. Histological Analysis
2.8. Primer Design for Quantitative PCR
2.9. Quantitative Real-Time PCR
2.10. Analysis of Fecal SCFAs
2.11. Microbial Community Analysis
2.12. Statistical Analysis
3. Results
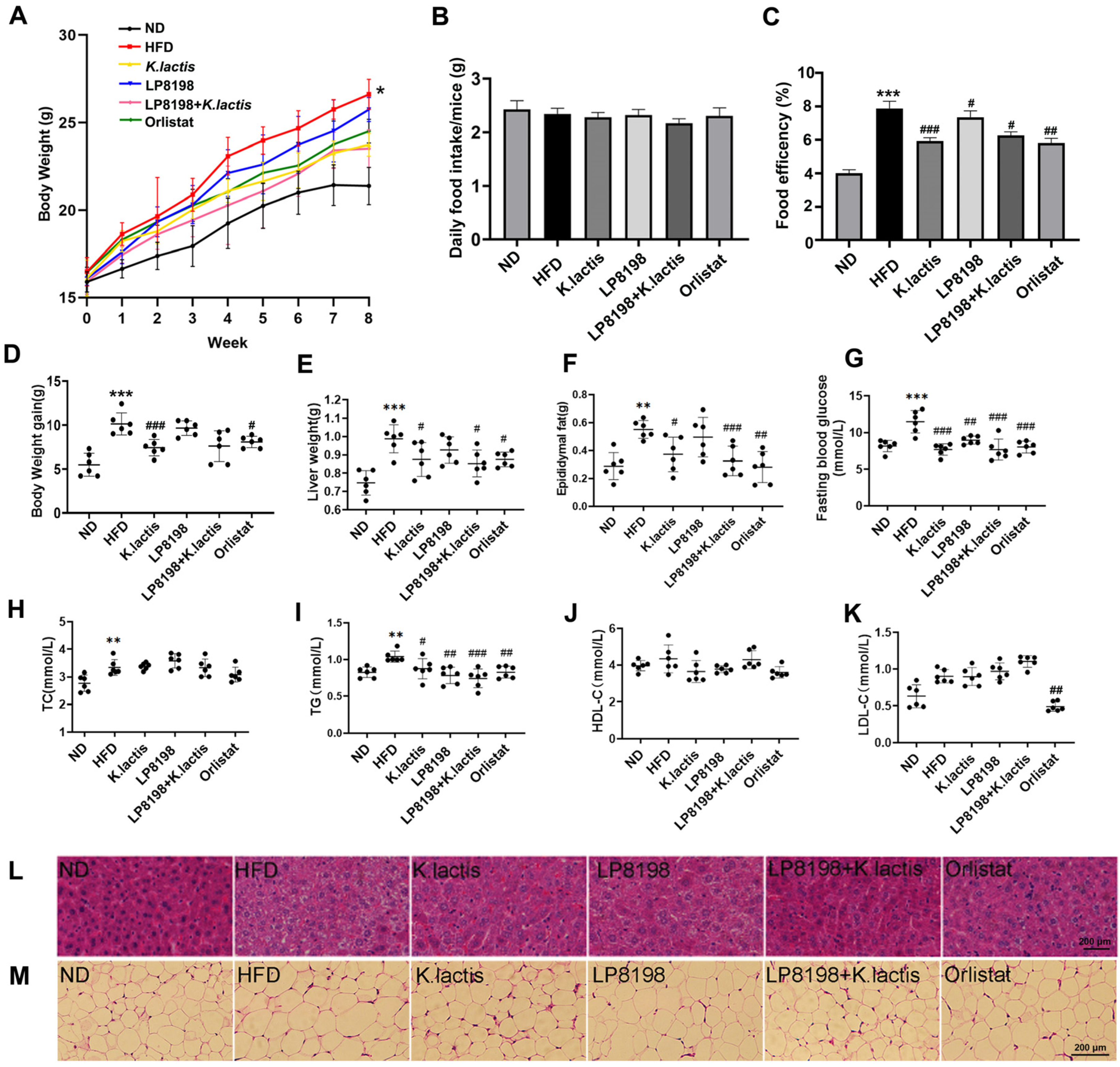
3.1. The Probiotic Strains K. lactis JSA18 and LP8198 Alleviated Body Weight Gain and Fat Accumulation in HFD-Fed Mice
3.2. The Probiotic Strains Showed an Alleviation of Physiological Changes Caused by a HFD
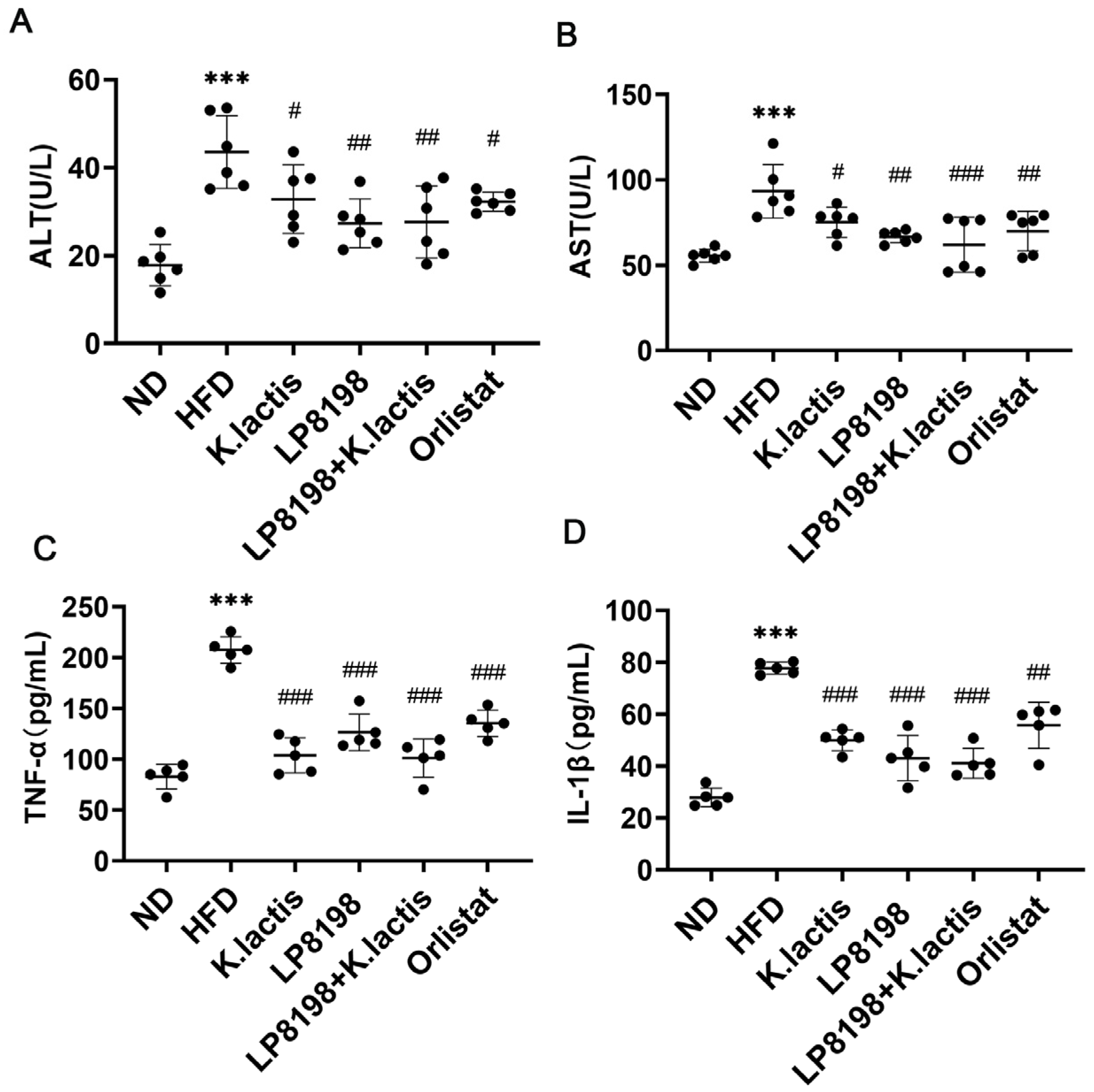
3.3. The Probiotic Strains Showed a Positive Effect on Liver Damage and Inflammation in the Serum Caused by a High-Fat Diet
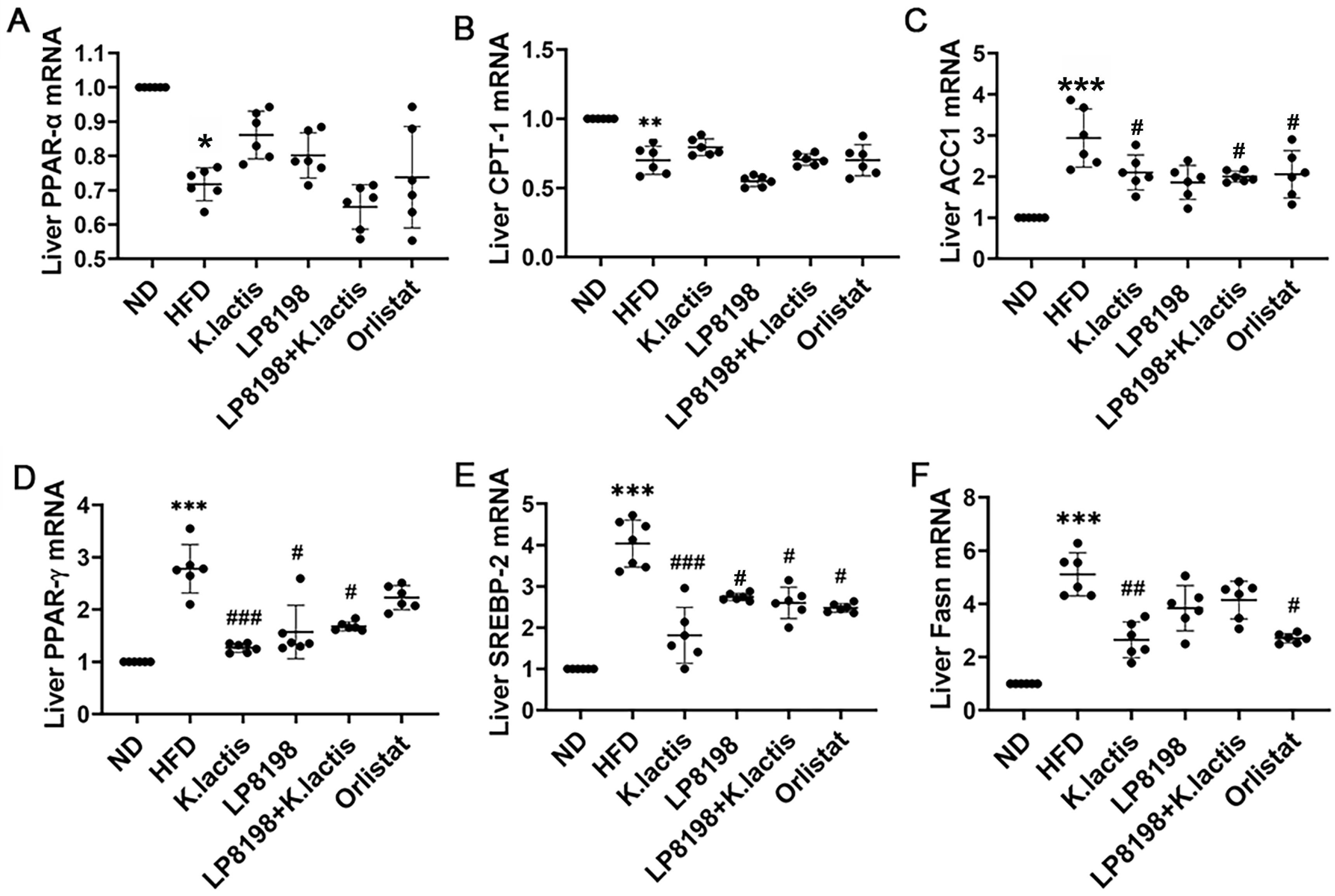
3.4. The Probiotic Strains Have an Influence on the Transcription of Genes Related to Lipid Metabolism
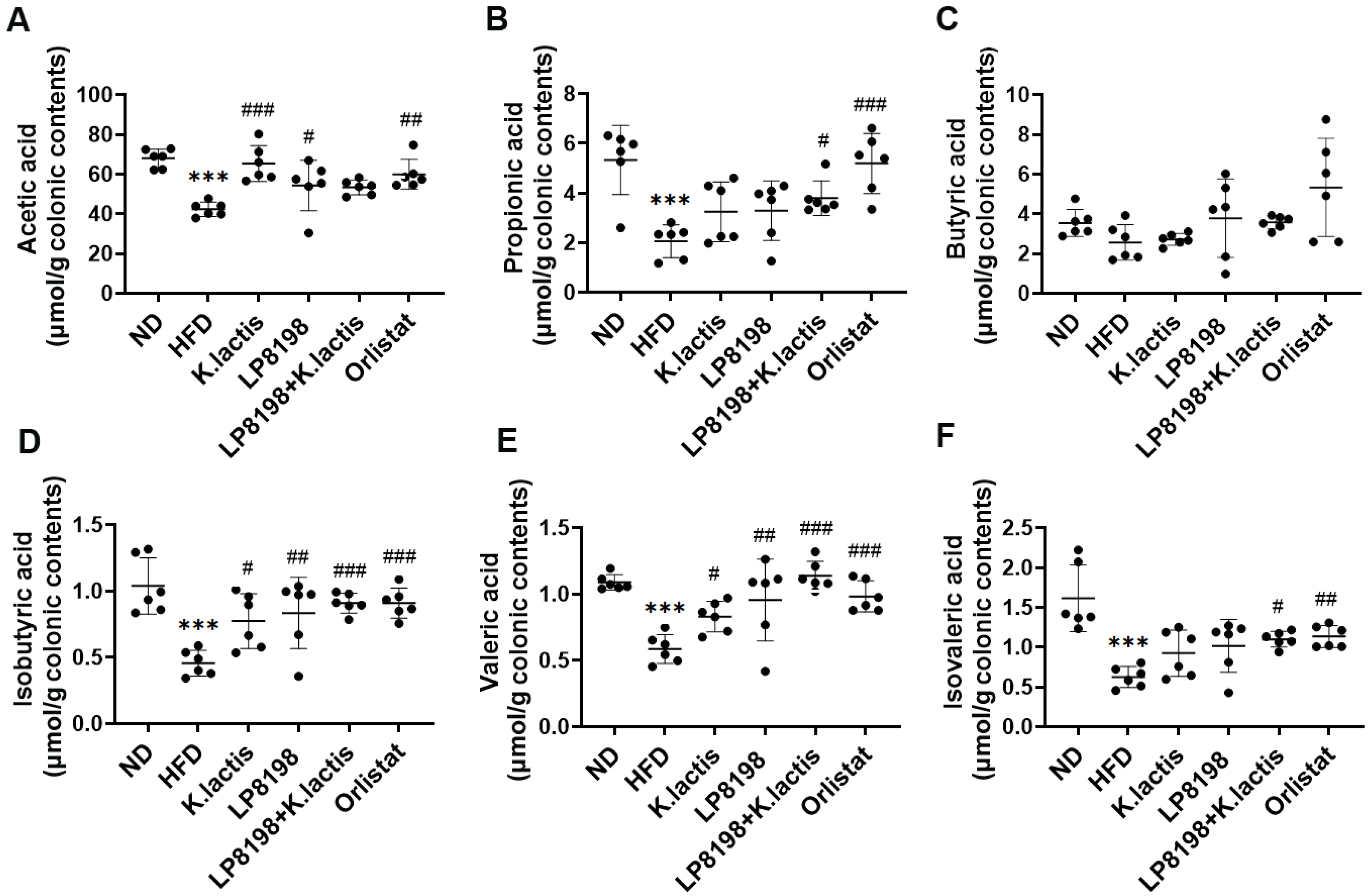
3.5. The Effects of Probiotic Strains on Short-Chain Fatty Acids (SCFAs) Metabolic Profiling
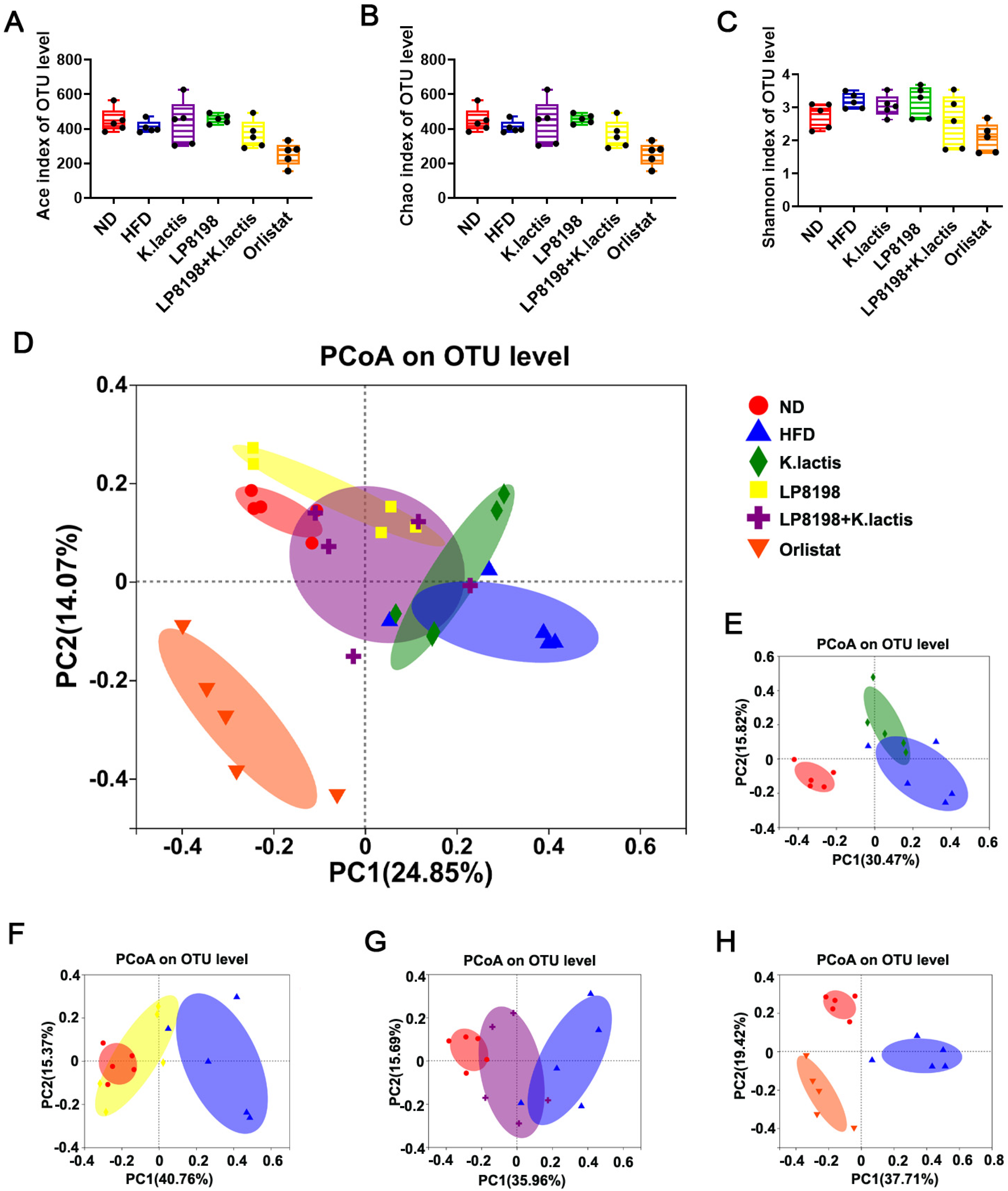
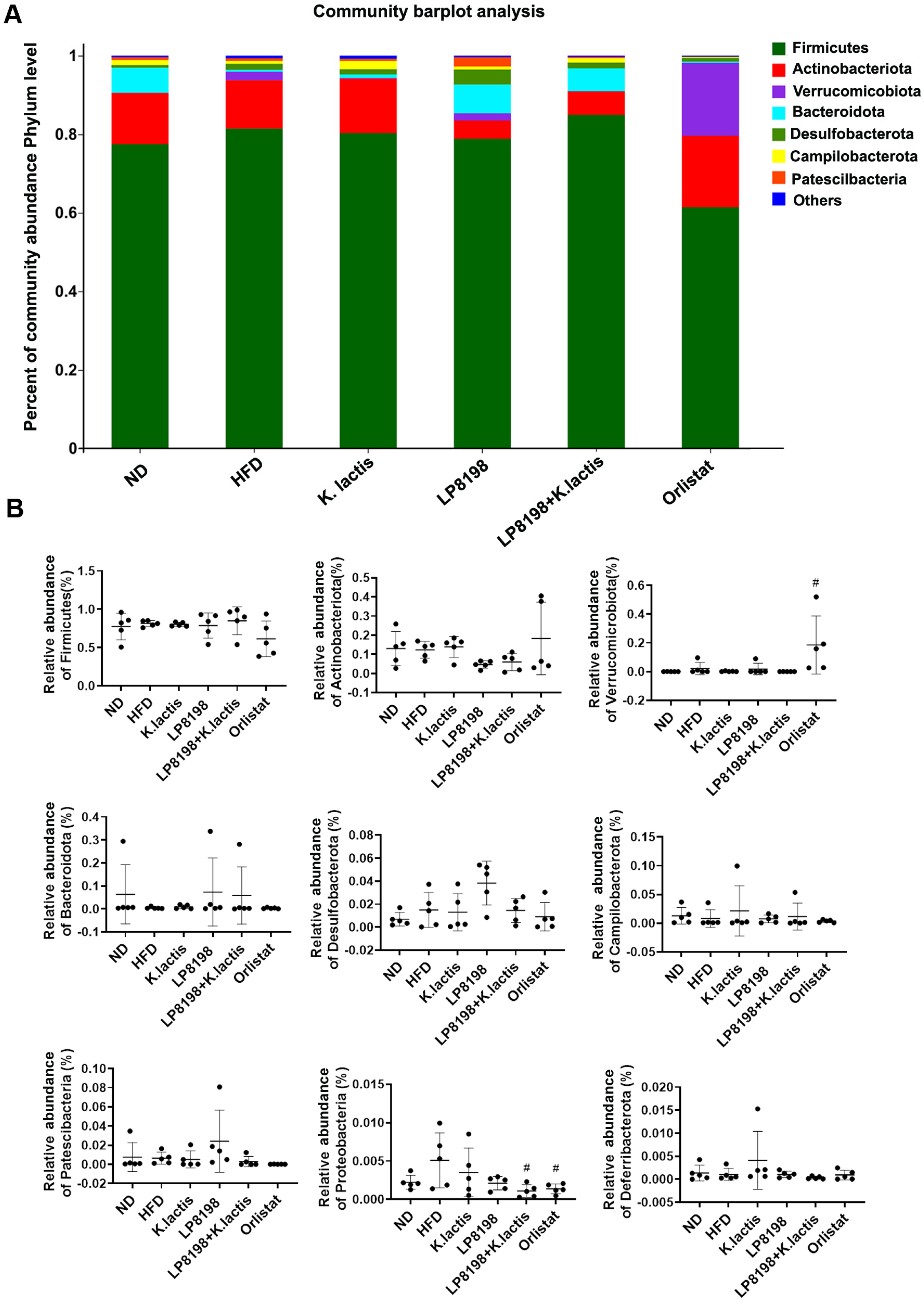
3.6. The Probiotic Strains Have an Effect on the Gut Microbial Composition of Mice That Are Fed with a High-Fat Diet
3.7. The Effect of HFD, K. lactis, and LP8198 Exposure on the Composition of the Intestinal Microbiome
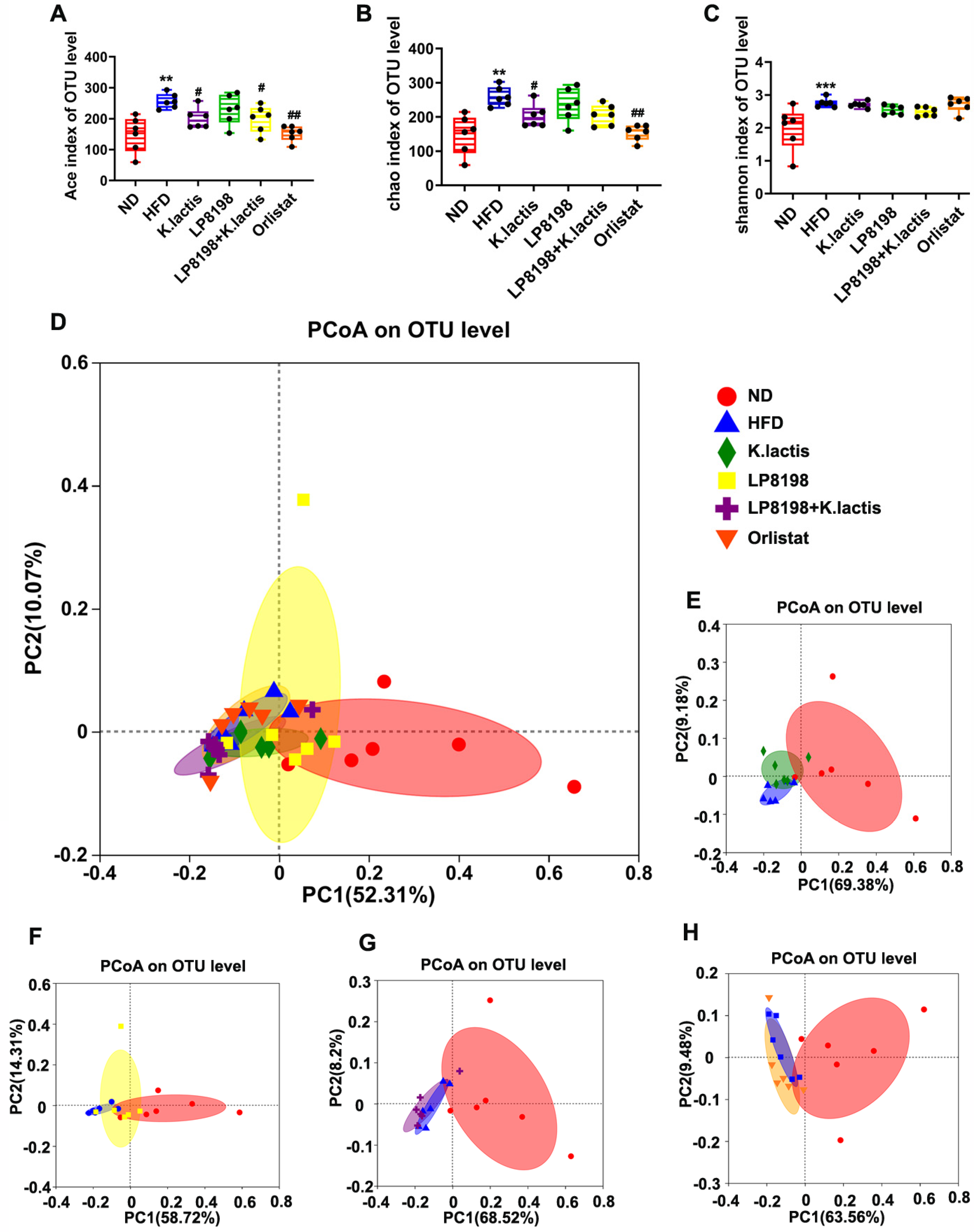
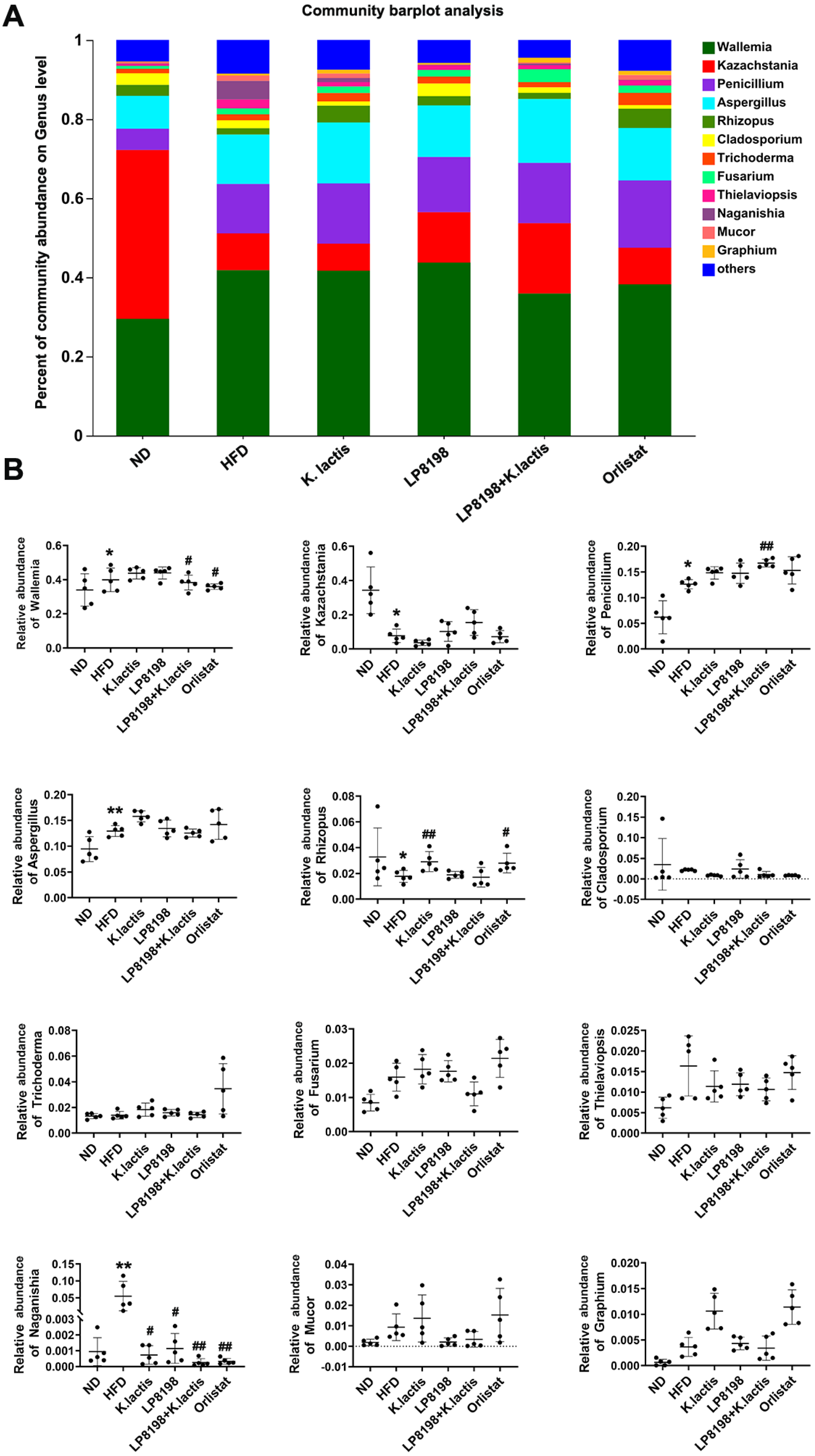
3.8. The Probiotic Bacteria Change the Gut Fungal Composition in Mice Consuming a High-Fat Diet
3.9. The Influence of Exposure to HFD, K. lactis, and LP8198 on the Composition of Intestinal Fungi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Apovian, C.M. Obesity: Definition, comorbidities, causes, and burden. Am. J. Manag. Care 2016, 22, s176–s185. [Google Scholar] [PubMed]
- Bray, G.A.; Kim, K.K.; Wilding, J.P.H.; World Obesity Federation. Obesity: A chronic relapsing progressive disease process. A position statement of the World Obesity Federation. Obes. Rev. 2017, 18, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Dubey, V.; Ghosh, A.R. Obesity: An overview of possible role(s) of gut hormones, lipid sensing and gut microbiota. Metab. Clin. Exp. 2016, 65, 48–65. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Vikram, N.K. Insulin resistance syndrome (metabolic syndrome) and obesity in Asian Indians: Evidence and implications. Nutrition 2004, 20, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, J.; Igarashi, M.; Watanabe, K.; Karaki, S.I.; Mukouyama, H.; Kishino, S.; Li, X.; Ichimura, A.; Irie, J.; Sugimoto, Y.; et al. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat. Commun. 2019, 10, 4007. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Ross, A.W.; Walker, A.W.; Morgan, P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 2017, 21, 1521–1533. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Ma, Y.; Zhang, F.; Zhao, C.; Nie, Y. Insights into the role of gut microbiota in obesity: Pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell 2018, 9, 397–403. [Google Scholar] [CrossRef]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef]
- Bleau, C.; Karelis, A.D.; St-Pierre, D.H.; Lamontagne, L. Crosstalk between intestinal microbiota, adipose tissue and skeletal muscle as an early event in systemic low-grade inflammation and the development of obesity and diabetes. Diabetes Metab. Res. Rev. 2015, 31, 545–561. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. The microbiota- gut-brain axis in obesity. Lancet Gastroenterol. Hepatol. 2017, 2, 747–756. [Google Scholar] [CrossRef]
- Li, Q.; Chen, H.; Zhang, M.; Wu, T.; Liu, R. Altered short chain fatty acid profiles induced by dietary fiber intervention regulate AMPK levels and intestinal homeostasis. Food Funct. 2019, 10, 7174–7187. [Google Scholar] [CrossRef] [PubMed]
- Nøhr, M.K.; Pedersen, M.H.; Gille, A.; Egerod, K.L.; Engelstoft, M.S.; Husted, A.S.; Sichlau, R.M.; Grunddal, K.V.; Poulsen, S.S.; Han, S.; et al. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 2013, 154, 3552–3564. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.H.; Chiou, J.; Tsai, T.Y. Effects of Lactobacillus plantarum TWK10-Fermented Soymilk on Deoxycorticosterone Acetate-Salt-Induced Hypertension and Associated Dementia in Rats. Nutrients 2016, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Liu, R.; Zhou, Z.; Xu, X.; Feng, S.; Kushmaro, A.; Marks, R.S.; Wang, D.; Sun, Q. Probiotic Characteristics of Lactiplantibacillus Plantarum N-1and Its Cholesterol-Lowering Effect in Hypercholesterolemic Rats. Probiotics Antimicrob. Proteins 2022, 14, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Ping, L.; Zhang, K.; Tang, H.; Liu, J.; Liu, D.; Zhao, L.; Evivie, S.E.; Li, B.; Huo, G. Immunomodulatory effects of mixed Lactobacillus plantarum on lipopolysaccharide-induced intestinal injury in mice. Food Funct. 2022, 13, 4914–4929. [Google Scholar] [CrossRef]
- Meng, Y.; Li, B.; Jin, D.; Zhan, M.; Lu, J.; Huo, G. Immunomodulatory activity of Lactobacillus plantarum KLDS1.0318 in cyclophosphamide-treated mice. Food Nutr. Res. 2018, 62. [Google Scholar] [CrossRef]
- Lee, Y.S.; Park, E.J.; Park, G.S.; Ko, S.H.; Park, J.; Lee, Y.K.; Kim, J.Y.; Lee, D.; Kang, J.; Lee, H.J. Lactiplantibacillus plantarum ATG-K2 Exerts an Anti-Obesity Effect in High-Fat Diet-Induced Obese Mice by Modulating the Gut Microbiome. Int. J. Mol. Sci. 2021, 22, 12665. [Google Scholar] [CrossRef]
- Lee, C.S.; Park, M.H.; Kim, B.K.; Kim, S.H. Antiobesity Effect of Novel Probiotic Strains in a Mouse Model of High-Fat Diet-Induced Obesity. Probiotics Antimicrob. Proteins 2021, 13, 1054–1067. [Google Scholar] [CrossRef]
- Won, S.M.; Chen, S.; Lee, S.Y.; Lee, K.E.; Park, K.W.; Yoon, J.H. Lactobacillus sakei ADM14 Induces Anti-Obesity Effects and Changes in Gut Microbiome in High-Fat Diet-Induced Obese Mice. Nutrients 2020, 12, 3703. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.W.; Pedroso, S.H.; Vieira, A.T.; Moreira, L.M.; França, C.S.; Cartelle, C.T.; Arantes, R.M.; Generoso, S.V.; Cardoso, V.N.; Neves, M.J.; et al. Saccharomyces cerevisiae UFMG A-905 treatment reduces intestinal damage in a murine model of irinotecan-induced mucositis. Benef. Microbes 2016, 7, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Pericolini, E.; Gabrielli, E.; Ballet, N.; Sabbatini, S.; Roselletti, E.; Cayzeele Decherf, A.; Pélerin, F.; Luciano, E.; Perito, S.; Jüsten, P.; et al. Therapeutic activity of a Saccharomyces cerevisiae-based probiotic and inactivated whole yeast on vaginal candidiasis. Virulence 2017, 8, 74–90. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.P.; Oliveira, D.R.; Lopes, A.C.A.; de Abreu, L.R.; Duarte, W.F. Survival of Kluyveromyces lactis and Torulaspora delbrueckii to simulated gastrointestinal conditions and their use as single and mixed inoculum for cheese production. Food Res. Int. 2019, 125, 108620. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.R.; Alencar Lopes, A.C.; Pereira, R.A.; Cardoso, P.G.; Duarte, W.F. Selection of potentially probiotic Kluyveromyces lactis for the fermentation of cheese whey–based beverage. Ann. Microbiol. 2019, 69, 1361–1372. [Google Scholar] [CrossRef]
- Psomas, E.I.; Fletouris, D.J.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Assimilation of cholesterol by yeast strains isolated from infant feces and feta cheese. J. Dairy. Sci. 2003, 86, 3416–3422. [Google Scholar] [CrossRef] [PubMed]
- Kunyeit, L.; Kurrey, N.K.; Anu-Appaiah, K.A.; Rao, R.P. Probiotic yeasts inhibit virulence of non-albicans Candida species. mBio 2019, 10, e02307-19. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Fontecilla, A.; Silva-Moreno, E.; Ganga, M.A.; Godoy, L. Evaluation of antimicrobial activity from native wine yeast against food industry pathogenic microorganisms. CyTA-J. Food 2017, 15, 457–465. [Google Scholar] [CrossRef]
- Fakruddin, M.; Hossain, M.N.; Ahmed, M.M. Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement. Altern. Med. 2017, 17, 64. [Google Scholar] [CrossRef]
- Saber, A.; Alipour, B.; Faghfoori, Z.; Khosroushahi, A.Y. Secretion metabolites of dairy Kluyveromyces marxianus AS41 isolated as probiotic, induces apoptosis in different human cancer cell lines and exhibit antipathogenic effects. J. Funct. Foods 2017, 34, 408–421. [Google Scholar] [CrossRef]
- Shamekhi, S.; Abdolalizadeh, J.; Ostadrahimi, A.; Mohammadi, S.A.; Barzegari, A.; Lotfi, H.; Bonabi, E.; Zarghami, N. Apoptotic effect of Saccharomyces cerevisiae on human colon cancer sw480 cells by regulation of Akt/NF-κB signaling pathway. Probiotics Antimicrob. Proteins 2020, 12, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Stier, H.; Bischoff, S.C. Saccharomyces boulardii CNCM I-745 beeinflusst das darmassoziierte Immunsystem. MMW-Fortschritte Med. 2017, 159 (Suppl. 5), 1–6. [Google Scholar]
- Rodríguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Utrilla, M.P.; Chueca, N.; García, F.; Rodríguez-Cabezas, M.E.; Gálvez, J. Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: Impact on microRNAs expression and gut microbiota composition. J. Nutr. Biochem. 2018, 61, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Fan, H.; Yao, M.; Yang, S.; Han, J. Oral administration of yeast β-glucan ameliorates inflammation and intestinal barrier in dextran sodium sulfate-induced acute colitis. J. Funct. Foods 2017, 35, 115–126. [Google Scholar] [CrossRef]
- Dong, Q.Q.; Hu, H.J.; Luo, X.G.; Wang, Q.T.; Gu, X.C.; Zhou, H.; Zhou, W.J.; Ni, X.M.; Zhang, T.C. Complete Genome Sequence of Lactobacillus plantarum CGMCC 8198. Genome Announc. 2017, 5, e01559-16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xue, J.; Shan, J.; Hong, Y.; Zhu, W.; Nie, Z.; Zhang, Y.; Ji, N.; Luo, X.; Zhang, T.; et al. Gut-Flora-Dependent Metabolite Trimethylamine-N-Oxide Promotes Atherosclerosis-Associated Inflammation Responses by Indirect ROS Stimulation and Signaling Involving AMPK and SIRT1. Nutrients 2022, 14, 3338. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Hsieh, Y.M.; Huang, C.C.; Tsai, C.C. Inhibitory Effects of Probiotic Lactobacillus on the Growth of Human Colonic Carcinoma Cell Line HT-29. Molecules 2017, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- Fan, J.G.; Kim, S.U.; Wong, V.W. New trends on obesity and NAFLD in Asia. J. Hepatol. 2017, 67, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Rokholm, B.; Baker, J.L.; Sorensen, T.I. The levelling off of the obesity epidemic since the year 1999-a review of evidence and perspectives. Obes. Rev. 2017, 18, 715–723. [Google Scholar] [CrossRef]
- Isomaa, B.; Almgren, P.; Tuomi, T.; Forsén, B.; Lahti, K.; Nissen, M.; Taskinen, M.R.; Groop, L. Cardiovascular morbidity and mortality associated with the metabolic syndrome. Diabetes Care 2001, 24, 683–689. [Google Scholar] [CrossRef]
- Bessesen, D.H.; Van Gaal, L.F. Progress and challenges in anti-obesity pharmacotherapy. Lancet Diabetes Endocrinol. 2018, 6, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kadooka, Y.; Sato, M.; Imaizumi, K.; Ogawa, A.; Ikuyama, K.; Akai, Y.; Okano, M.; Kagoshima, M.; Tsuchida, T. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur. J. Clin. Nutr. 2010, 64, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.T.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, D.A.; Puglisi, M.J.; Hasty, A.H. Impact of increased adipose tissue mass on inflammation, insulin resistance, and dyslipidemia. Curr. Diabetes Rep. 2009, 9, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Shimobayashi, M.; Albert, V.; Woelnerhanssen, B.; Frei, I.C.; Weissenberger, D.; Meyer-Gerspach, A.C.; Clement, N.; Moes, S.; Colombi, M.; Meier, J.A.; et al. Insulin resistance causes inflammation in adipose tissue. J. Clin. Investig. 2018, 128, 1538–1550. [Google Scholar] [CrossRef] [PubMed]
- Acosta Garcia, E. Obesity, adipose tissue and insulin resistance. Acta Bioquim. Clin. Latinoam. 2012, 46, 183–194. [Google Scholar]
- Do, G.M.; Oh, H.Y.; Kwon, E.Y.; Cho, Y.Y.; Shin, S.K.; Park, H.J.; Jeon, S.M.; Kim, E.; Hur, C.G.; Park, T.S.; et al. Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol. Nutr. Food Res. 2011, 55 (Suppl. 2), S173–S185. [Google Scholar] [CrossRef] [PubMed]
- Karpe, F.; Dickmann, J.R.; Frayn, K.N. Fatty acids, obesity, and insulin resistance: Time for a reevaluation. Diabetes 2011, 60, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Zhao, Y.H.; Vaziri, N.D.; Tang, D.D.; Chen, H.; Chen, H.; Khazaeli, M.; Tarbiat-Boldaji, M.; Hatami, L.; Zhao, Y.Y. Lipidomics Biomarkers of Diet-Induced Hyperlipidemia and Its Treatment with Poria cocos. J. Agric. Food Chem. 2016, 64, 969–979. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef]
- Siersbaek, R.; Nielsen, R.; Mandrup, S. PPARgamma in adipocyte differentiation and metabolism--novel insights from genome-wide studies. FEBS Lett. 2010, 584, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Wang, F.; Song, H.; Wang, Z.; Zuo, Z.; Cui, H.; Jia, Y.; Deng, J.; Yu, S.; Hu, Y.; et al. AMPK alpha pathway involved in hepatic triglyceride metabolism disorder in diet-induced obesity mice following Escherichia coli Infection. Aging 2018, 10, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Vasudevan, D.M. Monitoring oxidative stress in patients with non-alcoholic and alcoholic liver diseases. Indian. J. Clin. Biochem. 2005, 20, 24–28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knudsen, K.E.B. Microbial Degradation of Whole-Grain Complex Carbohydrates and Impact on Short-Chain Fatty Acids and Health. Adv. Nutr. 2015, 6, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Markowiak-Kopec, P.; Slizewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio 2019, 10, e02566-18. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Besten, G.D.; Bleeker, A.; Gerding, A.; Van Eunen, K.; Havinga, R.; Van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.-J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARγ-Dependent Switch from Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Boulangé, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Bratlie, M.; Hagen, I.V.; Helland, A.; Erchinger, F.; Midttun, Ø.; Ueland, P.M.; Rosenlund, G.; Sveier, H.; Mellgren, G.; Hausken, T.; et al. Effects of high intake of cod or salmon on gut microbiota profile, faecal output and serum concentrations of lipids and bile acids in overweight adults: A randomised clinical trial. Eur. J. Nutr. 2021, 60, 2231–2248. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; Da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohn’s Colitis 2016, 10, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.; Stewart, C.J.; A Kennedy, N.; Lodge, J.K.; Tremelling, M.; Probert, C.S.; Parkes, M.; Mansfield, J.C.; Smith, D.L.; Hold, G.L.; et al. The impact of NOD2 genetic variants on the gut mycobiota in Crohn’s disease patients in remission and in individuals without gastrointestinal inflammation. J. Crohn’s Colitis 2021, 15, 800–812. [Google Scholar] [CrossRef]
- Mar Rodríguez, M.; Pérez, D.; Javier Chaves, F.; Esteve, E.; Marin-Garcia, P.; Xifra, G.; Vendrell, J.; Jové, M.; Pamplona, R.; Ricart, W.; et al. Obesity changes the human gut mycobiome. Sci. Rep. 2015, 5, 14600. [Google Scholar] [CrossRef]
- Gouba, N.; Raoult, D.; Drancourt, M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS ONE 2013, 8, e59474. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Zou, R.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zheng, H. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders. J. Autism Dev. Disord. 2021, 51, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Fan, C.; Yao, A.; Xu, X.; Zheng, G.; You, Y.; Jiang, C.; Zhao, X.; Hou, Y.; Hung, M.-C.; et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity 2018, 49, 504–514e4. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, L.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol. Lett. 2022, 369, fnac072. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef] [PubMed]
- Lemoinne, S.; Kemgang, A.; Ben Belkacem, K.; Straube, M.; Jegou, S.; Corpechot, C.; Saint-Antoine, I.B.D.N.; Chazouilleres, O.; Housset, C.; Sokol, H. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020, 69, 92–102. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
| GenBank Accession Number | Gene | Sequences (5′-3′) | Melting Temperature | Amplicon Size |
|---|---|---|---|---|
| NM_007393.5 | Actb (β-Actin) | Forward CTGAACCCTAAGGCCAACCG | 59 °C | 113 |
| Reverse CGACCAGAGGCATACAGGGA | ||||
| NM_011146.4 | PPAR-γ | Forward GCAGGAGCAGAGCAAAGAGG | 59 °C | 196 |
| Reverse ATTCATCAGGGAGGCCAGCA | ||||
| NM_007988.3 | Fasn | Forward CTACAGCATCGACGCCAGTC | 58 °C | 108 |
| Reverse TTCCACACCAGGCACAGGTA | ||||
| NM_133360.3 | Acaca (ACC1) | Forward GCTGAGCTTCGGGGTGGTTC | 58 °C | 131 |
| Reverse CGGAATTTGATTTCTACTGT | ||||
| NM_033218.2 | SREBP-2 | Forward CCTCAAGTGCAAAGCCTCGT | 60 °C | 113 |
| Reverse AGTGTGCCATTGGCTGTCTG | ||||
| NM_013495.2 | Cpt1a (CPT1) | Forward GTGACTGGTGGGAGGAATAC | 56 °C | 83 |
| Reverse GAGCATCTCCATGGC GTAG | ||||
| NM_011144.6 | PPAR-α | ForwardAACATCGAGTGTCGAATATGTGG | 60 °C | 99 |
| Reverse CCGAATAGTTCGCCGAAAGAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Song, G.; Feng, X.; Niu, J.; Wang, L.; Yang, C.; Luo, X.; Zhou, S.; Ma, W. The Probiotic Kluyveromyces lactis JSA 18 Alleviates Obesity and Hyperlipidemia in High-Fat Diet C57BL/6J Mice. Foods 2024, 13, 1124. https://doi.org/10.3390/foods13071124
Hong Y, Song G, Feng X, Niu J, Wang L, Yang C, Luo X, Zhou S, Ma W. The Probiotic Kluyveromyces lactis JSA 18 Alleviates Obesity and Hyperlipidemia in High-Fat Diet C57BL/6J Mice. Foods. 2024; 13(7):1124. https://doi.org/10.3390/foods13071124
Chicago/Turabian StyleHong, Yingxiang, Guodong Song, Xiaoqian Feng, Jialei Niu, Lu Wang, Caini Yang, Xuegang Luo, Sa Zhou, and Wenjian Ma. 2024. "The Probiotic Kluyveromyces lactis JSA 18 Alleviates Obesity and Hyperlipidemia in High-Fat Diet C57BL/6J Mice" Foods 13, no. 7: 1124. https://doi.org/10.3390/foods13071124
APA StyleHong, Y., Song, G., Feng, X., Niu, J., Wang, L., Yang, C., Luo, X., Zhou, S., & Ma, W. (2024). The Probiotic Kluyveromyces lactis JSA 18 Alleviates Obesity and Hyperlipidemia in High-Fat Diet C57BL/6J Mice. Foods, 13(7), 1124. https://doi.org/10.3390/foods13071124







