Preparation, Isolation and Antioxidant Function of Peptides from a New Resource of Rumexpatientia L. ×Rumextianshanicus A. Los
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Hydrolyzed Peptides of RRL
2.3. Measurement of Hydrolyzed Peptide Concentration
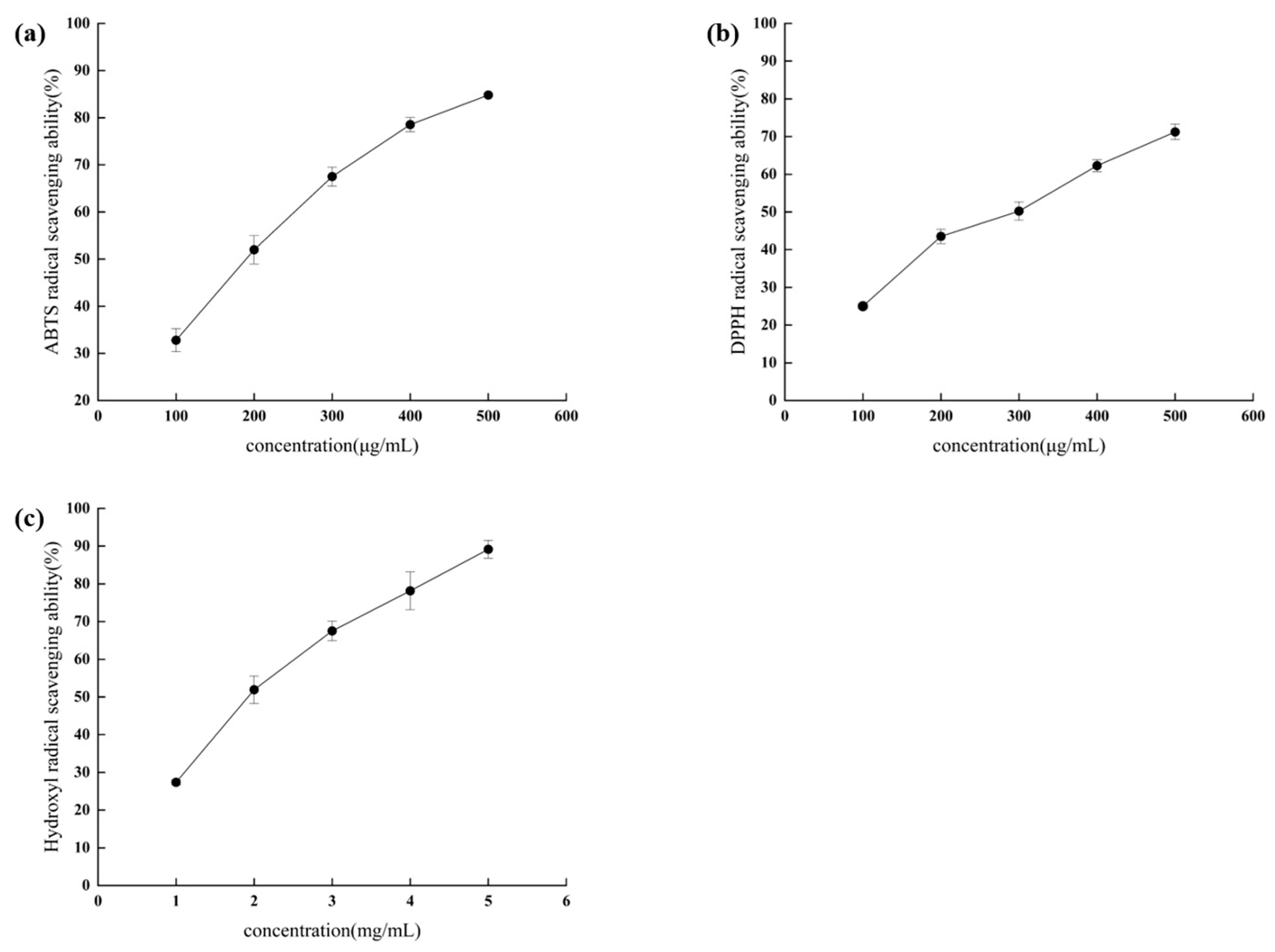
2.4. Measurement of ABTS Radical Scavenging Capacity
2.5. Measurement of DPPH Radical Scavenging Capacity
2.6. Determination of Hydroxyl Radical (·OH) Scavenging Capacity
2.7. Ultrafiltration
2.8. Gel Filtration Chromatography
2.9. Peptide Sequence Analysis by LC-MS/MS
2.10. Computer Analysis of Identified Peptides
2.11. Molecular Docking
2.12. Cell Culture and Cytotoxicity Assays
2.12.1. Establishment of an H2O2-Induced Oxidative Stress Model in HepG2 Cells
2.12.2. Effect of Novel Peptides on the Survival of HepG2 Cells
2.12.3. Effect of Novel Peptides on the Survival of Oxidatively Damaged HepG2 Cells by H2O2
2.12.4. Determination of ROS Levels in HepG2 Cells
2.12.5. Determination of MDA, SOD, GSH-Px, and CAT Activities in HepG2 Cells
2.13. Statistical Analysis
3. Results and Discussion
3.1. Chemical Antioxidant Activity of Hydrolyzed Peptides of RRL
3.2. RRL Hydrolyzed Peptide Ultrafiltration Fractions
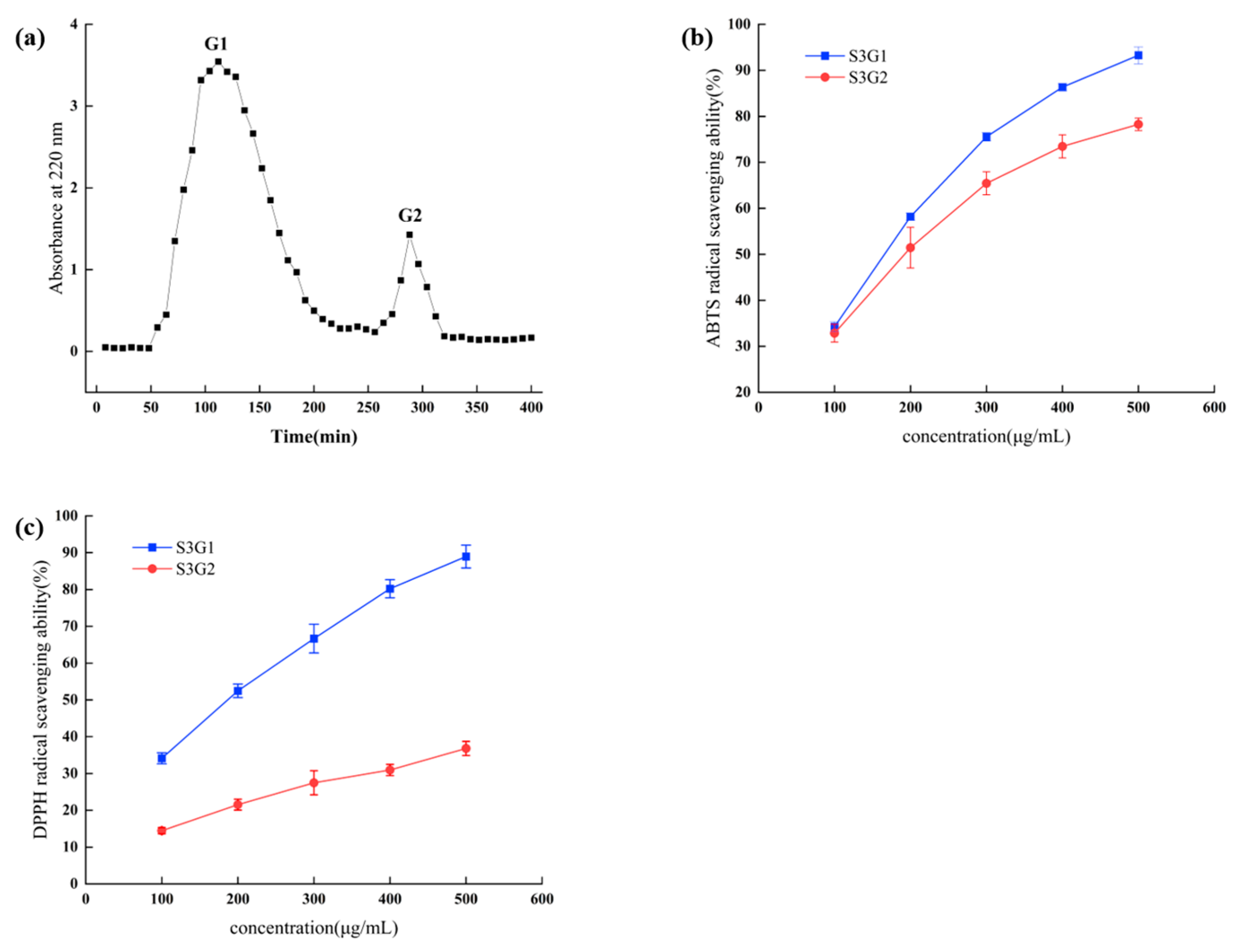
3.3. Gel Filtration Chromatography of the S3
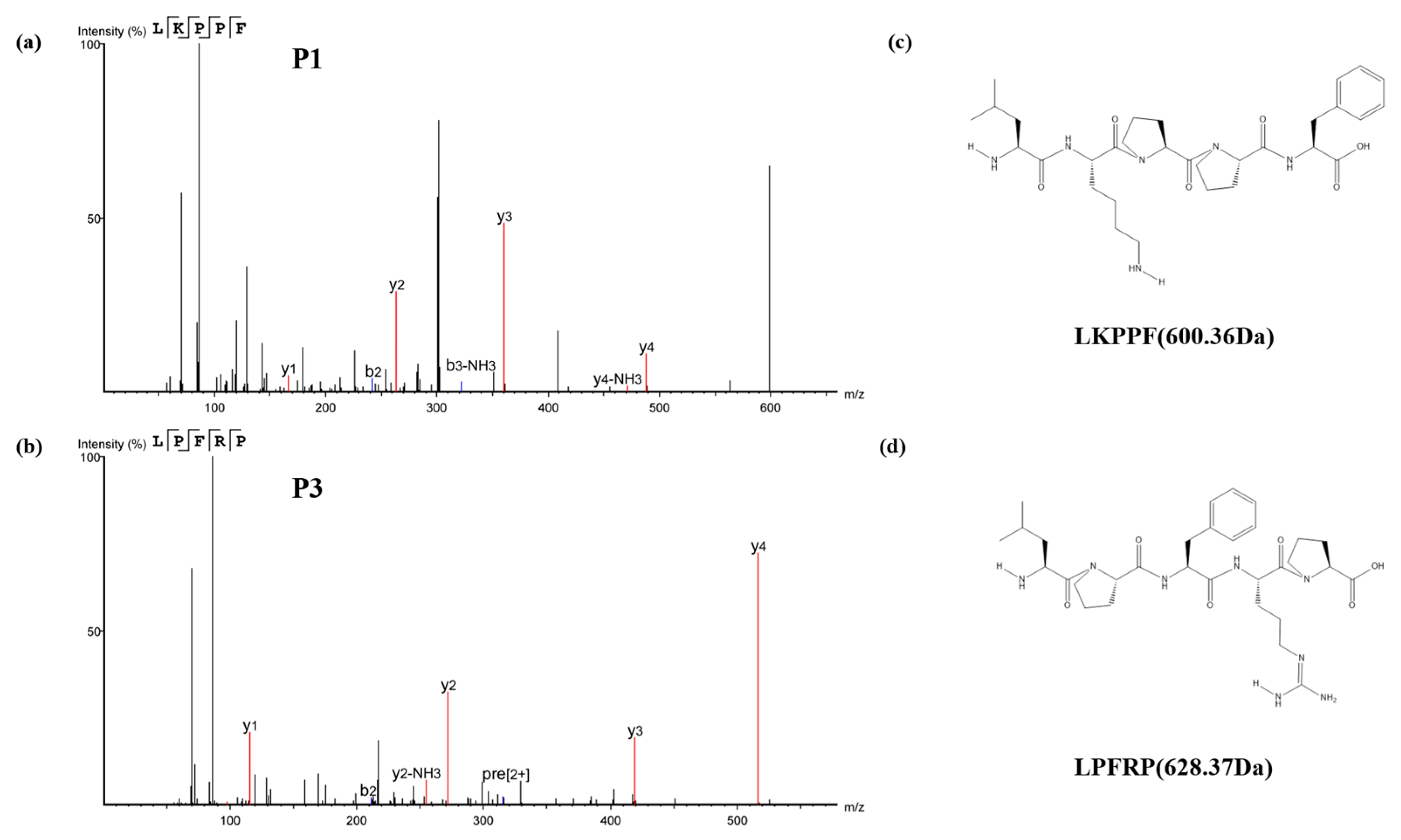
3.4. Identification of Antioxidant Peptides by LC-MS/MS
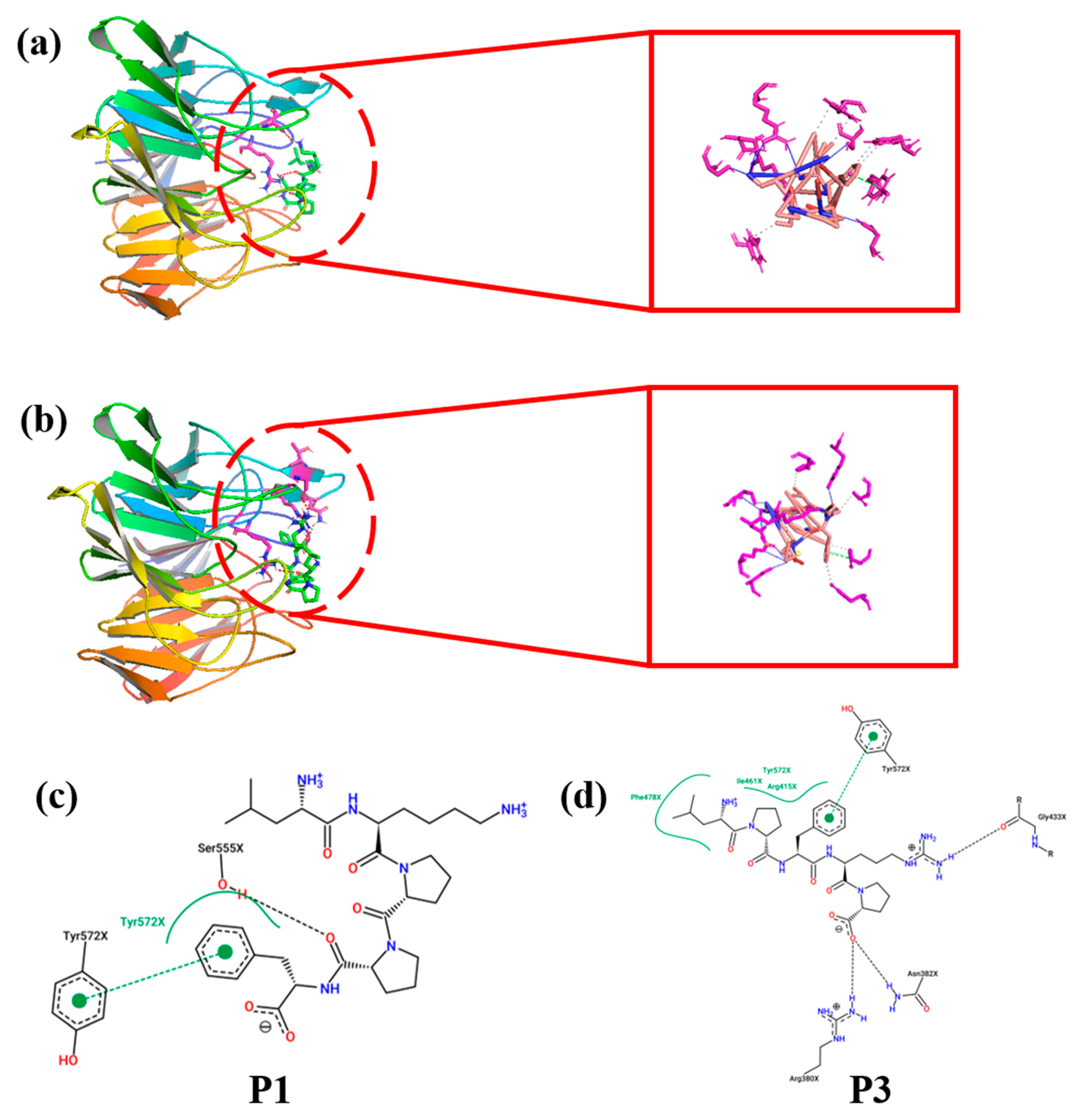
3.5. Molecular Docking
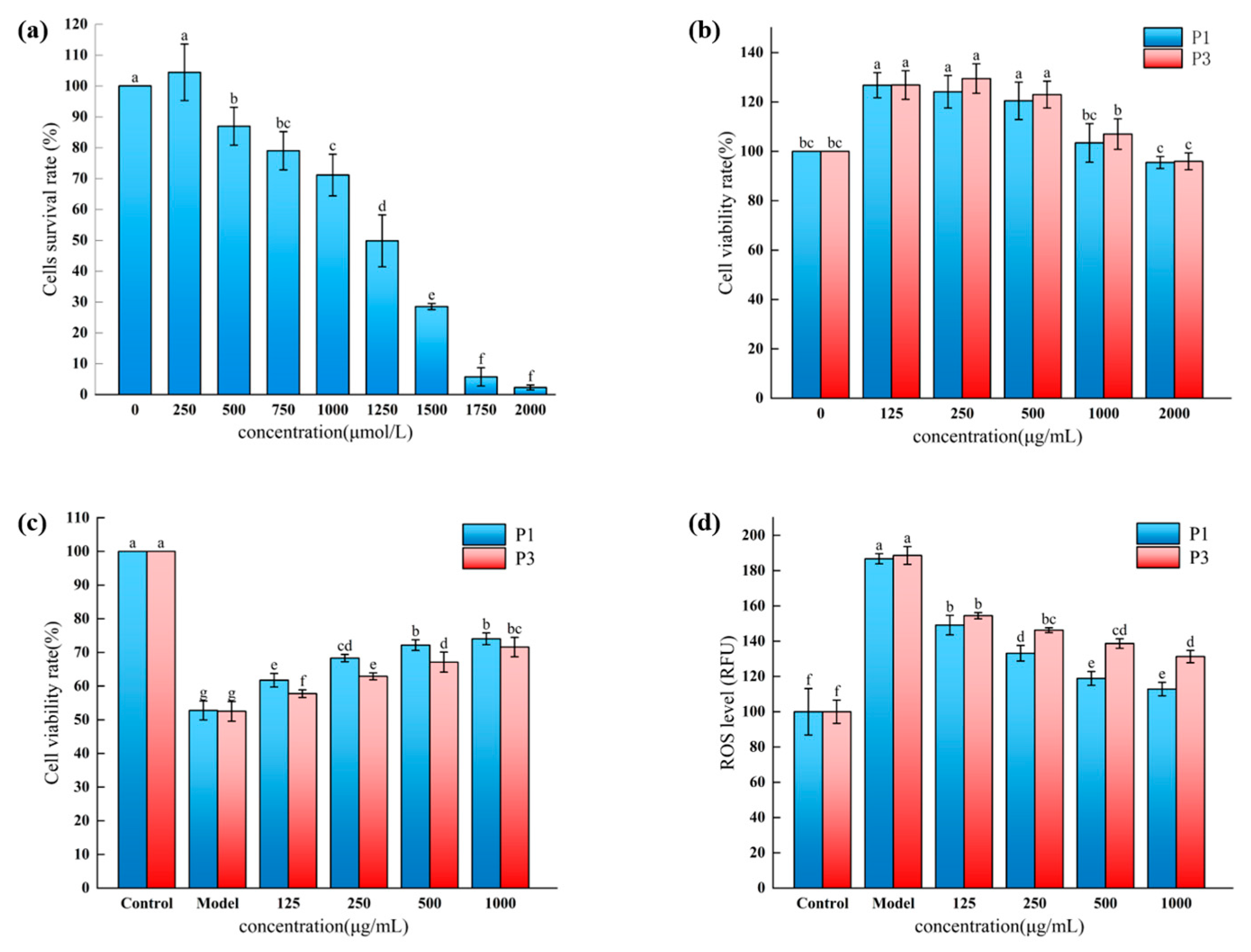
3.6. Effect of Novel Peptides on the Survival of Oxidatively Damaged HepG2 Cells by H2O2
3.7. Effect of Novel Peptides on ROS Levels in H2O2-Oxidatively Damaged HepG2 Cells
3.8. Effect of Novel Peptides on MDA Contents in H2O2-Oxidatively Damaged HepG2 cells
3.9. Effect of Novel Peptides on Antioxidant Enzymes in H2O2-Oxidatively Damaged HepG2 Cells
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kengne, I.C.; Feugap, L.D.T.; Njouendou, A.J.; Ngnokam, C.D.J.; Djamalladine, M.D.; Ngnokam, D.; Voutquenne-Nazabadioko, L.; Tamokou, J.-D.-D. Antibacterial, antifungal and antioxidant activities of whole plant chemical constituents of Rumex abyssinicus. BMC Complement. Med. Ther. 2021, 21, 164. [Google Scholar] [CrossRef] [PubMed]
- Vasas, A.; Orban-Gyapai, O.; Hohmann, J. The Genus Rumex: Review of traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2015, 175, 198–228. [Google Scholar] [CrossRef] [PubMed]
- Redei, D.; Kusz, N.; Rafai, T.; Bogdanov, A.; Burian, K.; Csorba, A.; Mandi, A.; Kurtan, T.; Vasas, A.; Hohmann, J. 14-Noreudesmanes and a phenylpropane heterodimer from sea buckthorn berry inhibit Herpes simplex type 2 virus replication. Tetrahedron 2019, 75, 1364–1370. [Google Scholar] [CrossRef]
- Quradha, M.M.; Khan, R.; Rehman, M.U.; Abohajeb, A. Chemical composition and in vitro anticancer, antimicrobial and antioxidant activities of essential oil and methanol extract from Rumex nervosus. Nat. Prod. Res. 2019, 33, 2554–2559. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-J.; Li, Y.-X.; Li, N.; Zhu, H.-T.; Wang, D.; Zhang, Y.-J. The genus Rumex (Polygonaceae): An ethnobotanical, phytochemical and pharmacological review. Nat. Prod. Bioprospect. 2022, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Berillo, D.; Kozhahmetova, M.; Lebedeva, L. Overview of the Biological Activity of Anthraquinons and Flavanoids of the Plant Rumex Species. Molecules 2022, 27, 1204. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, Y.; Ye, C.; Mou, D.; Li, X.; Zheng, Y.; Han, X. Research progress on the application of Rumex dapibus herba by in the field of animal husbandry. Today Anim. Husb. Vet. Med. 2021, 37, 69–70. [Google Scholar]
- Chen, W. Study on Nutritional Value and Development of Edible Leaf Weed and Research Progress on Edible Leaf Weed Product. Farm Prod. Process. 2018, 14, 63–64+67. [Google Scholar] [CrossRef]
- Zhu, X. Rumex dapibus herba by-Cultivation A New Model of Health. China High New Technol. 2019, 1, 45. [Google Scholar]
- Li, X.; He, T.; Yang, F.; Wang, C.; Zhou, Y.; Sha, R.; Mao, J. Analysis of Nutritional Components, Functional Components and Bioactivity of Edible Dock. Sci. Technol. Food Ind. 2023, 44, 307–315. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Barea, P.; Melgosa, R.; Illera, A.E.; Alonso-Riano, P.; de Cerio, E.D.; Benito-Roman, O.; Beltran, S.; Sanz, M.T. Production of small peptides and low molecular weight amino acids by subcritical water from fish meal: Effect of pressurization agent. Food Chem. 2023, 418, 135925. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Hamdan, N.; Ling, L.I.; Wong, S.L.; Nyakuma, B.B.; Khoo, S.C.; Ramachandran, H.; Jamaluddin, H.; Wong, K.Y.; Lee, T.H. Antioxidant and antidiabetic properties of bioactive peptides from soursop (Annona muricata) leaf biomass. Biomass Convers. Biorefinery 2023, 13, 353. [Google Scholar] [CrossRef]
- Tao, L.; Gu, F.; Liu, Y.; Yang, M.; Wu, X.Z.; Sheng, J.; Tian, Y. Preparation of antioxidant peptides from Moringa oleifera leaves and their protection against oxidative damage in HepG2 cells. Front. Nutr. 2022, 9, 1062671. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huo, X.; Zheng, Y.; Guo, Y.; Feng, C. ACE-Inhibitory Peptides Identified from Quinoa Bran Glutelin-2 Hydrolysates: In Silico Screening and Characterization, Inhibition Mechanisms of ACE, Coordination with Zinc Ions, and Stability. Plant Foods Hum. Nutr. 2023, 78, 419–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shang, Y.; Li, S.; Chen, Z.; Xia, J.; Tian, Y.; Jia, Y.; Ma, A. Molecular Docking Revealed the Potential Anti-Oxidative Stress Mechanism of the Walnut Polypeptide on HT22 Cells. Foods 2023, 12, 1554. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; He, Y.-L.; Liu, Y.; Hong, P.; Zhou, C.; Sun, S.; Qian, Z.-J. Comparative in silico and in vitro study of the stability and biological activity of an octapeptide from microalgae Isochrysis zhanjiangensis and its truncated short peptide. Food Funct. 2023, 14, 3659–3672. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Y.-J.; Bai, S.-S.; Yang, Z.-J.; Miao, C.; Megrous, S.; Aziz, T.; Sarwar, A.; Li, D.; Yang, Z.-N. Antioxidant Activity of Novel Casein-Derived Peptides with Microbial Proteases as Characterized via Keap1-Nrf2 Pathway in HepG2 Cells. J. Microbiol. Biotechnol. 2021, 31, 1163–1174. [Google Scholar] [CrossRef]
- Hu, Y.-M.; Lu, S.-Z.; Li, Y.-S.; Wang, H.; Shi, Y.; Zhang, L.; Tu, Z.-C. Protective effect of antioxidant peptides from grass carp scale gelatin on the H2O2-mediated oxidative injured HepG2 cells. Food Chem. 2022, 373, 131539. [Google Scholar] [CrossRef]
- He, Z.; Zhang, L.; Zhuo, C.; Jin, F.; Wang, Y. Apoptosis inhibition effect of Dihydromyricetin against UVA-exposed human keratinocyte cell line. J. Photochem. Photobiol. B-Biol. 2016, 161, 40–49. [Google Scholar] [CrossRef]
- Tan, J.; Li, P.; Xue, H.; Li, Q. Cyanidin-3-glucoside prevents hydrogen peroxide (H2O2)-induced oxidative damage in HepG2 cells. Biotechnol. Lett. 2020, 42, 2453–2466. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.M.; Silva, A.M.S. The Antioxidant Activity of Prenylflavonoids. Molecules 2020, 25, 696. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, R.W.; Macharia, J.M.; Wagara, I.N.; Bence, R.L. The antioxidant potential of different edible and medicinal mushrooms. Biomed. Pharmacother. 2022, 147, 112621. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Vashistha, V.K.; Das, D.K. Recent advances in the antioxidant activity and mechanisms of chalcone derivatives: A computational review. Free Radic. Res. 2022, 56, 378–397. [Google Scholar] [CrossRef] [PubMed]
- Ktari, N.; Jridi, M.; Bkhairia, I.; Sayari, N.; Ben Salah, R.; Nasri, M. Functionalities and antioxidant properties of protein hydrolysates from muscle of zebra blenny (Salaria basilisca) obtained with different crude protease extracts. Food Res. Int. 2012, 49, 747–756. [Google Scholar] [CrossRef]
- Hu, X.; Pan, C.; Cai, M.; Li, L.; Yang, X.; Xiang, H.; Chen, S. Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification. Foods 2022, 11, 1498. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-X.; Ruan, G.-R.; Jin, F.; Xu, J.; Wang, F.-J. Purification, identification, and synthesis of five novel antioxidant peptides from Chinese chestnut (Castanea mollissima Blume) protein hydrolysates. LWT-Food Sci. Technol. 2018, 92, 40–46. [Google Scholar] [CrossRef]
- Wali, A.; Gao, Y.; Ishimov, U.; Yili, A.; Aisa, H.A.; Salikhov, S. Isolation and Identification of Three Novel Antioxidant Peptides from the Bactrian Camel Milk Hydrolysates. Int. J. Pept. Res. Ther. 2020, 26, 641–650. [Google Scholar] [CrossRef]
- Rabiei, S.; Rezaei, M.; Asgharzade, S.; Nikoo, M.; Rafieia-kopai, M. Antioxidant and cytotoxic properties of protein hydrolysates obtained from enzymatic hydrolysis of Klunzinger’s mullet (Liza klunzingeri) muscle. Braz. J. Pharm. Sci. 2019, 55, e1830. [Google Scholar] [CrossRef]
- Paudel, P.; Seong, S.H.; Wagle, A.; Min, B.S.; Jung, H.A.; Choi, J.S. Antioxidant and anti-browning property of 2-arylbenzofuran derivatives from Morus alba Linn root bark. Food Chem. 2020, 309, 125739. [Google Scholar] [CrossRef]
- Ghassem, M.; Arihara, K.; Mohammadi, S.; Sani, N.A.; Babji, A.S. Identification of two novel antioxidant peptides from edible bird’s nest (Aerodramus fuciphagus) protein hydrolysates. Food Funct. 2017, 8, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Graziani, D.; Ribeiro, J.V.V.; Cruz, V.S.; Gomes, R.M.; Araujo, E.G.; Santos Junior, A.C.M.; Tomaz, H.C.M.; Castro, C.H.; Fontes, W.; Batista, K.A.; et al. Oxidonitrergic and antioxidant effects of a low molecular weight peptide fraction from hardened bean (Phaseolus vulgaris) on endothelium. Braz. J. Med. Biol. Res. 2021, 54, e10423. [Google Scholar] [CrossRef] [PubMed]
- Ajibola, C.F.; Fashakin, J.B.; Fagbemi, T.N.; Aluko, R.E. Effect of Peptide Size on Antioxidant Properties of African Yam Bean Seed (Sphenostylis stenocarpa) Protein Hydrolysate Fractions. Int. J. Mol. Sci. 2011, 12, 6685–6702. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Mao, X.; Wu, Q. Purification, Identification and Molecular Docking of Novel Antioxidant Peptides from Walnut (Juglans regia L.) Protein Hydrolysates. Molecules 2022, 27, 8423. [Google Scholar] [CrossRef] [PubMed]
- Indiano-Romacho, P.; Fernandez-Tome, S.; Amigo, L.; Hernandez-Ledesma, B. Multifunctionality of lunasin and peptides released during its simulated gastrointestinal digestion. Food Res. Int. 2019, 125, 108513. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef]
- Guo, H.; Kouzuma, Y.; Yonekura, M. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009, 113, 238–245. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food Protein-Derived Bioactive Peptides: Production, Processing, and Potential Health Benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Guo, J.; Lu, A.; Sun, Y.; Liu, B.; Zhang, J.; Zhang, L.; Huang, P.; Yang, A.; Li, Z.; Cao, Y.; et al. Purification and identification of antioxidant and angiotensin converting enzyme-inhibitory peptides from Guangdong glutinous rice wine. LWT-Food Sci. Technol. 2022, 169, 113953. [Google Scholar] [CrossRef]
- Liang, R.; Zhang, Z.; Lin, S. Effects of pulsed electric field on intracellular antioxidant activity and antioxidant enzyme regulating capacities of pine nut (Pinus koraiensis) peptide QDHCH in HepG2 cells. Food Chem. 2017, 237, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hu, Q.; Xie, M.; Liu, J.; Su, A.; Xu, H.; Yang, W. Protective effects of peptide KSPLY derived from Hericium erinaceus on H2O2-induced oxidative damage in HepG2 cells. Food Sci. Hum. Wellness 2023, 12, 1893–1904. [Google Scholar] [CrossRef]
- Liang, R.; Cheng, S.; Dong, Y.; Ju, H. Intracellular antioxidant activity and apoptosis inhibition capacity of PEF-treated KDHCH in HepG2 cells. Food Res. Int. 2019, 121, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-M.; Wang, Y.-M.; Zhao, Y.-Q.; Chi, C.-F.; Wang, B. Antioxidant Peptides from the Protein Hydrolysate of Monkfish (Lophius litulon) Muscle: Purification, Identification, and Cytoprotective Function on HepG2 Cells Damage by H2O2. Mar. Drugs 2020, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Luo, X.; Huang, M.; Wu, J.; Zhang, J.; Liu, X.; Li, H.; Sun, X. Identification and Antioxidant Activity of a Novel Peptide from Baijiu. Int. J. Pept. Res. Ther. 2020, 26, 1199–1210. [Google Scholar] [CrossRef]
- Igbokwe, C.J.; Feng, Y.; Louis, H.; Benjamin, I.; Quaisie, J.; Duan, Y.; Tuly, J.A.; Cai, M.; Zhang, H. Novel antioxidant peptides identified from coix seed by molecular docking, quantum chemical calculations and invitro study in HepG2 cells. Food Chem. 2024, 440, 138234. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ren, J.; Wu, H.; Zhang, X.; Han, L.; Gu, R. Inhibitory effects and action mechanism of five antioxidant peptides derived from wheat gluten on cells oxidative stress injury. Food Biosci. 2023, 56, 103236. [Google Scholar] [CrossRef]
- Ana, P.C.C.; Francisco, E.S.L.; Francisca, F.N.A.; Yago, O.P.; Claudia, R.A.; Felipe, P.M.; Gabrielly, O.S.; Cleverson, D.T.F.; Pedro, F.N.S. Antioxidant peptides from plants: A review. Phytochem. Rev. 2024, 23, 95–104. [Google Scholar] [CrossRef]
- Lopez-Garcia, G.; Dublan-Garcia, O.; Arizmendi-Cotero, D.; Gomez Olivan, L.M. Antioxidant and Antimicrobial Peptides Derived from Food Proteins. Molecules 2022, 27, 1343. [Google Scholar] [CrossRef]
- Federica, T.; Sara, C.; Federico, F.; Alessandro, G.; Marica, A.; Alessandra, F.; Stefania, F.; Agnese De, M.; Ilaria, P.; Cristina, M.; et al. Sunflower seed-derived bioactive peptides show antioxidant and anti-inflammatory activity: From in silico simulation to the animal model. Food Chem. 2024, 439, 138124. [Google Scholar] [CrossRef]
- Daodian, W.; Guangqiang, W.; Yanying, Y.; Yanling, Z.; Xiang, L.; Yanan, S.; Aixiang, H. Identification and molecular mechanism of novel bifunctional peptides from Duroc × (Landrace × Yorkshire) pig dry-cured ham: A peptidomics and in silico analysis. Food Res. Int. 2024, 180, 114066. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Chen, Z.; Chen, Y.; Lin, Q.; Liang, Y. Exogenous Bioactive Peptides Have a Potential Therapeutic Role in Delaying Aging in Rodent Models. Int. J. Mol. Sci. 2022, 23, 1421. [Google Scholar] [CrossRef] [PubMed]
- Zahedipour, F.; Hosseini, S.A.; Astaneh, M.; Kesharwani, P.; Jaafari, M.R.; Sahebkar, A. Application of VEGF/VEGFR peptide vaccines in cancer: A systematic review of clinical trials. Crit. Rev. Oncol. Hematol. 2023, 187, 104032. [Google Scholar] [CrossRef] [PubMed]


| No. | Peptide | Score | Length | RT | Area | No. | Peptide | Score | Length | RT | Area |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LAYKPPR | 99 | 7 | 11.12 | 5.22 × 108 | 62 | YKPPR | 97 | 5 | 6.48 | 3.10 × 107 |
| 2 | AFDEGPWPR | 99 | 9 | 32.62 | 2.39 × 108 | 63 | TDYPPLGR | 97 | 8 | 19.66 | 2.87 × 107 |
| 3 | FAPSLPEKN | 99 | 9 | 21.06 | 1.06 × 108 | 64 | LLYDDR | 97 | 6 | 18.22 | 2.72 × 107 |
| 4 | LSDPWHNT | 99 | 8 | 25.05 | 1.05 × 108 | 65 | LGRLSP | 97 | 6 | 14.36 | 2.53 × 107 |
| 5 | LFEEPVPGK | 99 | 9 | 23.61 | 1.00 × 108 | 66 | LKVPL | 97 | 5 | 26.99 | 1.95 × 107 |
| 6 | TVLLPR | 99 | 6 | 20.3 | 9.59 × 107 | 67 | LGNLRP | 97 | 6 | 13.4 | 1.68 × 107 |
| 7 | RPLVMH | 99 | 6 | 23.22 | 6.60 × 107 | 68 | LPKVP | 97 | 5 | 20.07 | 1.58 × 107 |
| 8 | FTGSNVKVA | 99 | 9 | 15.72 | 6.50 × 107 | 69 | LQVER | 97 | 5 | 9 | 1.51 × 107 |
| 9 | AFRVP | 99 | 5 | 23.53 | 3.99 × 107 | 70 | FVRLLG | 97 | 6 | 27.79 | 1.48 × 107 |
| 10 | VVRLP | 99 | 5 | 19.82 | 2.40 × 107 | 71 | LKAPA | 97 | 5 | 7.49 | 1.46 × 107 |
| 11 | LLPR | 99 | 4 | 10.81 | 2.25 × 107 | 72 | LGNNPAKGGL | 97 | 10 | 14.67 | 1.34 × 107 |
| 12 | LLSPDPATK | 99 | 9 | 15.21 | 1.77 × 107 | 73 | PFPPR | 97 | 5 | 18.32 | 1.30 × 107 |
| 13 | VSPLEVK | 99 | 7 | 16 | 1.35 × 107 | 74 | LFPRDPY | 97 | 7 | 28.53 | 1.29 × 107 |
| 14 | LGDAFYYGK | 99 | 9 | 29.33 | 1.30 × 107 | 75 | LVTGKGPLEN | 97 | 10 | 14.07 | 1.28 × 107 |
| 15 | FLDDVQVK | 99 | 8 | 28.11 | 1.31 × 107 | 76 | PPAPR | 97 | 5 | 12.07 | 1.24 × 107 |
| 16 | PLLRP | 98 | 5 | 15.62 | 6.32 × 108 | 77 | LLLPR | 97 | 5 | 23.17 | 1.02 × 107 |
| 17 | LWYGPDRP | 98 | 8 | 29.01 | 5.23 × 108 | 78 | LLKFE | 97 | 5 | 24.44 | 1.01 × 107 |
| 18 | VLLPR | 98 | 5 | 17.49 | 3.26 × 108 | 79 | WVPPEGK | 96 | 7 | 18.19 | 1.33 × 109 |
| 19 | LGKVYDY | 98 | 7 | 21.82 | 2.96 × 108 | 80 | LVVLGH | 96 | 6 | 17.87 | 5.31 × 108 |
| 20 | VTLPR | 98 | 5 | 14.51 | 1.43 × 108 | 81 | WYGPDRP | 96 | 7 | 22.97 | 1.87 × 108 |
| 21 | FDEGPWRP | 98 | 8 | 30.25 | 1.11 × 108 | 82 | SFRVTP | 96 | 6 | 22.05 | 8.02 × 107 |
| 22 | LTPSSSFKDA | 98 | 10 | 19.18 | 9.06 × 107 | 83 | WVPPEGR | 96 | 7 | 19.66 | 7.20 × 107 |
| 23 | LRLP | 98 | 4 | 22.05 | 7.11 × 107 | 84 | LKFE | 96 | 4 | 15.82 | 5.08 × 107 |
| 24 | FSEYPPLGR | 98 | 9 | 28.87 | 6.19 × 107 | 85 | VTGKGPFDNL | 96 | 10 | 31.92 | 4.63 × 107 |
| 25 | FRVP | 98 | 4 | 21.67 | 5.86 × 107 | 86 | LLDLH | 96 | 5 | 25.81 | 3.91 × 107 |
| 26 | LGTVPVGR | 98 | 8 | 15.21 | 5.70 × 107 | 87 | LLEGLPK | 96 | 7 | 24.37 | 3.81 × 107 |
| 27 | FVPGK | 98 | 5 | 8.5 | 5.05 × 107 | 88 | LFQGPPGH | 96 | 8 | 18.48 | 2.59 × 107 |
| 28 | LTKLGVK | 98 | 7 | 9 | 4.76 × 107 | 89 | LGDAFYYNK | 96 | 9 | 29.07 | 2.51 × 107 |
| 29 | LFPR | 98 | 4 | 15.62 | 4.53 × 107 | 90 | LLKVP | 96 | 5 | 21.03 | 2.33 × 107 |
| 30 | LRVP | 98 | 4 | 16.35 | 4.53 × 107 | 91 | TFSEYRGLPP | 96 | 10 | 30.99 | 2.29 × 107 |
| 31 | FRLP | 98 | 4 | 25.11 | 4.30 × 107 | 92 | LVMHDY | 96 | 6 | 18.07 | 2.22 × 107 |
| 32 | FTGKQPYDL | 98 | 9 | 27.35 | 3.52 × 107 | 93 | LLKFDP | 96 | 6 | 30.99 | 1.89 × 107 |
| 33 | WYGPDR | 98 | 6 | 18.22 | 3.08 × 107 | 94 | VVRLPYD | 96 | 7 | 27.02 | 1.75 × 107 |
| 34 | YKPP | 98 | 4 | 9.41 | 2.91 × 107 | 95 | LLTDFKP | 96 | 7 | 26.67 | 1.65 × 107 |
| 35 | AFRVTP | 98 | 6 | 22.05 | 2.69 × 107 | 96 | LELPR | 96 | 5 | 19.56 | 1.60 × 107 |
| 36 | LTPSSSFK | 98 | 8 | 15.21 | 2.53 × 107 | 97 | LLPVGR | 96 | 6 | 15.85 | 1.53 × 107 |
| 37 | VVRLYP | 98 | 6 | 28.18 | 2.44 × 107 | 98 | LLGFDNVRQ | 96 | 9 | 30.64 | 1.48 × 107 |
| 38 | VKTPW | 98 | 5 | 22.37 | 2.42 × 107 | 99 | LDPVLGR | 96 | 7 | 19.54 | 1.35 × 107 |
| 39 | LTPSSSFKD | 98 | 9 | 16.96 | 2.20 × 107 | 100 | YYDGR | 96 | 5 | 8.47 | 1.19 × 107 |
| 40 | LNLSSESGKY | 98 | 10 | 21.89 | 2.19 × 107 | 101 | LPRPRP | 96 | 6 | 8.99 | 1.19 × 107 |
| 41 | LPWKD | 98 | 5 | 22.05 | 1.89 × 107 | 102 | AAKWSPE | 96 | 7 | 14.58 | 1.17 × 107 |
| 42 | LSDPTHLGSP | 98 | 10 | 20.45 | 1.74 × 107 | 103 | LKPPF | 96 | 5 | 27.28 | 1.11 × 107 |
| 43 | FVPGR | 98 | 5 | 10.43 | 1.56 × 107 | 104 | LHEDVPHTP | 96 | 9 | 13.28 | 1.09 × 107 |
| 44 | LKAAP | 98 | 5 | 7.49 | 1.46 × 107 | 105 | LKFP | 95 | 4 | 24.6 | 9.92 × 107 |
| 45 | LDVLPHPQ | 98 | 8 | 28.14 | 1.24 × 107 | 106 | VGYPGKY | 95 | 7 | 17.33 | 5.27 × 107 |
| 46 | LFKDPF | 98 | 6 | 36.75 | 1.18 × 107 | 107 | YYYDGR | 95 | 6 | 15.85 | 4.30 × 107 |
| 47 | LRPP | 98 | 4 | 10.59 | 1.10 × 107 | 108 | LLPH | 95 | 4 | 11.03 | 3.75 × 107 |
| 48 | LLFGEK | 98 | 6 | 25.68 | 1.02 × 107 | 109 | LLPPVDLK | 95 | 8 | 29.33 | 3.29 × 107 |
| 49 | LKFPD | 97 | 5 | 23.26 | 1.86 × 108 | 110 | LLLNR | 95 | 5 | 15.47 | 2.92 × 107 |
| 50 | VTLRP | 97 | 5 | 14.51 | 1.43 × 108 | 111 | HHSPGYYDGR | 95 | 10 | 6.92 | 2.44 × 107 |
| 51 | LWYGDPR | 97 | 7 | 25.11 | 1.34 × 108 | 112 | LNREPPEYRP | 95 | 10 | 15.85 | 2.05 × 107 |
| 52 | YTPDYEPH | 97 | 8 | 17.72 | 1.27 × 108 | 113 | KFPERAGP | 95 | 8 | 10.43 | 1.96 × 107 |
| 53 | LSNPTKY | 97 | 7 | 14.83 | 1.10 × 108 | 114 | VTLPPR | 95 | 6 | 12.87 | 1.96 × 107 |
| 54 | LPFRP | 97 | 5 | 22.28 | 9.76 × 107 | 115 | LLLRP | 95 | 5 | 22.27 | 1.61 × 107 |
| 55 | LLLGDR | 97 | 6 | 18.45 | 8.40 × 107 | 116 | LRFGP | 95 | 5 | 22.02 | 1.55 × 107 |
| 56 | STLPGNKY | 97 | 8 | 14.64 | 8.22 × 107 | 117 | RPLVMHDY | 95 | 8 | 32.3 | 1.48 × 107 |
| 57 | LVDPKGPF | 97 | 8 | 27.7 | 7.71 × 107 | 118 | PGYPGKY | 95 | 7 | 16.16 | 1.45 × 107 |
| 58 | LVDPKGFP | 97 | 8 | 27.7 | 7.71 × 107 | 119 | WYGPDRKP | 95 | 8 | 15.21 | 1.33 × 107 |
| 59 | LDGLPPAPR | 97 | 9 | 22.65 | 6.59 × 107 | 120 | LLDALPK | 95 | 7 | 26.48 | 1.25 × 107 |
| 60 | LLHF | 97 | 4 | 28.27 | 4.91 × 107 | 121 | PLRPRP | 95 | 6 | 9 | 1.19 × 107 |
| 61 | FTGKQPYD | 97 | 8 | 13.44 | 3.67 × 107 |
| Fraction No. | Sequence | Peptide Ranker Score | Toxin | Estimated Solubility | Hydrophobic | Allergen | Peptide |
|---|---|---|---|---|---|---|---|
| P1 | LKPPF | 0.898368 | Non-Toxin | Good | 80.00% | Probable non-allergen | Novel peptide |
| P2 | PFPPR | 0.963098 | Non-Toxin | Good | 80.00% | Probable allergen | Novel peptide |
| P3 | LPFRP | 0.941112 | Non-Toxin | Good | 80.00% | Probable non-allergen | Novel peptide |
| P4 | PPAPR | 0.803504 | Non-Toxin | Good | 80.00% | Probable allergen | Novel peptide |
| Intermolecular Forces | No. | Residue | AA |
|---|---|---|---|
| Hydrophobic Interactions | 1 | 334X | Tyr |
| 2 | 334X | Tyr | |
| 3 | 478X | Phe | |
| 4 | 572X | Tyr | |
| 5 | 577X | Phe | |
| 6 | 577X | Phe | |
| Hydrogen Bonds | 1 | 380X | Arg |
| 2 | 380X | Arg | |
| 3 | 414X | Asn | |
| 4 | 415X | Arg | |
| 5 | 431X | Ser | |
| 6 | 431X | Ser | |
| 7 | 530X | Gln | |
| 8 | 602X | Ser | |
| π-Stacking | 1 | 572X | Tyr |
| Salt Bridges | 1 | 415X | Arg |
| Intermolecular Forces | No. | Residue | AA |
|---|---|---|---|
| Hydrophobic Interactions | 1 | 415X | Arg |
| 2 | 478X | Phe | |
| 3 | 525X | Tyr | |
| 4 | 572X | Tyr | |
| 5 | 572X | Tyr | |
| 6 | 577X | Phe | |
| 7 | 577X | Phe | |
| Hydrogen Bonds | 1 | 380X | Arg |
| 2 | 380X | Arg | |
| 3 | 380X | Arg | |
| 4 | 382X | Asn | |
| 5 | 414X | Asn | |
| 6 | 415X | Arg | |
| 7 | 431X | Ser | |
| 8 | 433X | Gly | |
| 9 | 483X | Arg | |
| π-Stacking | 1 | 572X | Tyr |
| Salt Bridges | 1 | 380X | Arg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Wang, J.; Hong, D.; Chen, Z.; Li, S.; Ma, A.; Jia, Y. Preparation, Isolation and Antioxidant Function of Peptides from a New Resource of Rumexpatientia L. ×Rumextianshanicus A. Los. Foods 2024, 13, 981. https://doi.org/10.3390/foods13070981
Liu C, Wang J, Hong D, Chen Z, Li S, Ma A, Jia Y. Preparation, Isolation and Antioxidant Function of Peptides from a New Resource of Rumexpatientia L. ×Rumextianshanicus A. Los. Foods. 2024; 13(7):981. https://doi.org/10.3390/foods13070981
Chicago/Turabian StyleLiu, Chang, Jianing Wang, Dan Hong, Zhou Chen, Siting Li, Aijin Ma, and Yingmin Jia. 2024. "Preparation, Isolation and Antioxidant Function of Peptides from a New Resource of Rumexpatientia L. ×Rumextianshanicus A. Los" Foods 13, no. 7: 981. https://doi.org/10.3390/foods13070981
APA StyleLiu, C., Wang, J., Hong, D., Chen, Z., Li, S., Ma, A., & Jia, Y. (2024). Preparation, Isolation and Antioxidant Function of Peptides from a New Resource of Rumexpatientia L. ×Rumextianshanicus A. Los. Foods, 13(7), 981. https://doi.org/10.3390/foods13070981






