Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains Isolation and Growth Conditions
2.2. Bacterial Isolate Identification
2.2.1. Identification of Selected Isolates by MALDI Biotyper Sirius GP System
2.2.2. Molecular Identification
16S rRNA Amplification with Colony PCR
Sanger Sequencing Analysis
Phylogenetic and Taxonomic Analysis of 16S rRNA Sequences
| Category | Target Gene | Abbreviation | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) | Amplicon Size (bp) | Tm | References |
|---|---|---|---|---|---|---|---|
| Molecular identification | 16S rRNA | 16S rRNA | AGAGTTTGATCCTGGCTCAG | ACGGCTACCTTGTTACGACTT | 1485 | 56 | [40] |
| Genotyping | ERIC | ERIC | ATGTAAGCTCCTGGGGATTCAC | AAGTAAGTGACTGGGGTGAGCG | / | 52 | [41,42] |
| (GTG)5 | (GTG)5 | GTGGTGGTGGTGGTG | / | 45 | [43,44] | ||
| BOX-PCR | BOX-PCR | CTACGGCAAGGCGACGCTGACG | / | 53 | [44,45] | ||
| Enterocin genes | Enterocin A | entA | GGTACCACTCATAGTGGAAA | CCCTGGAATTGCTCCACCTAA | 138 | 55 | [46] |
| Enterocin B | entB | CAAAATGTAAAAGAATTAAGTACG | AGAGTATACATTTGCTAACCC | 201 | 56 | [46] | |
| Enterocin L50A | entL50A | ATGGGAGCAATCGCAAAATTA | TTTGTTAATTGCCCATCCTTC | 135 | 56 | [47] | |
| Enterocin L50B | entL50B | ATGGGAGCAATCGCAAAATTA | TAGCCATTTTTCAATTTGATC | 274 | 58 | [46] | |
| Enterocin P | entP | ATGAGAAAAAAATTATTTAGTTT | TTAATGTCCCATACCTGCCAAACCAG | 216 | 41 | [48] | |
| Enterocin Q | entQ | GGAATAAGAGTAGTAGTGGAATACTGATATGAGAC | AAAGACTGCTCTTCCGAGCAGCC | 653 | 60 | [48] | |
| Enterocin AS-48 | entAS-48 | GAGGAGTATCATGGTTAAAGA | ATATTGTTAAATTACCAA | 339 | 56 | [48] | |
| Enterocin 31 | ent31 | CCTACGTATTACGGAAATGGT | GCCATGTTGTACCCAACCATT | 130 | 58 | [48] | |
| Cytolysin | cyl | GGCGGTATTTTTACTGGAGT | CCTACTCCTAAGCCTATGGTA | 248 | 56 | [47] | |
| Enterocin CRL35 | entCRL35 | GCAAACCGATAAGAATGTGGGAT | TATACATTGTCCCCACAACC | 490 | 55 | [49] | |
| Mundticin KS | munKS | TGAGAGAAGGTTTAAGTTTTGAAGAA | TCCACTGAAATCCATGAATGA | 380 | 55 | [49] | |
| Virulence factors | Enterococcal surface protein | esp | ACGTGGATGTAGAGTTTGC | GAATATGTCACTACAACCGTAC | 5791 | 57 | [50] |
| Gelatinase E | gelE | ACCCCGTATCATTGGTTT | ACGCATTGCTTTTCCATC | 419 | 54 | [51] | |
| Hyaluronidase | hyl | ACAGAAGAGCTGCAGGAAATG | GACTGACGTCCAAGTTTCCAA | 276 | 55 | [42] | |
| Antibiotic resistance genes | Vancomycin A | vanA | CATGAATAGAATAAAAGTTGCAATA | CCCCTTTAACGCTAATACGATCAA | 1030 | 58 | [52] |
| Vancomycin B | vanB | GTGACAAACCGGAGGCGAGGA | CCGCCATCCTCCTGGAAAAAA | 433 | 64 | [52] | |
| Erythromycin Ribosomal Methylase A | ermA | TCTAAAAAGCATGTAAAAGAA | TGATTATAATTATTTGATAGCTTC | 645 | 50 | [53] | |
| Erythromycin Ribosomal Methylase B | ermB | GAAAAGGTACTCAACCAAATA | AGTAACGGTACTTAAATTGTTTAC | 639 | 54 | [53] | |
| Aminoglycosidase Acetyltransferase (6′)-Aminoglycoside Phosphotransferase (2″) | aac(6′)-Ie-alph(2″) | CCAAGAGCAATAAGGGCATA | CACTATCATAACCACTACCG | 220 | 44 | [54] | |
2.2.3. Genotyping and Molecular Profiling Using REP-PCR
2.3. Bacteriocinogenic Strain Screening and Assays
2.4. Molecular Screening of Enterocin Genes, Virulence Factors, and Antibiotic Resistance
2.5. Statistical Analyses
3. Results
3.1. Lactic Acid Bacteria Isolation
3.2. Identification of Enterococcus Strains
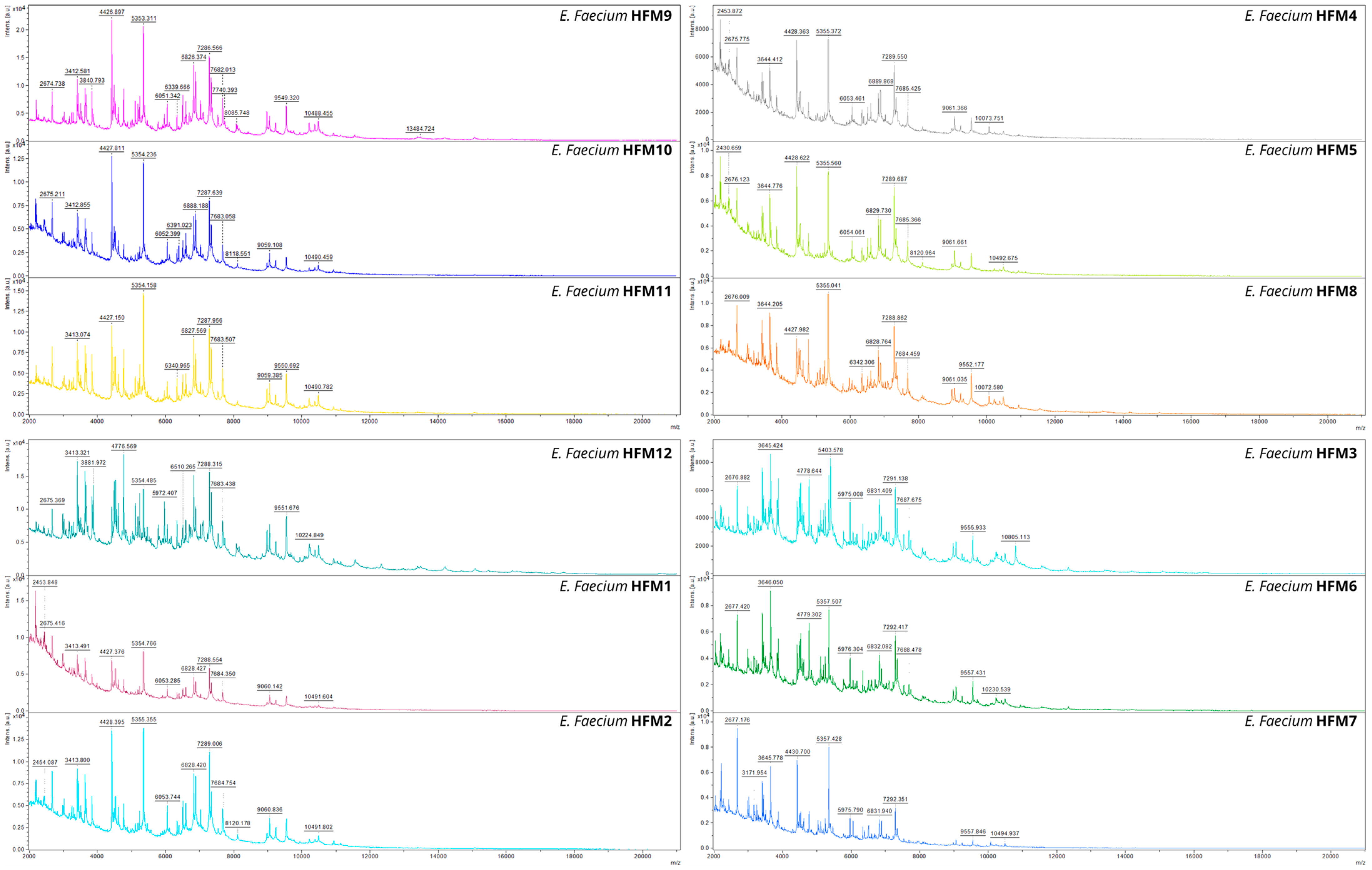
3.2.1. MALDI-TOF MS Analysis
3.2.2. 16S rRNA Gene Sequencing
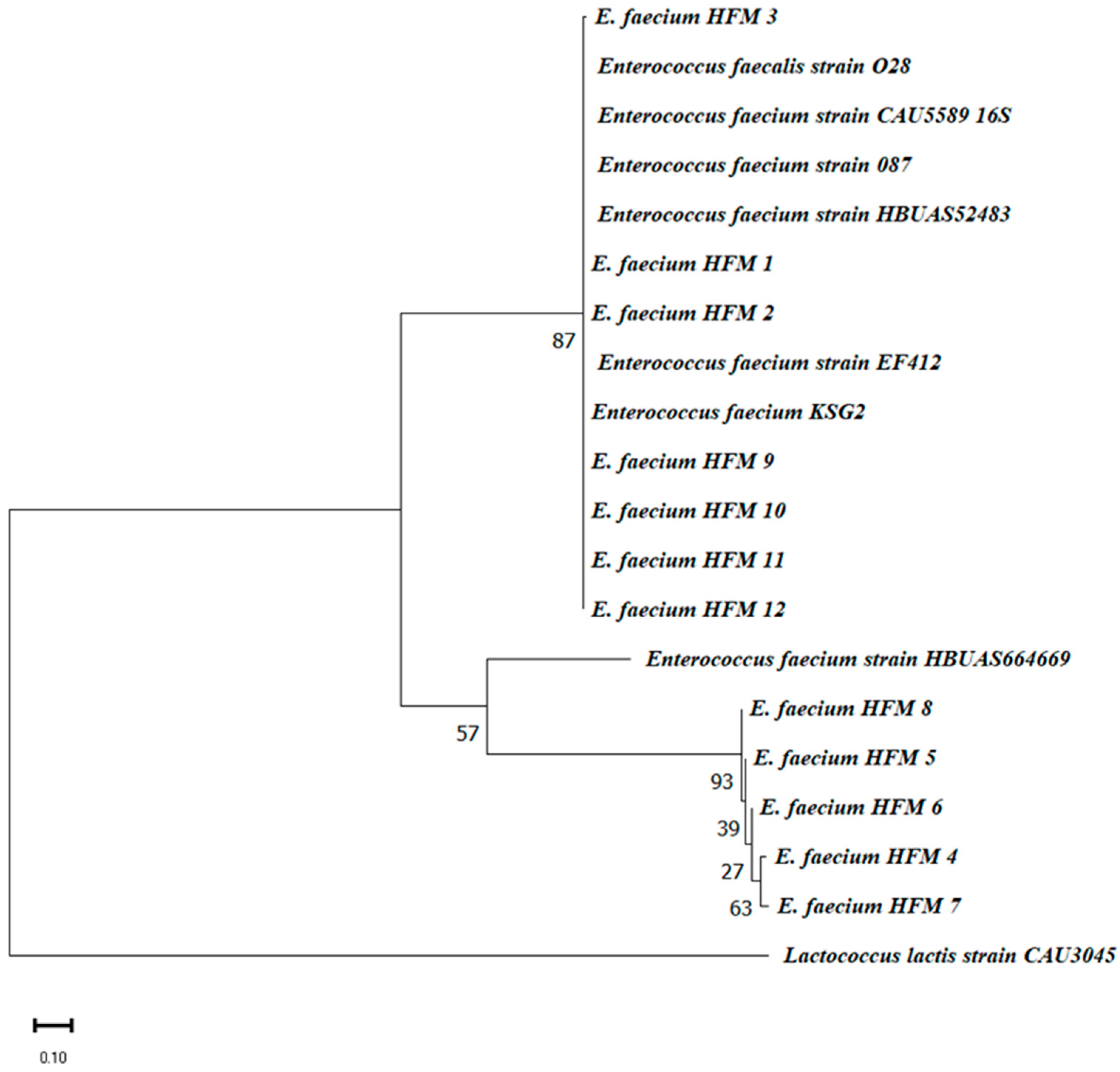
3.2.3. Phylogenetic Analysis
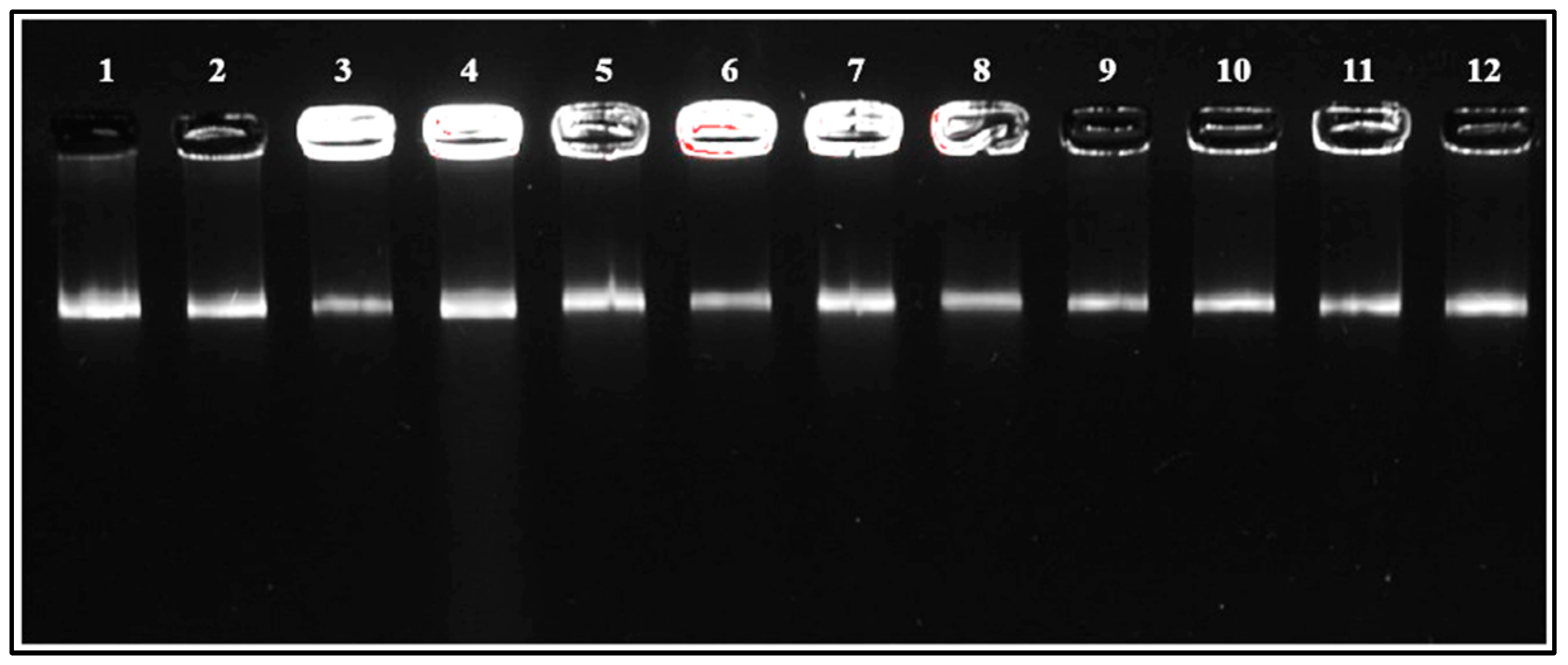
3.2.4. Genotyping Analysis
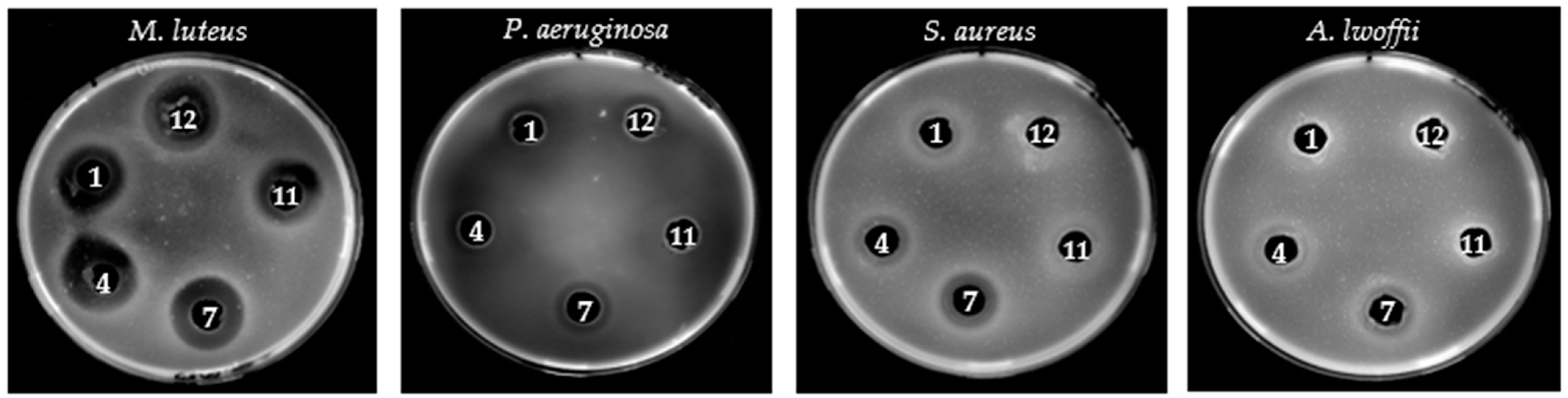
3.3. Screening of Bacteriocinogenic Strains and Bacteriocin-like Activity Validation
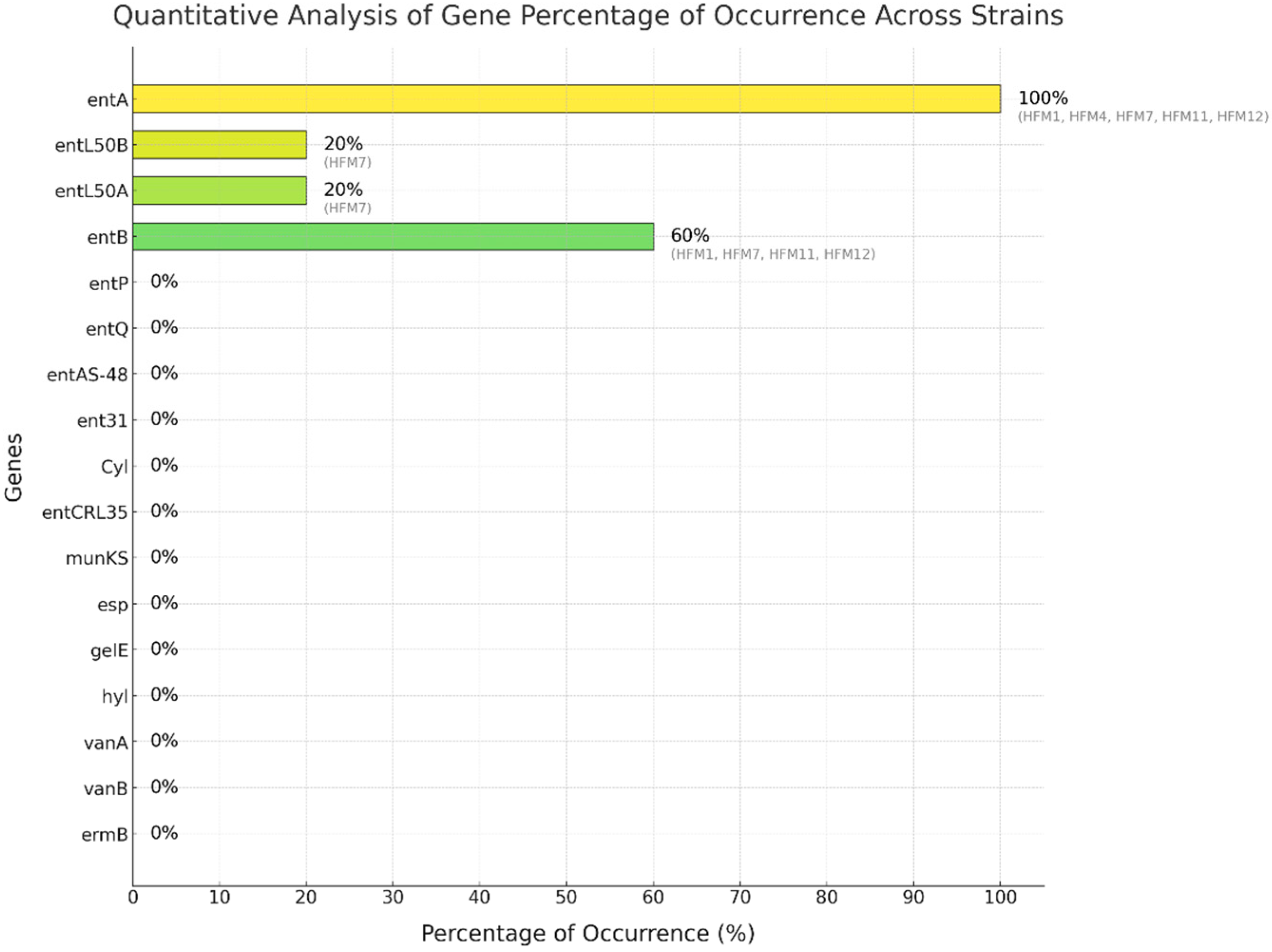
3.4. Screening of Enterocin Genes, Virulence Factors, and Antibiotic Resistance Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Todorov, S.D.; Wachsman, M.; Tomé, E.; Dousset, X.; Destro, M.T.; Dicks, L.M.T.; de Melo Franco, B.D.G.; Vaz-Velho, M.; Drider, D. Characterisation of an Antiviral Pediocin-like Bacteriocin Produced by Enterococcus faecium. Food Microbiol. 2010, 27, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Holzapfel, W.H.; Tagg, J.R. Bacteriocinogenic Enterococcus casseliflavus Isolated from Fresh Guava Fruit (Psidium guajava): Characterization of Bacteriocin ST192Gu and Some Aspects of Its Mode of Action on Listeria spp. and Enterococcus spp. Fermentation 2023, 9, 226. [Google Scholar] [CrossRef]
- Amenu, D.; Bacha, K. Probiotic Potential and Safety Analysis of Lactic Acid Bacteria Isolated from Ethiopian Traditional Fermented Foods and Beverages. Ann. Microbiol. 2023, 73, 37. [Google Scholar] [CrossRef]
- Javed, A.; Masud, T.; Ul Ain, Q.; Imran, M.; Maqsood, S. Enterocins of Enterococcus faecium, Emerging Natural Food Preservatives. Ann. Microbiol. 2011, 61, 699–708. [Google Scholar] [CrossRef]
- Pujato, S.A.; Mercanti, D.J.; Briggiler Marcó, M.; Capra, M.L.; Quiberoni, A.; Guglielmotti, D.M. Bacteriocins from Lactic Acid Bacteria: Strategies for the Bioprotection of Dairy Foods. Front. Food Sci. Technol. 2024, 4, 1439891. [Google Scholar] [CrossRef]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef] [PubMed]
- Zapaśnik, A.; Sokołowska, B.; Bryła, M. Role of Lactic Acid Bacteria in Food Preservation and Safety. Foods 2022, 11, 1283. [Google Scholar] [CrossRef] [PubMed]
- Anumudu, C.K.; Miri, T.; Onyeaka, H. Multifunctional Applications of Lactic Acid Bacteria: Enhancing Safety, Quality, and Nutritional Value in Foods and Fermented Beverages. Foods 2024, 13, 3714. [Google Scholar] [CrossRef]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocins: Novel Solutions to Age Old Spore-Related Problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef]
- Laranjo, M.; Potes, M.E.; Elias, M. Role of Starter Cultures on the Safety of Fermented Meat Products. Front. Microbiol. 2019, 10, 853. [Google Scholar] [CrossRef] [PubMed]
- Ismael, M.; Huang, M.; Zhong, Q. The Bacteriocins Produced by Lactic Acid Bacteria and the Promising Applications in Promoting Gastrointestinal Health. Foods 2024, 13, 3887. [Google Scholar] [CrossRef]
- Marco, M.L.; Sanders, M.E.; Gänzle, M.; Arrieta, M.C.; Cotter, P.D.; De Vuyst, L.; Hill, C.; Holzapfel, W.; Lebeer, S.; Merenstein, D.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on Fermented Foods. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 196–208. [Google Scholar] [CrossRef]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented Foods in a Global Age: East Meets West. Comp. Rev. Food Sci. Food Safe. 2020, 19, 184–217. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, A.K.; Islam, M.; Edirisinghe, I.; Burton-Freeman, B. Phytochemical Composition and Health Benefits of Figs (Fresh and Dried): A Review of Literature from 2000 to 2022. Nutrients 2023, 15, 2623. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, Y.; Guo, Y.; Jiang, Y.; Wen, L.; Yang, B. New Insights of Fig (Ficus carica L.) as a Potential Function Food. Trends Food Sci. Technol. 2023, 140, 104146. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-E.; Heo, S.; Lee, G.; Park, H.-J.; Jeong, D.-W. Novel Starter Strain Enterococcus faecium DMEA09 from Traditional Korean Fermented Meju. Foods 2023, 12, 3008. [Google Scholar] [CrossRef]
- Ucak, S.; Yurt, M.N.Z.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Soyucok, A.; Aydin, A.; Ozalp, V.C.; Sudagidan, M. Identification of Bacterial Communities of Fermented Cereal Beverage Boza by Metagenomic Analysis. LWT 2022, 153, 112465. [Google Scholar] [CrossRef]
- Valledor, S.J.D.; Bucheli, J.E.V.; Holzapfel, W.H.; Todorov, S.D. Exploring Beneficial Properties of the Bacteriocinogenic Enterococcus faecium ST10Bz Strain Isolated from Boza, a Bulgarian Cereal-Based Beverage. Microorganisms 2020, 8, 1474. [Google Scholar] [CrossRef] [PubMed]
- Portilha-Cunha, M.F.; Macedo, A.C.; Malcata, F.X. A Review on Adventitious Lactic Acid Bacteria from Table Olives. Foods 2020, 9, 948. [Google Scholar] [CrossRef]
- Randazzo, C.L.; Rajendram, R.; Caggia, C. Lactic Acid Bacteria in Table Olive Fermentation. In Olives and Olive Oil in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2010; pp. 369–376. ISBN 978-0-12-374420-3. [Google Scholar]
- Cavicchioli, V.Q.; Camargo, A.C.; Todorov, S.D.; Nero, L.A. Novel Bacteriocinogenic Enterococcus hirae and Pediococcus pentosaceus Strains with Antilisterial Activity Isolated from Brazilian Artisanal Cheese. J. Dairy Sci. 2017, 100, 2526–2535. [Google Scholar] [CrossRef]
- Moraes, P.M.; Perin, L.M.; Todorov, S.D.; Silva, A.; Franco, B.D.G.M.; Nero, L.A. Bacteriocinogenic and Virulence Potential of Enterococcus Isolates Obtained from Raw Milk and Cheese. J. Appl. Microbiol. 2012, 113, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Toğay, S.Ö.; Ay, M.; Güneşer, O.; Yüceer, Y.K. Investigation of Antimicrobial Activity and entA and entB Genes in Enterococcus faecium and Enterococcus faecalis Strains Isolated from Naturally Fermented Turkish White Cheeses. Food Sci. Biotechnol. 2016, 25, 1633–1637. [Google Scholar] [CrossRef]
- Todorov, S.D.; Favaro, L.; Gibbs, P.; Vaz-Velho, M. Enterococcus faecium Isolated from Lombo, a Portuguese Traditional Meat Product: Characterisation of Antibacterial Compounds and Factors Affecting Bacteriocin Production. Benef. Microbes 2012, 3, 319–330. [Google Scholar] [CrossRef]
- Todorov, S.D.; Stojanovski, S.; Iliev, I.; Moncheva, P.; Nero, L.A.; Ivanova, I.V. Technology and Safety Assessment for Lactic Acid Bacteria Isolated from Traditional Bulgarian Fermented Meat Product “Lukanka”. Braz. J. Microbiol. 2017, 48, 576–586. [Google Scholar] [CrossRef]
- Rwubuzizi, R.; Carneiro, K.O.; Holzapfel, W.H.; Vaz-Velho, M.; Todorov, S.D. Bacteriocin and Antioxidant Production, a Beneficial Properties of Lactic Acid Bacteria Isolated from Fermented Vegetables of Northwest Bulgaria. Probiot. Antim. Prot. 2023, 17, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Belgacem, Z.B.; Abriouel, H.; Omar, N.B.; Lucas, R.; Martínez-Canamero, M.; Gálvez, A.; Manai, M. Antimicrobial Activity, Safety Aspects, and Some Technological Properties of Bacteriocinogenic Enterococcus faecium from Artisanal Tunisian Fermented Meat. Food Control 2010, 21, 462–470. [Google Scholar] [CrossRef]
- Goa, T.; Beyene, G.; Mekonnen, M.; Gorems, K. Isolation and Characterization of Lactic Acid Bacteria from Fermented Milk Produced in Jimma Town, Southwest Ethiopia, and Evaluation of Their Antimicrobial Activity against Selected Pathogenic Bacteria. Int. J. Food Sci. 2022, 2022, 2076021. [Google Scholar] [CrossRef]
- Zater, Z.Y.; Merzoug, M.; Baltaci, M.O.; Todorov, S.D.; Adiguzel, A.; Roudj, S. Impact of a Novel Caseinolytic Protease Single Mutation on Lactiplantibacillus pentosus Growth Performance. Process Biochem. 2024, 145, 145–152. [Google Scholar] [CrossRef]
- McElvania TeKippe, E.; Shuey, S.; Winkler, D.W.; Butler, M.A.; Burnham, C.-A.D. Optimizing Identification of Clinically Relevant Gram-Positive Organisms by Use of the Bruker Biotyper Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry System. J. Clin. Microbiol. 2013, 51, 1421–1427. [Google Scholar] [CrossRef] [PubMed]
- Haider, A.; Ringer, M.; Kotroczó, Z.; Mohácsi-Farkas, C.; Kocsis, T. The Importance of Protein Fingerprints in Bacterial Identification: The MALDI-TOF Technique. J. Environ. Geogr. 2023, 16, 38–45. [Google Scholar] [CrossRef]
- Bergkessel, M.; Guthrie, C. Colony PCR. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 529, pp. 299–309. ISBN 0076-6879. [Google Scholar]
- Bell, J.R. A Simple Way to Treat PCR Products Prior to Sequencing Using ExoSAP-IT®. Biotechniques 2008, 44, 834. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.; Fernandes, N.; Silva, B.N.; Gonzales-Barron, U.; Cadavez, V. Molecular Identification and Phylogenetic Inference of Lactic Acid Bacteria Isolated from Goat’s Raw Milk Cheese. Biol. Life Sci. Forum 2023, 26, 39. [Google Scholar] [CrossRef]
- Pertsemlidis, A.; Fondon, J.W. Having a BLAST with Bioinformatics (and Avoiding BLASTphemy). Gen. Biol. 2001, 2, 1–10. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Katsura, Y.; Stanley Jr, C.E.; Kumar, S.; Nei, M. The Reliability and Stability of an Inferred Phylogenetic Tree from Empirical Data. Mol. Biol. Evol. 2017, 34, 718–723. [Google Scholar] [CrossRef]
- El-Zamkan, M.A.; Mohamed, H.M.A. Antimicrobial Resistance, Virulence Genes and Biofilm Formation in Enterococcus Species Isolated from Milk of Sheep and Goat with Subclinical Mastitis. PLoS ONE 2021, 16, e0259584. [Google Scholar] [CrossRef]
- Saengsuwan, P.; Singkhamanan, K.; Madla, S.; Ingviya, N.; Romyasamit, C. Molecular Epidemiology of Vancomycin-Resistant Enterococcus faecium Clinical Isolates in a Tertiary Care Hospital in Southern Thailand: A Retrospective Study. Peer J. 2021, 9, e11478. [Google Scholar] [CrossRef]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of Repetitive DNA Sequences in Eubacteria and Application to Finerpriting of Bacterial Enomes. Nucl. Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Pino, A.; Russo, N.; Solieri, L.; Sola, L.; Caggia, C.; Randazzo, C.L. Microbial Consortia Involved in Traditional Sicilian Sourdough: Characterization of Lactic Acid Bacteria and Yeast Populations. Microorganisms 2022, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J. Genomic Fingerprinting of Bacteria Using Repetitive Sequence-Based Polymerase Chain Reaction. Methods Mol. Cell Biol. 1994, 5, 25–40. [Google Scholar]
- Bilung, L.M.; Pui, C.F.; Su’ut, L.; Apun, K. Evaluation of BOX-PCR and ERIC-PCR as Molecular Typing Tools for Pathogenic Leptospira. Dis. Mark. 2018, 2018, 1351634. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Rocha, K.R.; Terra, M.R.; Furlaneto, M.C.; Maia, L.F. Screening of the Enterocin-Encoding Genes and Antimicrobial Activity in Enterococcus Species. J. Microbiol. Biotechnol. 2016, 26, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Foulquie Moreno, M.R.; Callewaert, R.; Devreese, B.; Van Beeumen, J.; De Vuyst, L. Isolation and Biochemical Characterisation of Enterocins Produced by Enterococci from Different Sources. J. Appl. Microbiol. 2003, 94, 214–229. [Google Scholar] [CrossRef]
- Choeisoongnern, T.; Sirilun, S.; Waditee-Sirisattha, R.; Pintha, K.; Peerajan, S.; Chaiyasut, C. Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation. Foods 2021, 10, 2264. [Google Scholar] [CrossRef]
- Toplu, M.S.; Tuncer, B.Ö. Evaluation of the Functional Properties and Safety of Enterocin-Producing Enterococcus faecium BT29.11 Isolated from Turkish Beyaz Cheese and Its Inhibitory Activity against Listeria monocytogenes in UHT Whole Milk. Ital. J. Food Sci. 2023, 35, 54–70. [Google Scholar] [CrossRef]
- Oancea, C. Conjugative Transfer of the Virulence Gene, esp, among Isolates of Enterococcus faecium and Enterococcus Faecalis. J. Antimicrob. Chemother. 2004, 54, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Avci, M.; Tuncer, B.Ö. Safety Evaluation of Enterocin Producer Enterococcus Sp. Strains Isolated from Traditional Turkish Cheeses. Pol. J. Microbiol. 2017, 66, 223–233. [Google Scholar] [CrossRef]
- Biswas, P.; Dey, S.; Sen, A.; Adhikari, L. Molecular Characterization of Virulence Genes in Vancomycin-Resistant and Vancomycin-Sensitive Enterococci. J. Glob. Infect. Dis. 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Juda, M.; Chudzik-Rzad, B.; Malm, A. The Prevalence of Genotypes That Determine Resistance to Macrolides, Lincosamides, and Streptogramins B Compared with Spiramycin Susceptibility among Erythromycin-Resistant Staphylococcus epidermidis. Memórias Do Inst. Oswaldo Cruz 2016, 111, 155–160. [Google Scholar] [CrossRef]
- Xu, J.; Shi, C.; Song, M.; Xu, X.; Yang, P.; Paoli, G.; Shi, X. Phenotypic and Genotypic Antimicrobial Resistance Traits of Foodborne Staphylococcus aureus Isolates from Shanghai. J. Food Sci. 2014, 79, M635–M642. [Google Scholar] [CrossRef] [PubMed]
- Ahamdi, F.; Siasi Torbati, E.; Amini, K. A Comparative Study of ERIC-PCR and BOX-PCR Methods for Evaluation of Genomic Polymorphism among Multidrug-Resistant Enterococcus faecium Clinical Isolates. Med. Lab. J. 2023, 17, 13–19. [Google Scholar] [CrossRef]
- Åvec, P.; Vancanneyt, M.; Seman, M.; Snauwaert, C.; Lefebvre, K.; Sedlacek, I.; Swings, J. Evaluation of (GTG)5 -PCR for Identification of Enterococcus spp. FEMS Microbiol. Lett. 2005, 247, 59–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Vuyst, L.; Camu, N.; De Winter, T.; Vandemeulebroecke, K.; Van de Perre, V.; Vancanneyt, M.; De Vos, P.; Cleenwerck, I. Validation of the (GTG)5-Rep-PCR Fingerprinting Technique for Rapid Classification and Identification of Acetic Acid Bacteria, with a Focus on Isolates from Ghanaian Fermented Cocoa Beans. Int. J. Food Microbiol. 2008, 125, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Furtula, V.; Farrell, E.G.; Barrett, J.B.; Hiott, L.M.; Chambers, P. A Comparison of BOX-PCR and Pulsed-Field Gel Electrophoresis to Determine Genetic Relatedness of Enterococci from Different Environments. Microb. Ecol. 2012, 64, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ–a Tool for Analyzing DNA Fingerprint Gel Images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Barry, A.L. Standardization of Antimicrobial Susceptibility Testing. Clin. Lab. Med. 1989, 9, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-H.; Pan, T.-M. Characterization of an Antimicrobial Substance Produced by Lactobacillus plantarum NTU 102. J. Microbiol. Immunol. Infect. 2019, 52, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Ruxton, G.D.; Beauchamp, G. Time for Some a Priori Thinking about Post Hoc Testing. Behav. Ecol. 2008, 19, 690–693. [Google Scholar] [CrossRef]
- Alharthy, N.; Bawazir, A. Effects of The Mixture Dried Figs (Ficus carica) And Olive Oil on Amnesia Model of Alzheimer’s Induced By Scopolamine in Male Albino Rats. Pharmacophore 2019, 10, 62–67. [Google Scholar]
- Debib, A.; Tir-Touil, M.; Meddah, B.; Hamaidi-Chergui, F.; Menadi, S.; Alsayadi, M. Evaluation of Antimicrobial and Antioxidant Activities of Oily Macerates of Algerian Dried Figs (Ficus carica L.). Int. Food Res. J. 2018, 25, 351. [Google Scholar]
- El Dessouky Abdel-Aziz, M.; Samir Darwish, M.; Mohamed, A.H.; El-Khateeb, A.Y.; Hamed, S.E. Potential Activity of Aqueous Fig Leaves Extract, Olive Leaves Extract and Their Mixture as Natural Preservatives to Extend the Shelf Life of Pasteurized Buffalo Milk. Foods 2020, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.A.H.M.; Sharma, S.P.; Chung, H.-J.; Kim, H.-J.; Hong, S.-T.; Lee, S. Ultra-High Efficient Colony PCR for High Throughput Screening of Bacterial Genes. Ind. J. Microbiol. 2017, 57, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Uzuriaga, M.; Guillén-Grima, F.; Rua, M.; Leiva, J.; Yuste, J.R. Accelerated Bacterial Identification with MALDI-TOF MS Leads to Fewer Diagnostic Tests and Cost Savings. Antibiotics 2024, 13, 1163. [Google Scholar] [CrossRef] [PubMed]
- Carbonnelle, E.; Raskine, L. MALDI-TOF Mass Spectrometry Tools for Bacterial Identification in Clinical Microbiology Laboratory. Bio. Trib. Magaz. 2011, 39, 35–42. [Google Scholar] [CrossRef]
- Kim, S.-H.; Chon, J.-W.; Jeong, H.-W.; Song, K.-Y.; Kim, D.-H.; Bae, D.; Kim, H.; Seo, K.-H. Identification and Phylogenetic Analysis of Enterococcus Isolates Using MALDI-TOF MS and VITEK 2. AMB Expr. 2023, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, E.; Cai, Y.; Marchioni, E.; Ennahar, S. Genetic Identification of the Bacteriocins Produced by Enterococcus faecium IT62 and Evidence That Bacteriocin 32 Is Identical to Enterocin IT. Antimicrob. Agents Chemother. 2009, 53, 1907–1911. [Google Scholar] [CrossRef]
- Vimont, A.; Fernandez, B.; Hammami, R.; Ababsa, A.; Daba, H.; Fliss, I. Bacteriocin-Producing Enterococcus faecium LCW 44: A High Potential Probiotic Candidate from Raw Camel Milk. Front. Microbiol. 2017, 8, 865. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, B.R.; Broersma, K.; Mazumder, A. Comparison of Five Rep-PCR Genomic Fingerprinting Methods for Differentiation of Fecal Escherichia coli from Humans, Poultry and Wild Birds. FEMS Microbiol. Lett. 2007, 277, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Santos, A.C.; Novais, C.; Peixe, L.; Freitas, A.R. Enterococcus spp. as a Producer and Target of Bacteriocins: A Double-Edged Sword in the Antimicrobial Resistance Crisis Context. Antibiotics 2021, 10, 1215. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as Probiotics and Their Implications in Food Safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial Activities of Bacteriocins: Application in Foods and Pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing Innate Immunity for Food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Casaus, P.; Nilsen, T.; Cintas, L.M.; Nes, I.F.; Hernández, P.E.; Holo, H. Enterocin B, a New Bacteriocin from Enterococcus faecium T136 Which Can Act Synergistically with Enterocin A. Microbiology 1997, 143, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Merzoug, M.; Mosbahi, K.; Walker, D.; Karam, N.-E. Screening of the Enterocin-Encoding Genes and Their Genetic Determinism in the Bacteriocinogenic Enterococcus faecium GHB21. Probiot. Antim. Prot. 2019, 11, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Sonsa-Ard, N.; Rodtong, S.; Chikindas, M.L.; Yongsawatdigul, J. Characterization of Bacteriocin Produced by Enterococcus faecium CN-25 Isolated from Traditionally Thai Fermented Fish Roe. Food Control 2015, 54, 308–316. [Google Scholar] [CrossRef]
- Merzoug, M.; Dalache, F.; Zadi Karam, H.; Karam, N.-E. Isolation and Preliminary Characterisation of Bacteriocin Produced by Enterococcus faecium GHB21 Isolated from Algerian Paste of Dates “Ghars”. Ann. Microbiol. 2016, 66, 795–805. [Google Scholar] [CrossRef]
- O’Shea, E.F.; O’Connor, P.M.; Raftis, E.J.; O’Toole, P.W.; Stanton, C.; Cotter, P.D.; Ross, R.P.; Hill, C. Production of Multiple Bacteriocins from a Single Locus by Gastrointestinal Strains of Lactobacillus salivarius. J. Bacteriol. 2011, 193, 6973–6982. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L. Screening for Enterocins and Detection of Hemolysin and Vancomycin Resistance in Enterococci of Different Origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Furlaneto-Maia, L.; Ramalho, R.; Rocha, K.R.; Furlaneto, M.C. Antimicrobial Activity of Enterocins against Listeria sp. and Other Food Spoilage Bacteria. Biotechnol. Lett. 2020, 42, 797–806. [Google Scholar] [CrossRef]
- Strateva, T.; Dimov, S.G.; Atanasova, D.; Petkova, V.; Savov, E.; Mitov, I. Molecular Genetic Study of Potentially Bacteriocinogenic Clinical and Dairy Enterococcus spp. Isolates from Bulgaria. Ann. Microbiol. 2016, 66, 381–387. [Google Scholar] [CrossRef]
- Gyurova, A.; Vladimirova, A.; Peykov, S.; Dimitrov, M.; Strateva, T.; Dimov, S.G. Characterization of Enterococcus durans EDD2, a Strain from Beehives with Inhibitory Activity Against Paenibacillus Larvae. J. Apicult. Res. 2023, 62, 1183–1196. [Google Scholar] [CrossRef]
- Strompfová, V.; Lauková, A.; Simonová, M.; Marciňáková, M. Occurrence of the Structural Enterocin A, P, B, L50B Genes in Enterococci of Different Origin. Vet. Microbiol. 2008, 132, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Ahmadova, A.; Todorov, S.D.; Choiset, Y.; Rabesona, H.; Mirhadi Zadi, T.; Kuliyev, A.; Franco, B.D.G.D.M.; Chobert, J.-M.; Haertlé, T. Evaluation of Antimicrobial Activity, Probiotic Properties and Safety of Wild Strain Enterococcus faecium AQ71 Isolated from Azerbaijani Motal Cheese. Food Control 2013, 30, 631–641. [Google Scholar] [CrossRef]
- Rehaiem, A.; Belgacem, Z.B.; Edalatian, M.R.; Martínez, B.; Rodríguez, A.; Manai, M.; Guerra, N.P. Assessment of Potential Probiotic Properties and Multiple Bacteriocin Encoding-Genes of the Technological Performing Strain Enterococcus faecium MMRA. Food Control 2014, 37, 343–350. [Google Scholar] [CrossRef]
- García-Vela, S.; Ben Said, L.; Soltani, S.; Guerbaa, R.; Fernández-Fernández, R.; Ben Yahia, H.; Ben Slama, K.; Torres, C.; Fliss, I. Targeting Enterococci with Antimicrobial Activity against Clostridium perfringens from Poultry. Antibiotics 2023, 12, 231. [Google Scholar] [CrossRef] [PubMed]
- Cintas, L.M.; Casaus, P.; Holo, H.; Hernandez, P.E.; Nes, I.F.; Håvarstein, L.S. Enterocins L50A and L50B, Two Novel Bacteriocins from Enterococcus faecium L50, Are Related to Staphylococcal Hemolysins. J. Bacteriol. 1998, 180, 1988–1994. [Google Scholar] [CrossRef] [PubMed]
- Ferchichi, M.; Sebei, K.; Boukerb, A.M.; Karray-Bouraoui, N.; Chevalier, S.; Feuilloley, M.G.; Connil, N.; Zommiti, M. Enterococcus spp.: Is It a Bad Choice for a Good Use—A Conundrum to Solve? Microorganisms 2021, 9, 2222. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus Spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Nami, Y.; Vaseghi Bakhshayesh, R.; Mohammadzadeh Jalaly, H.; Lotfi, H.; Eslami, S.; Hejazi, M.A. Probiotic Properties of Enterococcus Isolated From Artisanal Dairy Products. Front. Microbiol. 2019, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Rehaiem, A.; Martínez, B.; Manai, M.; Rodríguez, A. Production of Enterocin A by Enterococcus faecium MMRA Isolated from ‘Rayeb’, a Traditional Tunisian Dairy Beverage. J. Appl. Microbiol. 2010, 108, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Ananou, S.; Lotfi, S.; Azdad, O.; Nzoyikorera, N. Production, Recovery and Characterization of an Enterocin with Anti-Listerial Activity Produced by Enterococcus hirae OS1. Appl. Food Biotechnol. 2020, 7, 103–114. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merzoug, M.; Bendida, K.; Aireche, M.; Zater, Z.Y.; Brakna, C.N.; Hammadi, A.I.; Saidi, Y.; Todorov, S.D.; Saidi, D. Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations. Foods 2025, 14, 766. https://doi.org/10.3390/foods14050766
Merzoug M, Bendida K, Aireche M, Zater ZY, Brakna CN, Hammadi AI, Saidi Y, Todorov SD, Saidi D. Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations. Foods. 2025; 14(5):766. https://doi.org/10.3390/foods14050766
Chicago/Turabian StyleMerzoug, Mohamed, Keltoum Bendida, Marwa Aireche, Zohra Yasmine Zater, Chaimaa Naila Brakna, Amaria Ilhem Hammadi, Yasmine Saidi, Svetoslav Dimitrov Todorov, and Djamal Saidi. 2025. "Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations" Foods 14, no. 5: 766. https://doi.org/10.3390/foods14050766
APA StyleMerzoug, M., Bendida, K., Aireche, M., Zater, Z. Y., Brakna, C. N., Hammadi, A. I., Saidi, Y., Todorov, S. D., & Saidi, D. (2025). Isolation and Characterization of Enterocin-Producing Enterococcus faecium Strains from Algerian Traditional Food “Dried Figs Marinated in Olive Oil”: Functional and Safety Evaluations. Foods, 14(5), 766. https://doi.org/10.3390/foods14050766







