Co-Analysis of Transcriptome and Metabolome Reveals Flavonoid Biosynthesis in Macadamia Pericarp Across Developmental Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Metabolite Profiling and Data Analysis
2.3. Transcriptome Analysis
2.4. Quantitative Real-Time PCR
2.5. Statistical Analysis
3. Results
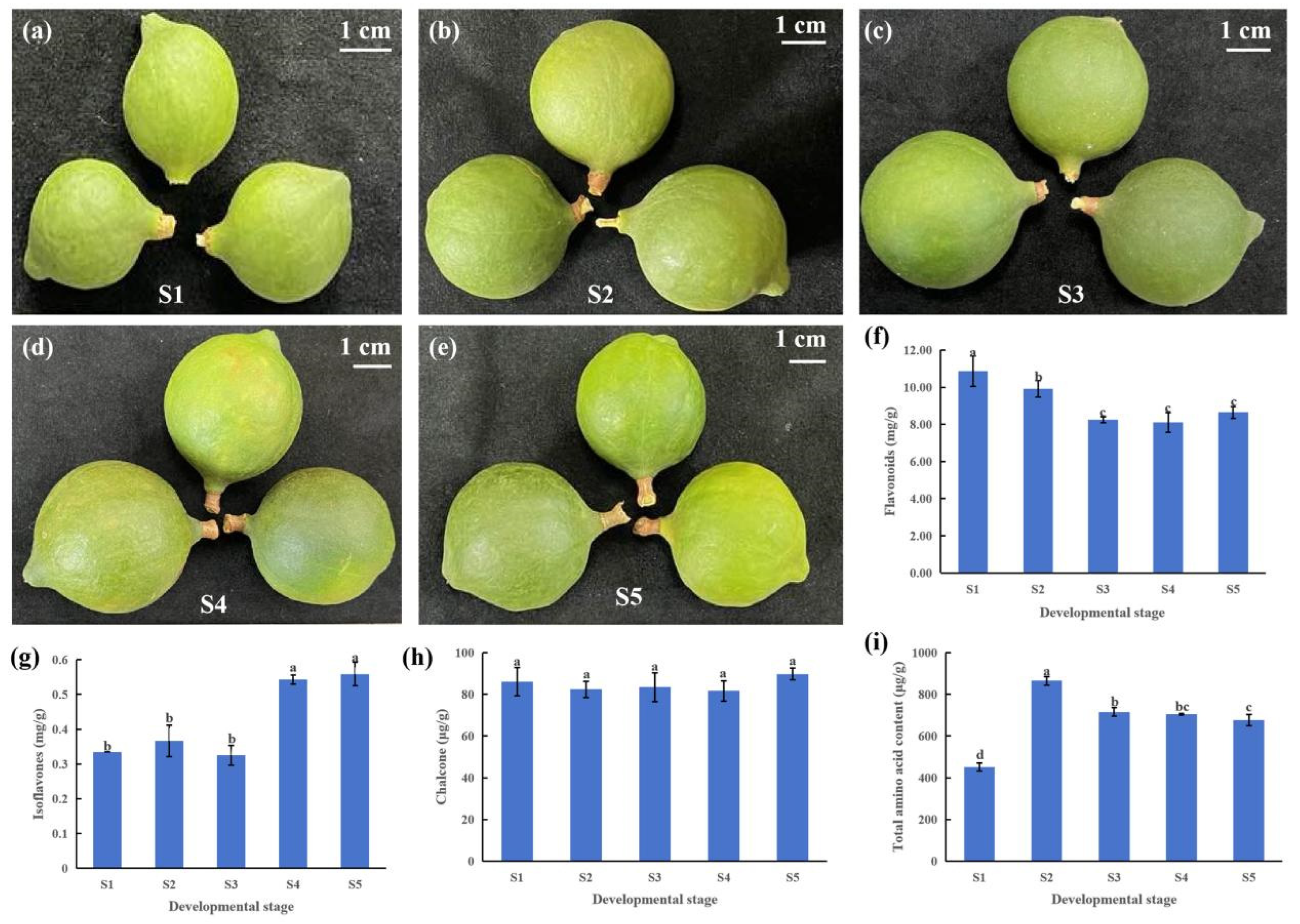
3.1. Changes in Flavonoid Content in the Pericarp of Macadamia
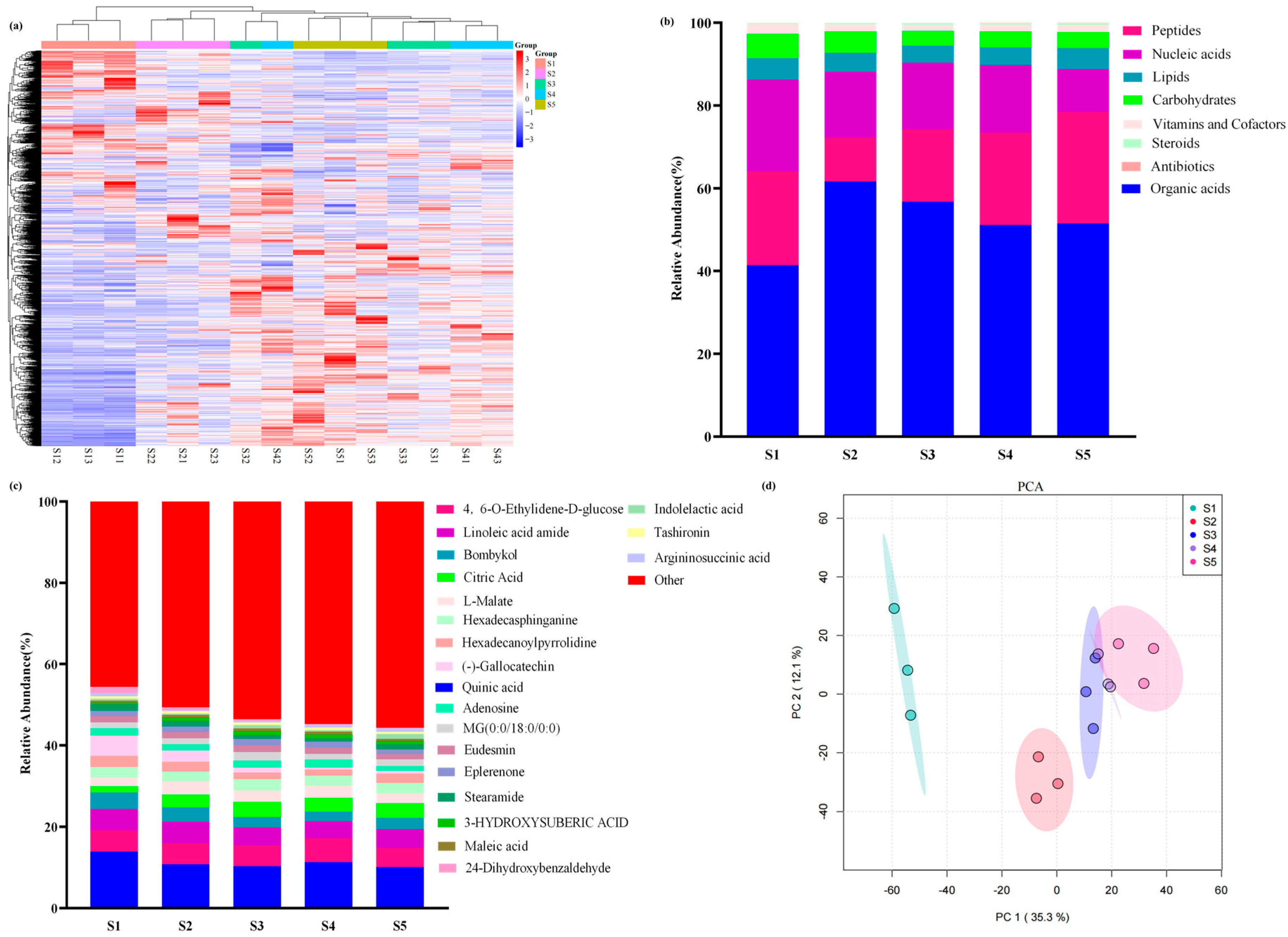
3.2. Metabolomic Analysis of Pericarp at Different Developmental Stages
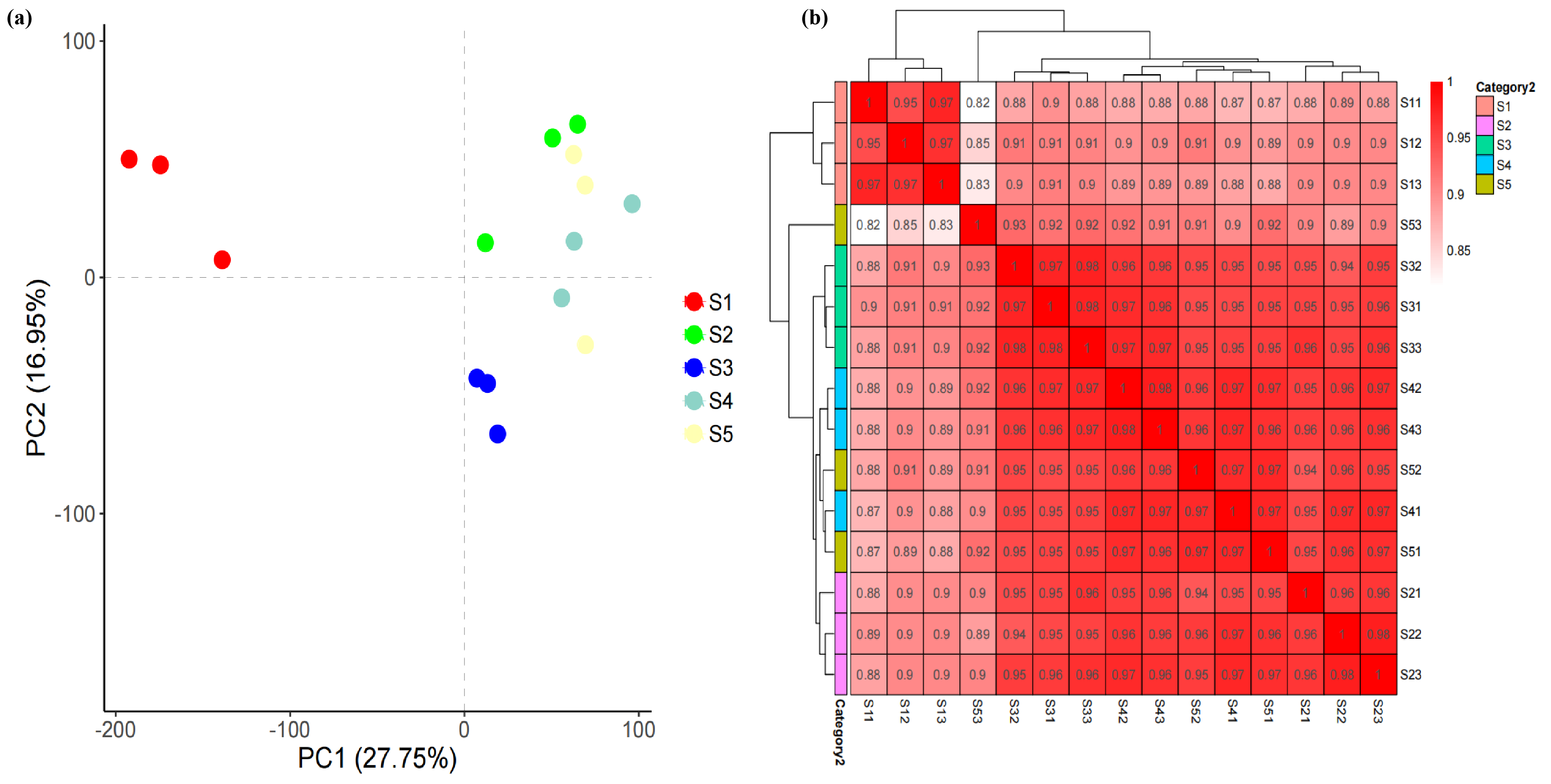
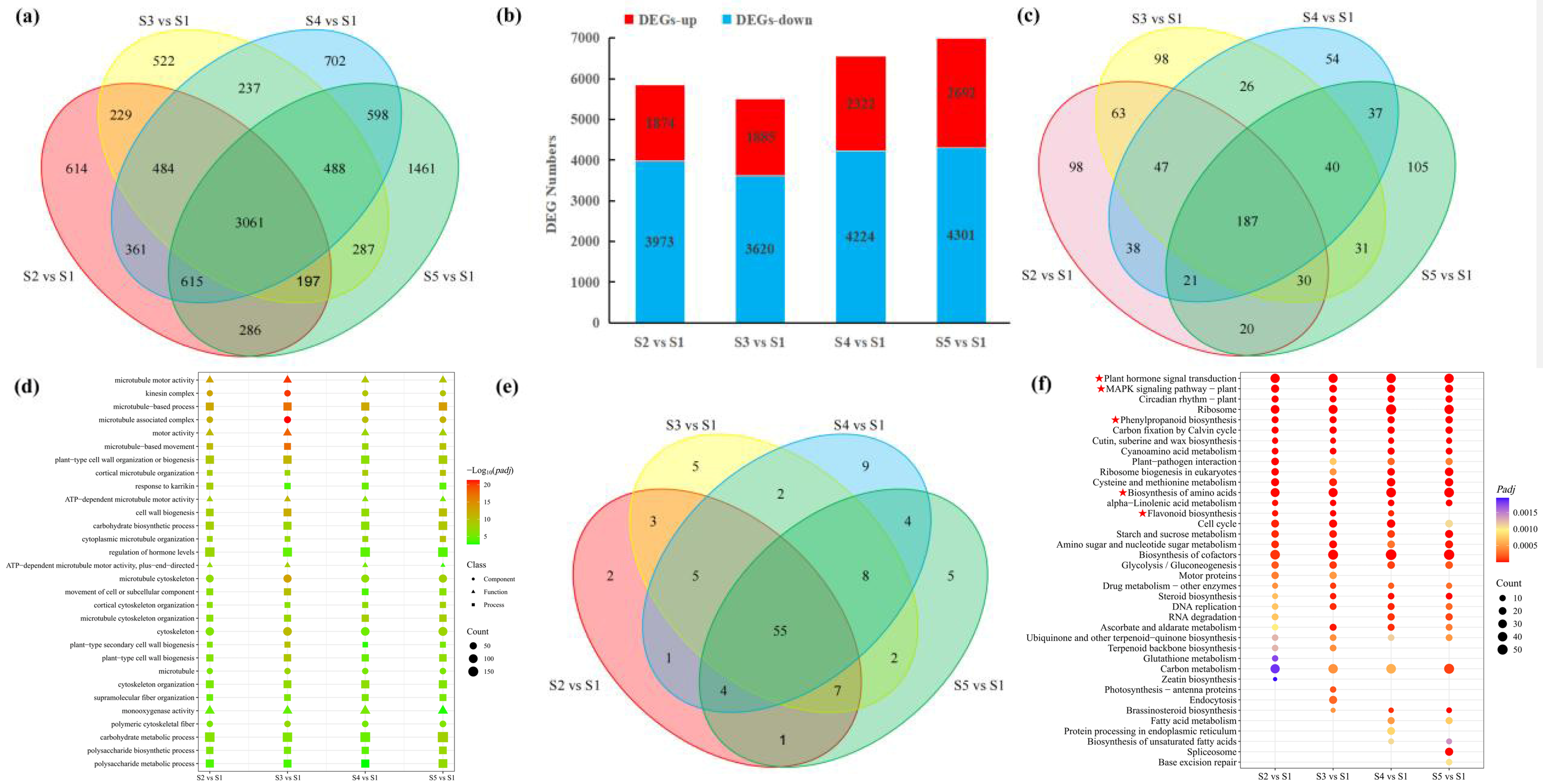
3.3. Transcriptome Analysis of the Pericarp at Different Developmental Stages of Macadamia
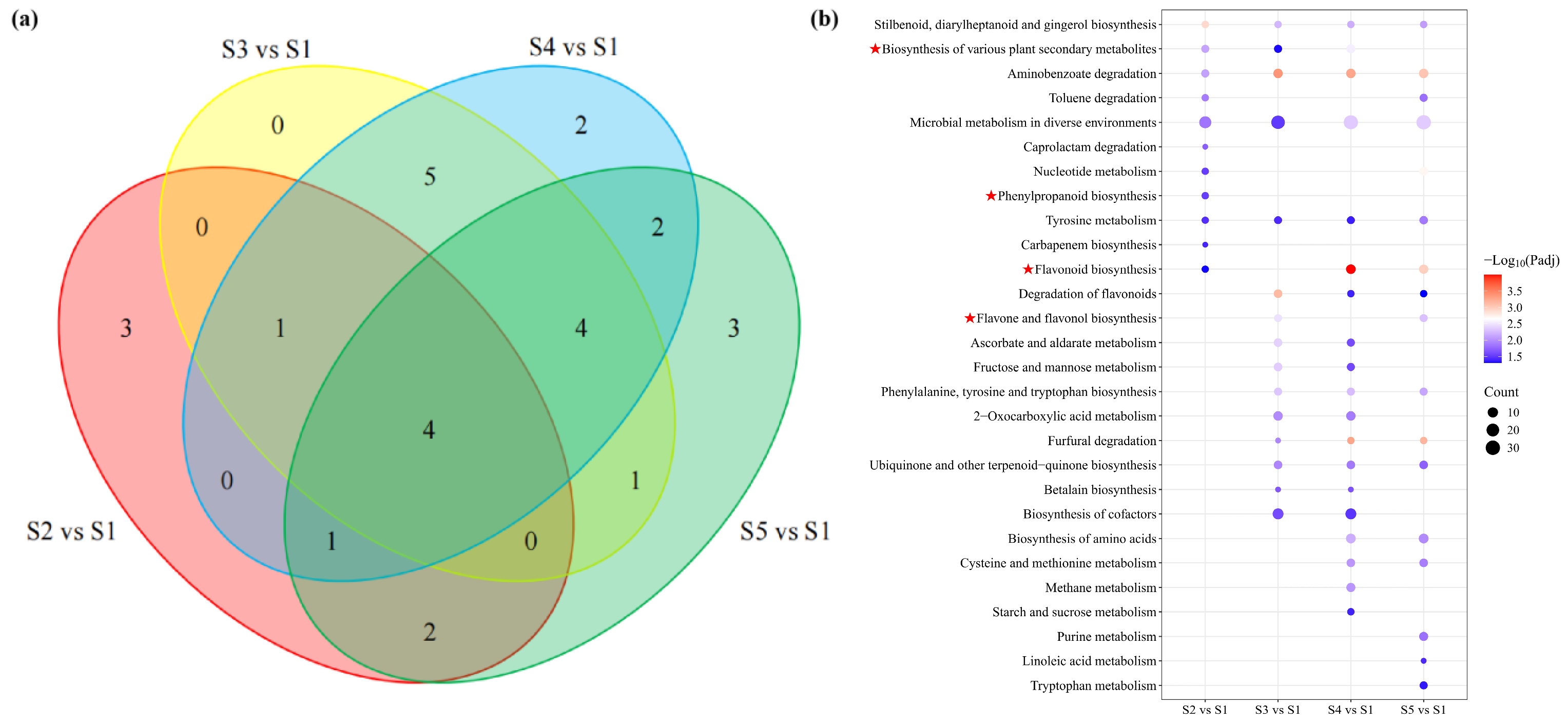
3.4. Identification and Functional Annotation of DEGs in Pericarp Across Developmental Stages
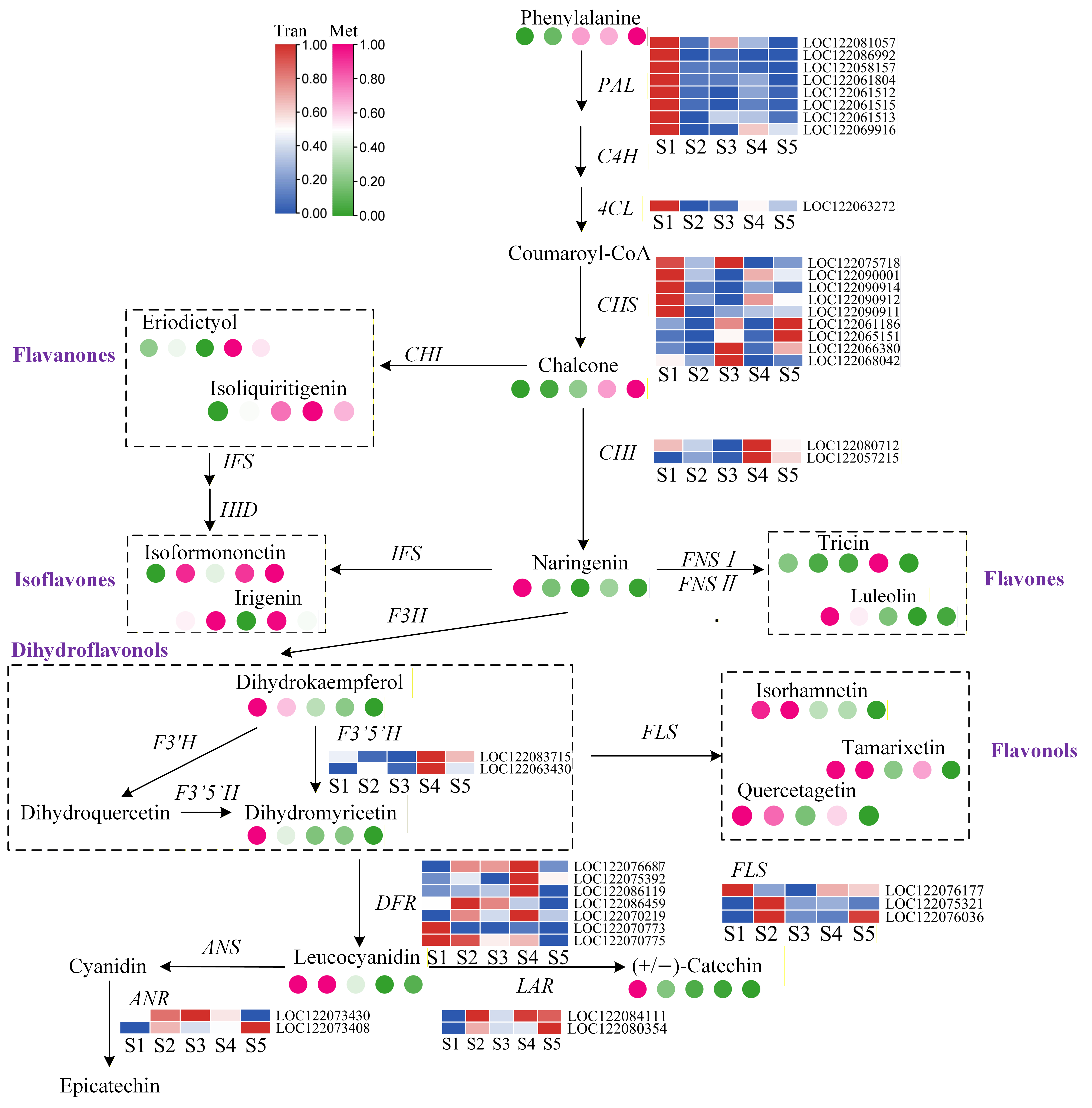
3.5. Integrated Analysis of Transcriptome and Metabolome Profiles Across Different Developmental Stages of Macadamia Pericarp
3.6. Correlation of Flavonoid Biosynthesis and Plant Hormone Signal Transduction Pathway
4. Discussion
4.1. The Important Role of Flavonoids in Macadamia Pericarp Development
4.2. The Influence of Plant Hormones on Flavonoid Biosynthesis in Macadamia Pericarp
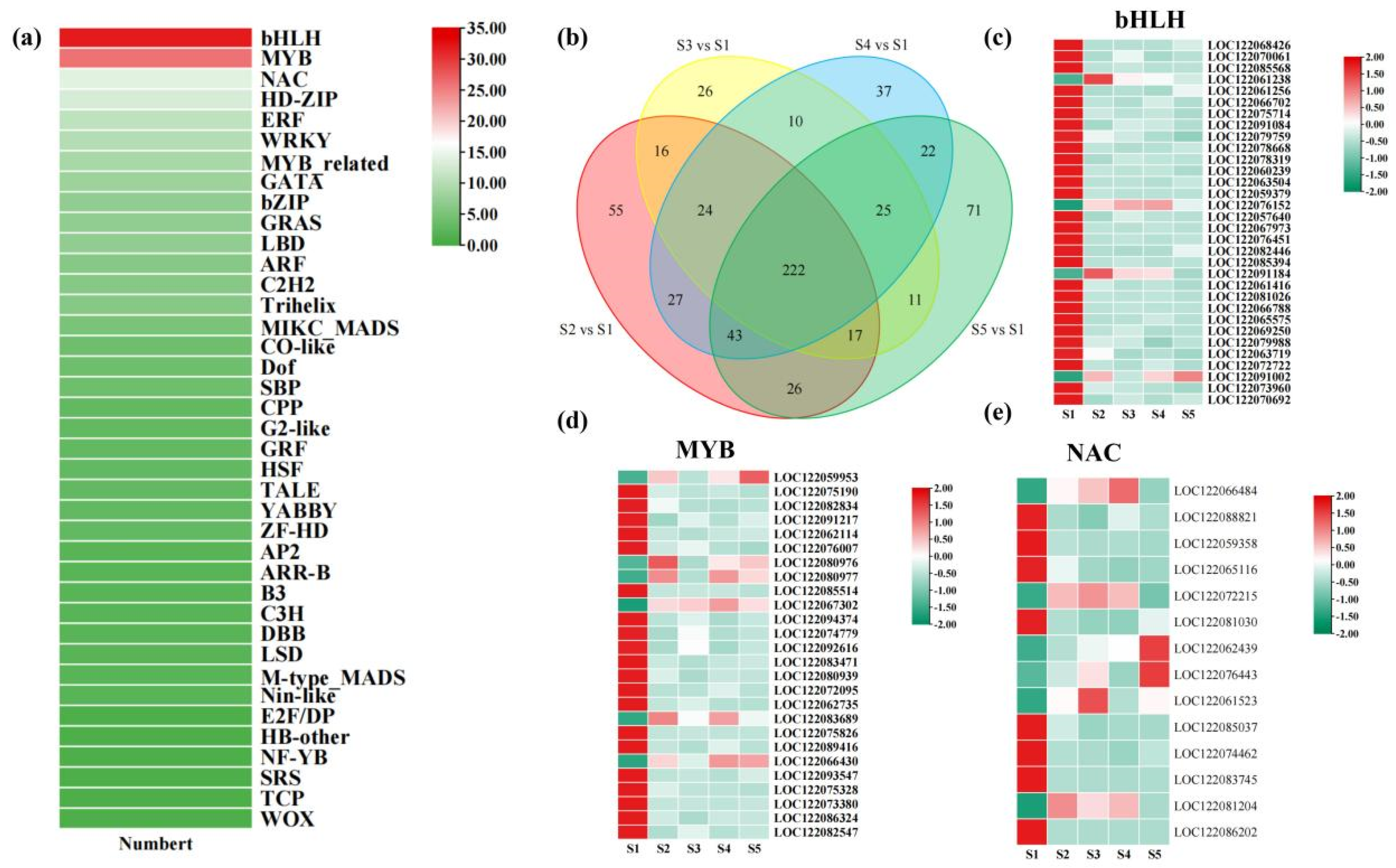
4.3. The Impact of Transcription Factors on Flavonoid Biosynthesis in Macadamia Pericarp
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, H.Y.; Xiao, H.Y.; Zhang, T.; He, P.; Zheng, S.; Xu, P.; Wei, Y.; Wang, W. Preliminary Study on Organic Fertilizer Production by Fermentation of Macadamia Nut Peel. Chin. Agric. Sci. Bull. 2022, 38, 38–44. (In Chinese) [Google Scholar]
- Shi, L.; Wang, J.H.; Xiong, Z.; Wei, M.; Li, X.; Liu, X. Composition Analysis of Cellulose and Lignin in Macadamia Nut Shell. Hubei Agric. Sci. 2009, 48, 2846–2848. (In Chinese) [Google Scholar]
- Mao, Y.; Luo, J.; Cai, Z. Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review. Plants 2025, 14, 1847. [Google Scholar] [CrossRef]
- Ninfali, P.; Antonelli, A.; Magnani, M.; Scarpa, E.S. Antiviral Properties of Flavonoids and Delivery Strategies. Nutrients 2020, 12, 2534. [Google Scholar] [CrossRef]
- Andrade, A.W.L.; Machado, K.d.C.; Machado, K.d.C.; Figueiredo, D.D.R.; David, J.M.; Islam, M.T.; Uddin, S.J.; Shilpi, J.A.; Costa, J.P. In vitro antioxidant properties of the biflavonoid agathisflavone. Chem. Cent. J. 2018, 12, 75. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Heo, M.Y.; Kim, H.P. Flavonoids: Broad Spectrum Agents on Chronic Inflammation. Biomolecules. Therapeutics 2019, 27, 241–253. [Google Scholar] [CrossRef]
- Ying, Z.P.; Li, B.; Zhang, A.; Wang, R.; Xiong, Z.; Shi, R. Analysis of Phenolic Acids in Macadamia Nut Pericarp at Different Developmental Stages. South China Fruits 2023, 52, 66–71. (In Chinese) [Google Scholar]
- Xu, M.Y. Study on Chemical Constituents and Biological Activities of Total Flavonoids from Purple Passion Fruit Peel Produced in Guangxi. Master’s Thesis, Minzu University of China, Beijing, China, 2021. (In Chinese). [Google Scholar] [CrossRef]
- Vella, F.M.; Cautela, D.; Laratta, B. Characterization of Polyphenolic Compounds in Cantaloupe Melon By-Products. Foods 2019, 8, 196. [Google Scholar] [CrossRef]
- Yang, F. Chemotaxonomy Study of Plants from the Family Proteaceae Based on Its Natural Product Profile (Alkaloid). Ph.D. Thesis, Griffith University, Brisbane, Australia, 2017. [Google Scholar]
- Gadea, A.; Khazem, M.; Gaslonde, T. Current knowledge on chemistry of Proteaceae family, and biological activities of their bis-5-alkylresorcinol derivatives. Phytochemistry 2022, 21, 1969–2005. [Google Scholar] [CrossRef]
- Kang, Z.M.; Guo, G.Z.; Wang, D.G.; He, F.; Zeng, H.; Wang, W.; Tu, X. Analysis of component differences between macadamia nut pericarp and kernel based on widely targeted metabolomics. South China Fruits 2025, 54, 62–69+75. (In Chinese) [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, X.; Zeng, F. The regulatory network composed of phytohormones, transcription factors and non-coding RNAs is involved in the flavonoids biosynthesis of fruits. Hortic. Plant J. 2025; in press. [Google Scholar]
- Sun, H.; Cui, H.; Zhang, J.; Kang, J.; Wang, Z.; Li, M.; Yi, F.; Yang, Q.; Long, R. Gibberellins Inhibit Flavonoid Biosynthesis and Promote Nitrogen Metabolism in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 9291. [Google Scholar] [CrossRef]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, Å.; Qian, M.; Li, J.; Li, Y.; Shen, J.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef]
- Flores, G.; Blanch, G.P.; del Castillo, M.L.R. Postharvest treatment with (−) and (+)-methyl jasmonate stimulates anthocyanin accumulation in grapes. Food Sci. Technol./Lebensm.-Wiss. Und-Technol. 2015, 62 Pt 2, 6. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Ahmad, N.; Sheng, X.; Iqbal, B.; Naeem, M.; Wang, N.; Li, F.; Yao, N.; Liu, X. Unraveling the functional characterization of a jasmonate-induced flavonoid biosynthetic CYP45082G24 gene in Carthamus tinctorius. Funct. Integr. Genom. 2023, 23, 172. [Google Scholar] [CrossRef] [PubMed]
- Lama, K.; Harlev, G.; Shafran, H.; Peer, R.; Flaishman, M.A. Anthocyanin accumulation is initiated by abscisic acid to enhance fruit color during fig (Ficus carica L.) ripening. J. Plant Physiol. 2020, 251, 153192. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 2020, 228, 1864–1879. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Zhao, T.; Huang, C.; Li, N.; Ge, Y.; Wang, L.; Tang, Y.; Wang, Y.; Li, Y.; Zhang, C. Ubiquitin ligase VvPUB26 in grapevine promotes proanthocyanidin synthesis and resistance to powdery mildew. Plant Physiol. 2024, 195, 2891–2910. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Liao, L.; Deng, L.; Jin, Z.; Huang, Z.; Sun, G.; Xiong, B.; Wang, Z. Combined Transcriptome and Metabolome Analyses Reveal Candidate Genes Involved in Tangor (Citrus reticulata × Citrus sinensis) Fruit Development and Quality Formation. Int. J. Mol. Sci. 2022, 23, 5457. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Y.; Liu, Z.; Chen, W.; Wang, Y.; Wang, X.; Liu, Y.; Zheng, Y. Combined analysis of the transcriptome and metabolome provides insights into the fleshy stem expansion mechanism in stem lettuce. Front. Plant Sci. 2022, 13, 1101199. [Google Scholar] [CrossRef]
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; King, G.J. Chromosome-scale assembly and annotation of the macadamia genome (Macadamia integrifolia HAES 741). G3 Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, D.; Wei, R.; Hong, Y.; Zhang, W.; Pan, X. Genome-wide characterization, identification, and function analysis of candidate JsMYB genes involved in regulating flavonol biosynthesis in Juglans sigillata Dode. Sci. Hortic. 2023, 317, 112044. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef]
- Ramaroson, M.L.; Koutouan, C.; Helesbeux, J.J.; Le Clerc, V.; Hamama, L.; Geoffriau, E.; Briard, M. Role of Phenylpropanoids and Flavonoids in Plant Resistance to Pests and Diseases. Molecules 2022, 27, 8371. [Google Scholar] [CrossRef]
- Liu, L.; Gregan, S.; Winefield, C.; Jordan, B. From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry. Plant Cell Environ. 2015, 38, 905–919. [Google Scholar] [CrossRef]
- Li, T.; Li, F.; Liu, X.; Liu, J.; Li, D. Synergistic anti-inflammatory effects of quercetin and catechin via inhibiting activation of TLR4-MyD88-mediated NF-κB and MAPK signaling pathways. Phytother. Res. 2019, 33, 756–767. [Google Scholar] [CrossRef]
- da Silva, A.B.; Coelho, P.L.C.; Oliveira, M.d.N.; Oliveira, J.L.; Amparo, J.A.O.; da Silva, K.C.; Soares, J.R.P.; Pitanga, B.P.S.; Souza, C.d.S.; Lopes, G.P.d.F.; et al. The flavonoid rutin and its aglycone quercetin modulate the microglia inflammatory profile improving antiglioma activity. Brain Behav. Immun. 2020, 85, 170–185. [Google Scholar] [CrossRef]
- Han, S.; Luo, L.; Lu, J.; He, E.; Lu, Z. Fruit development dynamics and model establishment of macadamia nut in Guizhou. Seed 2023, 42, 79–85+97. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Pu, Z.; Wang, J.; Zheng, Y.; Li, Y.; Wei, Y. Regulation, evolution, and functionality of flavonoids in cereal crops. Biotechnol. Lett. 2013, 35, 1765–1780. [Google Scholar] [CrossRef]
- Yan, L.; Yang, H.; Ye, Q.; Huang, Z.; Zhou, H.; Cui, D. Metabolome and transcriptome profiling reveal regulatory network and mechanism of flavonoid biosynthesis during color formation of Dioscorea cirrhosa L. PeerJ 2022, 10, e13659. [Google Scholar] [CrossRef]
- Günther, C.S.; Plunkett, B.J.; Cooney, J.M.; Jensen, D.J.; Trower, T.M.; Elborough, C.; Nguyen, H.M.; Deng, C.H.; Lafferty, D.J.; Albert, N.W.; et al. Biotic stress-induced and ripening-related anthocyanin biosynthesis are regulated by alternate phytohormone signals in blueberries. Environ. Exp. Bot. 2022, 203, 105065. [Google Scholar] [CrossRef]
- Chung, S.W.; Yu, D.J.; Oh, H.D.; Ahn, J.H.; Huh, J.H.; Lee, H.J. Transcriptional regulation of abscisic acid biosynthesis and signal transduction, and anthocyanin biosynthesis in ‘Bluecrop’ highbush blueberry fruit during ripening. PLoS ONE 2019, 14, e0220015. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Yang, S.; Chai, L.; Wu, C.; Zhou, J.; Liu, Y.; Xue, Z. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, L.; Yuan, C.; Guan, J. Molecular Characterization of Ethylene-Regulated Anthocyanin Biosynthesis in Plums During Fruit Ripening. Plant Mol. Biol. Rep. 2016, 34, 777–785. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153–S164. [Google Scholar] [CrossRef]
- Flores, G.; del Castillo, M.L.R. Influence of preharvest and postharvest methyl jasmonate treatments on flavonoid content and metabolomic enzymes in red raspberry. Postharvest Biol. Technol. 2014, 97, 77–82. [Google Scholar] [CrossRef]
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of Phytohormone Signaling: A Primary Function of Flavonoids in Plant-Environment Interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Zhai, R.; Wang, Z.; Zhang, S.W.; Meng, G.; Song, L.Y.; Wang, Z.G.; Li, P.M.; Ma, F.W.; Xu, L.F. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 2016, 67, 1275–1284. [Google Scholar] [CrossRef]
- Weerawanich, K.; Halbwirth, H.; Sirikantaramas, S. A novel MYB transcription factor from durian (Durio zibethinus), DzMYB1, regulates flavonoid biosynthesis in fruit pulp. Sci. Hortic. 2024, 333, 113246. [Google Scholar] [CrossRef]
- Chandler, V.L. Two Regulatory Genes of the Maize Anthocyanin Pathway Are Homologous: Isolation of B Utilizing R Genomic Sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef]
- Fan, Y.; Peng, J.; Wu, J.; Zhou, P.; He, R.; Allan, A.C.; Zeng, L. NtbHLH1, a JAF13-like bHLH, interacts with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus. BMC Plant Biol. 2021, 21, 275. [Google Scholar] [CrossRef]
- Li, M.; Sun, L.; Gu, H.; Cheng, D.; Guo, X.; Chen, R.; Wu, Z.; Jiang, J.; Fan, X.; Chen, J. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 2021, 11, 6863. [Google Scholar] [CrossRef]
- Heendeniya, R.G.; Gruber, M.Y.; Lei, Y.; Yu, P. Biodegradation Profiles of Proanthocyanidin-Accumulating Alfalfa Plants Coexpressing Lc-bHLH and C1-MYB Transcriptive Flavanoid Regulatory Genes. J. Agric. Food Chem. 2019, 67, 4793–4799. [Google Scholar] [CrossRef]
- Li, X.; Cao, L.; Jiao, B.; Yang, H.; Ma, C.; Liang, Y. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion(Allium cepa L.). Hortic. Res. 2022, 9, uhac128. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Kong, W.; Peng, R.; Liu, Q.; Yao, Q. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 2016, 244, 59–73. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, L.; Long, Q.; Chen, J.; Zhang, Q.; Guo, G.; He, F.; Cai, H.; Geng, J.; Song, X.; Zeng, H.; et al. Co-Analysis of Transcriptome and Metabolome Reveals Flavonoid Biosynthesis in Macadamia Pericarp Across Developmental Stages. Foods 2025, 14, 3618. https://doi.org/10.3390/foods14213618
Tao L, Long Q, Chen J, Zhang Q, Guo G, He F, Cai H, Geng J, Song X, Zeng H, et al. Co-Analysis of Transcriptome and Metabolome Reveals Flavonoid Biosynthesis in Macadamia Pericarp Across Developmental Stages. Foods. 2025; 14(21):3618. https://doi.org/10.3390/foods14213618
Chicago/Turabian StyleTao, Liang, Qingyi Long, Jinyan Chen, Qin Zhang, Guangzheng Guo, Fengping He, Hu Cai, Jianjian Geng, Ximei Song, Hui Zeng, and et al. 2025. "Co-Analysis of Transcriptome and Metabolome Reveals Flavonoid Biosynthesis in Macadamia Pericarp Across Developmental Stages" Foods 14, no. 21: 3618. https://doi.org/10.3390/foods14213618
APA StyleTao, L., Long, Q., Chen, J., Zhang, Q., Guo, G., He, F., Cai, H., Geng, J., Song, X., Zeng, H., Wang, W., Yang, F., Kang, Z., & Tu, X. (2025). Co-Analysis of Transcriptome and Metabolome Reveals Flavonoid Biosynthesis in Macadamia Pericarp Across Developmental Stages. Foods, 14(21), 3618. https://doi.org/10.3390/foods14213618




