Listeria monocytogenes and Listeriosis: The Global Enigma
Abstract
1. Introduction
2. Distinctive Features of L. monocytogenes Relevant for Survival in Varied Habitats
2.1. Biofilm Formation
2.1.1. Implications of Biofilm Formation
Antibiotic Tolerance/Resistance
| Description of Tested Samples | Countries | Prevalence Rates (%) of Antibiotic-Resistant L. monocytogenes | Antibiotics to Which Isolates Are Resistant | Genes Conferring Resistance and Virulence Genes | References |
|---|---|---|---|---|---|
| RTE foods (cooked red meat, cooked chicken, seafood, vegetarian, and baked egg products, raw lettuce, fruit salad, vegetable salad, dairy products, mayonnaise-based salad, deli salad, and desserts with milk) (Total samples = 201) | Turkey (Ankara) | 35.3–94.1 | Oxacillin, kanamycin, levofloxacin, teicoplanin, amoxicillin, rifampicin, ciprofloxacin 100% MDR | Not completed Virulence genes: hly A Listeriolysin | Sanlibaba et al. [90] |
| Artisanal foods (minimal and moderate processing) (Total samples = 400) | Chile | 12.5–25 | Ampicillin, trimethoprim-sulfamethoxazole | Not completed Virulence genes: hylA, prfA, inlA | Bustamante et al. [91] |
| Retail foods (RTE foods and raw foods) Total samples = 3354) | China (Zhejiang province) | 11.0 | Tetracycline | Not completed Virulence genes: prfA, hlyA, plcA, plcB, mpl, actA, genes, LIPI-1, inlA, inlB, inlC, inlJ, LIPI-2, LIPI-3, LIPI-4 | Zhang et al. [92] |
| Cake, raw meat, ice cream, minced beef, fish, unpasteurized milk, pizza (Total = 384) | Ethiopia (Gondar town) | 16.6–66.7 | Penicillin, nalidixic acid, tetracycline, chloramphenicol 16.7% MDR | Not completed | Garedew et al. [87] |
| Bovine milk (Total samples = 161) | China (Yunnan Province) | 12.5–100 | Ampicillin, tetracycline, trimethoprim-sulfamethoxazole, erythromycin, vancomycin, ciprofloxacin, meropenem 75% MDR | novA, kdpE, NmcR, drfG, facT, norB, fusA, van RM, sul4, tetA, tetB, tetD, tetM, tetS (amongst others, 99) Virulence genes (83): actA, hlyA, inlA, inlB, inlC, inlJ, mpl, plcA, plcB, prfA, fliE, flgK, etc. | Su et al. [93] |
| RTE foods including salads (lettuce, carrot, cabbage, sweetcorn, mayonnaise as options), meat pies (potatoes, minced meat, carrots as options), fried snails, and edible worms (Total samples = 411) | Southern Nigeria | 47.77–100 | Amoxicillin, cloxacillin, Augmentin, ceftazidime | Not completed Virulence genes: hlyA, inlA, iap | Ebakota et al. [94] |
| Raw fish, open-air market environment (Total samples = 862) Caesar salad, Olivier salad, burger, schnitzel, sushi, sausage (Total samples = 110) | Iran Iran (Tehran) | 16.3–27.9 0–100 | Tetracycline, ampicillin, cephalothin, penicillin, streptomycin Oxacillin, streptomycin, cotrimoxazole, clindamycin, cefoxitin, erythromycin | TetA, tetM, ampC, penA Virulence genes: inlA, inlB, inlC, inlJ, actA, hlyA, iap, plcA, prfA. ermA, ermB, cfxA, mecA Virulence genes: hlyA and prfA | Jamali et al. [9] Mirzaei et al. [95] |
| Milk samples (Total samples = 65) | South Africa (Eastern Cape) | 42.86–71.43 | Sulfamethoxazole, trimethoprim, erythromycin, cefotetan, oxytetracycline 85.71% MDR | blaTEM, blaSHV, blaZ, tetA, tetD, tetG, tetM, tetK, aph(3)-IIa (aph12)a, sul2, sul1 Virulence genes: prfA, plcA, plcB, inlA, inlC, hylA, mpl, actA, inlJ, inB | Kayode and Okoh [96] |
| Poultry meat (Total samples = 250) | Egypt (Mansoura city) | 58.33–91.67 | Tetracycline, oxytetracycline, penicillin, amoxicillin, augmentin, and ceftazidime 79.2% MDR, 16.7% XDR | Not completed Virulence genes: hlyA, actA, iap | Zakaria and Sebala [97] |
| Beef and chicken (Total samples = 90) | Iran (Zanjan city) | 91.7–100 | Trimethoprim-sulfamethoxazole, tetracycline, penicillin, and gentamycin. | Not completed Virulence genes: hlyA | Farhoumand et al. [98] |
| Retail RTE foods (cheese, cooked meats, pre-processed fruits and vegetables, mixed dishes with raw and/or cooked ingredients) (Total samples = 436) | Chile (Maule region) | 21.43–100 | Ampicillin, tetracycline | fosX, lin, norB, mprF, tetA, tetC Virulence genes: hlyA, prfA, inlA, | Parra-Flores et al. [99] |
| RTE products: Processed dairy, bovine meat, and poultry products. Processed pork meat (sausage, ham and bacon) and fish products (Total samples = 8151) | Romania (North-Western region) | 23.07–26.92 | Oxacillin, trimethoprim-sulfamethoxazole, penicillin, tetracycline 23.07% MDR | tetC, tetM, tetK, ampC, drfD Virulence genes: hlyA, prfA | Duma et al. [100] |
| Meat, seafood, dairy, confectionary products, sauces, RTE dishes, food-processing environment (Total = 269) | Italy (Lazio region) | 78.44–88.48 | Oxacillin, fosfomycin, flumenique 87.36% MDR | Not completed | Rippa et al. [101] |
| Processed raw meat products (Total samples = 270) | Jordan (Amman) | 5–56.6 | Neomycin, tetracycline, kanamycin, erythromycin | Not completed | Al-Nabulsi et al. [60] |
| Locally processed fermented foods, e.g., garri, Qunu, zobo (Total samples = 80) | Nigeria (Ethiope, Delta State) | 62.5–100 | Penicillin, clindamycin | Not completed | Beshiru and Uwhuba [102] |
| RTE foods (Total samples = 105) | India (Tamil Nadu) | 24–52 | Methicillin, clindamycin, lincomycin, azithromycin, carbenicillin, amoxicillin | Not completed | Elavarasi et al. [103] |
| Raw kebab and hamburger (Total samples = 100) | Iran | 66.7–100 | Amoxicillin, penicillin, cefalexin | Mec A Virulence genes: Not completed | Rajei et al. [104] |
| Raw meat (Total samples = 190) | Turkey (Ankara) | 86.90–100 | Ampicillin, fosmycin, nalidixic acid, linezolid, clindamycin, piperacillin. 73.91% MDR | Not completed Virulence genes: hlyA | Sanlibaba et al. [105] |
| Raw milk, ice cream, minced meat, fish fillet, sausage (Total samples = 250) | Egypt (Menoufiya governorate) | 41.2–76.4 | Oxytetracycline. Trimethoprim-sulfamethoxazole, chloramphenicol, doxycycline, levofloxacin, azithromycin 100% MDR | Not completed Virulence genes: hlyA, iap, actA | Abdeen et al. [106] |
| RTE foods including meat-free cig kofte, kavurma, pastrami, doner, salad, dessert, cheese, ice cream (Total samples = 300) | Turkey | 46.7–80 | Fusidic acid, ceftriaxone, clindamycin 40% MDR | Not completed Virulence genes: actA, iap, inlA, inlB, inlC, inlJ, plcA, prfA | Arslan and Özdemir [107] |
| Retailed beef and beef products (Total samples = 400) | South Africa (Gauteng Province) | 62.5–100 | Clindamycin, penicillin, nalidixic, cefotaxime 75.7% MDR | Not completed | Gana et al. [108] |
Persistence of L. monocytogenes in the Environment (Food Industry) and Its Implication
- The microbial species employs biofilm production as a survival strategy. The biofilm-associated bacterial cells are difficult to remove via mechanical means off/from surfaces, and they demonstrate a noticeable reduction in their sensitivity to chemical disinfectants, thus allowing them to resist traditional cleaning methods [139].
- Owing to the formation of biofilms, bacterial cells can adapt to other environmental stress conditions that usually occur in the food-processing environment such as high salinity and temperature, acidic pH, and UV light [47].
- Biofilm cells might be more equipped to sequester toxins, cooperate metabolically, exchange nutrients, as well as become more capable of obtaining novel genetic traits, e.g., antibiotic resistance genes, facilitating the survival of the organism in the said milieu [139].
2.1.2. Control of Biofilms in Food Industries
2.2. Acid Tolerance
2.3. Thermotolerance
2.4. Osmotic Shock
3. Virulence Factors, Strain Variation, and Pathogenesis of L. monocytogenes
| Virulence Factors | Descriptions | Genes Responsible | Functions | References |
|---|---|---|---|---|
| Internalins InlA | This is a major virulence factor of the bacterium of molecular weight (80-KDa), attached to the cell wall. | InlA-InB locus | It mediates the uptake of L. monocytogenes into non-phagocytic cells and promotes the adhesion and invasion of the intestinal epithelium by interacting with E-cadherin receptors. | Dellafiora et al. [209]; Ireton et al. [210]. |
| InlB | Together with inlA, inlB is equally a major adhesion protein that is attached to the cell wall and interacts via non-covalent bonds with the teichoic acid component of the cell wall. | InlB-InlA locus | The protein helps in the adherence of the pathogen and invasion of the intestinal barrier. | Ireton et al. [210]. |
| Listeria adhesion protein (LAP) | A cell wall protein (104-KDa) described as an alcohol acetaldehyde dehydrogenase that is produced predominantly as a cytosolic protein. It is an essential enzyme that occurs ubiquitously in all Listeria species. | Not applicable | In pathogenic Listeria species, LAP is translocated to the surface of the cell via the SecA2 secretory system to enable the adhesion of the pathogenic species to the intestinal cells. It equally helps in the translocation of the pathogenic cell across the intestinal epithelium. | Burkholder et al. [211]; Drolia et al. [212]. |
| Fibronectin-binding protein (FbpA) | This is a 570-amino acid polypeptide that is attached to the cell wall but exposed on the cell surface with no signal peptide. It is similar in homology to Fbp in Streptococcus. The Fbps are widely distributed in Gram-positive bacteria. | Not applicable | They recognise and bind to fibronectin, forming a three-component bridge, enabling the adhesion of the bacterial cells to the host cells. | Henderson et al. [213]; Hymes et al. [214]. |
| Actin polymerisation protein (ActA) | This is a surface protein that is attached through its hydrophobic C-terminal domain to the cell membrane of the bacterium, whereas its N-terminal domain is exposed to the cytoplasm host cell. | ActA | The protein demonstrates an asymmetrical distribution, influencing the directionality of the bacterium’s motility. Secondly, it recruits a host of vasodilator-stimulated phosphoprotein (VASP) and actin-related proteins-2 and 3 (Arp2/3) complex to facilitate filament formation and actin nucleation. | Suárez et al. [215]; Kühn and Enninga [216]. |
| Listeriolysis (LLO) | LLO is a 56-kDa pore-forming cytotoxin, belonging to the cholesterol-dependent cytolysin (CDC) family. | Hly gene | Responsible in the lysis of internalisation vacuole that leads to the discharge of the pathogen into the cytosol of the host cells. | Hamon et al. [217]; Phelps et al. [218]. |
| Phospholipases (two types) Phosphatidylinositol-specific phospholipase C (PI-PLC) Phosphatidylcholine phospholipase C (PC-PLC) | PI-PLC | PlcA gene | PI-PLC complements LLO in the lysis of the primary and secondary vacuole after the internalisation of the pathogen. It provokes the splitting of the phosphatidylinositol membrane into inositol phosphate and diacylglycerol. | Pizarro-Cerdá et al. [219]. |
| PC-PLC is a 29-KDa enzyme, broad ranged in nature, and it is formed from a precursor of 33-KDa through cleavage but requiring a zinc-dependent metalloprotease for maturation. | PlcB gene | In an LLO-deficient milieu, PC-PLC is involved in the lysis of the double-membrane secondary vacuole and the primary vacuole. | Coffey et al. [220]; Gründling et al. [221]. | |
| Positive regulatory factor (PrfA) | PrfA is formed from three promoters, including Sigma B (SigB). | LIPI-1 | It induces the transcription of LIPI-1, the predominant virulence regulon, acting as the major regulator of virulence factors that enable intracellular replication and bacterial spread to neighbouring cells. | Quereda et al. [142]; Tiensuu et al. [222]. |
Strain Variation/Diversity in L. monocytogenes
4. Epidemiology and Transmission of L. monocytogenes
4.1. Policy and Standard Compliance: Analysing Compliance with the Regulatory Framework Through Linking Monitoring Strategies to HACCP
4.2. Prevalence of L. monocytogenes in Ready-to-Eat (RTE) Food Products and Listeriosis
4.2.1. Developing Countries
4.2.2. Developed Countries
| Food Samples | Prevalence Rates (%) | Methods Used for the Recovery of L. monocytogenes from Samples | Regions/Countries | References | |
|---|---|---|---|---|---|
| 384 RTE foods consisting of raw and pasteurized milk, cheese, cream cakes, ice cream, minced meat, pizza, and fish | 6.2 | Fraser broth + ferric ammonium citrate as supplement (primary and secondary enrichment with incubation at 30 °C for 24 h and 37 °C for 48 h, respectively). Polymixin acriflavine Lithium Chloride ceftazidime Aesculin (PALCAM) (cultivation and isolation). Confirmation obtained by transferring presumed Listeria sp onto tryptone soya yeast extract agar, incubation at 37 °C for 24 h. Further confirmation by Gram staining, haemolysis, motility, catalase, CAMP test, and sugar fermentation. | Gondar Town (Egypt) | Garedew et al. [87] | |
| 250 chicken carcasses | 9.6 | Primary and secondary enrichment performed in Fraser broth, and both incubations carried out at 30 °C for 24 h. Fraser broth cultures were plated on PALCAM, incubated at 37 °C for 48 h. Confirmation via catalase, oxidase, and sugar fermentation tests plus haemolysis type. Isolates further verified API Listeria test (BioMerieux). | Mansoura city (Egypt) | Zakaira and Sabala [97] | |
| 90 chicken and beef | 45 | Samples homogenised in 0.1% buffered peptone water and serially diluted; 1 mL serial dilution inoculated onto Listeria CHROM agar and incubated at 37 °C for 24 h. Blue colonies with halos were confirmed using Gram staining. Confirmed colonies were transferred into Brain Heart Infusion broth, incubated at 37 °C for 24 h, and further confirmed via polymerase chain reaction. | Zanjan city (Iran) | Farhoumand et al. [98] | |
| 65 raw milk, pasteurized/fresh milk, cheese | 18.46 | Primary and secondary enrichment of samples were performed in Fraser broth based with Frazer selective supplement (SR0166E, Oxoid, UK) of different strengths. Initial incubation at 30 °C for 24 h and secondary at 37 °C for 24–48 h. Broth cultures were plated on chromogenic Listeria agar (ISO) base mixed with OCLA (ISO) differential (SR 0244E) and selective (SR 0226E) supplement in addition to Brilliance Listeria Agar Base, seeded with Brilliance differential (SR0228E) and selective (SR0227E) supplements. All supplements procured from Oxoid, Ltd., UK and incubation carried out at 37 °C for 24–48 h. | Amathole, Chris Hani and Sarah Baartman District Municipalities, Eastern Cape Province (South Africa) | Kayode and Okoh [96] | |
| 100 raw kebab and hamburger (RTE Food) | 50% and 22%, respectively | Samples were enriched in Listeria enrichment broth and incubated at 30 °C for 4 h. Listeria selective enrichment supplement was introduced into the broth and incubated further for 44 h. A loopful of the enrichment broth was plated onto PALCAM Listeria selective agar and incubated for 48 h at 35 °C. Suspected colonies were confirmed via Gram staining, motility, catalase, urea, haemolysis, CAMP, and sugar fermentation tests. | Tabriz city (Iran) | Rajaei et al. [104] | |
| 750 meats (chicken, chevon, pork, and beef), milk, and milk products (curd and paneer) | 6.4 | Fixed quantity of samples mixed with PALCAM broth plus Listeria supplement and incubated for 24 h at 30 °C. Broth culture (loopful) streaked onto PALCAM agar for 24 h at 30 °C. Suspected Listeria colonies were confirmed preliminarily using Gram staining, catalase, oxidase tests. Typical colonies of Listeria were transferred into BHI broth and incubated at 25 °C overnight. Motility and catalase-positive and oxidase-negative isolates are further confirmed, biochemically and by PCR. | Guwahati (India) | Deka et al. [298] | |
| 30,016 RTE foods (meat, fish, culinary, pastry, fruit and vegetable products, gravy sauce, mixed salads, and meals) | 3.6 | Primary and secondary enrichment in both half- and full-strength Fraser broth. Initial incubation for 24 hat 30 °C and 37 °C for 48 h. Both broth cultures were plated on ALOA and PALCAM agar and incubated at 37 °C for 24–48 h. Presumed L. monocytogenes colonies were streaked on tryptone soya yeast extract agar and incubated a 37 °C for 24 h. Confirmation test via catalase test, Gram staining, microscopy, motility, CAMP tests, and sugar fermentation. | Estonia | Koskar et al. [299] | |
| 1000 RTE foods (bacon, chorizo paisa, grilled hamburger meat, mortadella, and salami) | 16.3 | ISO 11290-1. A portion of sample was homogenised in half-strength Fraser broth and incubated at 30 °C for 24 h for primary enrichment. Secondary enrichment was performed in full-strength Fraser broth and incubated for 24 h at 37 °C. L. monocytogenes colonies were isolated and identified on Listeria chromogenic agar. | Quevedo (Ecuador) | Meta-Bone et al. [268] | |
| 132 RTE seafoods (smoked fish, salted fish, dried fish, raw marinated fish, cooked marinated cephalopods, surimi crab sticks) | 6.1 | ISO 11290-1/A1 procedure was employed. A fixed portion of sample was homogenized in half-strength Fraser broth and incubated at 20 °C for 1 h to recover stress organisms. For primary enrichment, the homogenate was supplemented with Fraser half-selective supplements and incubated for 24 h at 30 °C. An aliquot of primary enrichment culture was transferred into Fraser broth and incubated for 48 h at 30 °C for secondary enrichment. Subsequently, both primary and secondary broth cultures (a loopful) were streaked onto ALOA and Oxford Agar and examined for growth after 24 and 48 h at 37 °C. Suspected colonies were purified by growing on tryptone soya yeast extract agar. Pure cultures were subjected to multiplex PCR for bacterial identification. | Thessaloniki (Northern Greece) | Soultos et al. [263] | |
| 6000 poultry, pork, and beef | 2.1 | technique was employed. For primary enrichment, sponges obtained from samples were dipped in half-strength Fraser broth and incubated for 24 h at 30 °C. Sponge was squeezed firmly multiple times into the bag and secondary enrichment performed in full-strength Fraser broth; incubation occurred at 37 °C for 48 h. Reductive inoculation of cultures was carried out on Chromocult Listeria Selective Agar (ALOA) and incubated at 37 °C for 24 h. Suspected colonies were transferred to Columbia agar with 5% sheep blood. Confirmatory tests included haemolysis type and polymerase chain reaction. | Poland | Skowron et al. [289] | |
| 184 meat samples (chicken, quail, duck, turkey, and pork) | 10.32 | Primary enrichment of samples was performed in half-strength Fraser broth and incubated at 30 °C for 24 h. Subsequently, secondary enrichment was completed in Fraser broth and incubated at 37 °C for 24 h. Cultures plated on ALOA and incubated for 24–48 h at 37 °C. Suspected colonies were purified in tryptone soy agar and BHI broth. Purified isolates were identified via matrix-assisted laser desorption/ionization–time of flight Mass spectrometry (MALDI-TOF MS) Biotyper. | La Rioja (Spain) | Martinez-Laorden et al. [300] | |
| 200 minced meat, poultry meat, tilapia fish, and raw milk | 10 | Enrichment was carried out in both half- and full-strength Fraser broth and plated on Oxford and Agar Listeria Ottaviani and Agosti (ALOA) agar. Colonies were purified on tryptone soya yeast extract agar and suspected colonies were confirmed by beta-haemolysis, triple iron sugar, and oxidase tests. Further confirmation by multiplex PCR. | Sharkia Province (Egypt) | EL-Demerdash and Raslan [297] | |
| 1096 (dairy products, bovine meat products, pastry, salads, poultry meat products, chickpeas cooked with eggs, and mayonnaise | 1.5 | Enrichment steps and selective media were employed. Fraser broth was used to grow the bacterium, while plating was carried out on selective media (e.g., ALOA). Isolates were identified biochemically using API-Listeria system and PCR serogrouping. | Tetouan, North-Western (Morocco) | Amajoud et al. [301] | |
| 443 pasteurised milk, raw milk, and yoghurt | 5.6 | Enrichment of samples was performed in Listeria broth-LEB and incubated for 4 h and medium later, seeded with Listeria selective supplements, and further incubated for 48 h at 30 °C. Bacterial growth was streaked on Oxford Agar plates following 24 and 48 h of incubation and maintained at 35 °C. Suspected colonies were transferred into tryptic soy yeast extract agar and incubated at 30 °C for 24 to 48 h. Confirmatory tests included microscopy, catalase, sugar fermentation, and CAMP tests. | Addis Ababa, Ethiopia | Seyoum et al. [302] | |
| 400 raw beef, RTE food products, milled beef, offal, and organs | 8.3 | Meat sample homogenized in ONE Broth-Listeria and incubated at 35 °C for 48 h. Enriched broth samples inoculated onto Chromogenic Brilliance Listeria agar and incubated at 35 °C for 48 h. Suspected colonies identified using phenotypic and molecular methods. | Mpumalanga (South Africa) | Moabelo et al. [303] | |
| 567 retailed raw foods (fishery products, raw/fresh meat, frozen food, edible fungi, and vegetables | 22 | The National Food Safety Standard of China—Food microbiological examination: L. monocytogenes. Samples were homogenized in Listeria enrichment broth I (LBI) and incubated for 24 h at 30 °C. Secondary enrichment was carried out in LB2 and incubated for 24 h at 30 °C. Enrichment broth (LB2) culture (a loopful) was inoculated onto Chromagar Listeria selective agar plates and incubated for 48 h at 37 °C. Suspected colonies were identified via the Microgen ID Listeria identification system. | Guangzhou city (South China) | Chen et al. [304] | |
| 3171 food samples (frozen, deli meat, RTE, and cheese. | 11.2 | Primary and secondary enrichment conducted as above in half and full strength fraser broth. Real-time PCR Dupont Qualicon BAXR system was employed. Positive real-time PCR samples were streaked on Oxford medium base with modified Oxford antimicrobic supplement and on BBLTM CHROM agar TM Listeria (BD) and both incubated at 35–37 °C for 24–48 h. Suspected colonies were identified using API Listeria kit, beta-haemolysis, halo production, catalase reaction, bile esculin, and Christie–Atkins–Munch–Petersen (CAMP) test. | Montevidio city (Uruguay) | Braga et al. [265] | |
| 122 RTE smoked and gravad fish (retailed products) 63 RTE soft and semi-soft cheese and 60 heat-treated meat products | 12.3 0 | A fixed portion of sample was homogenized in half-strength Fraser broth and incubated at 20 °C for 1 h to recover stress organisms. For primary enrichment, the homogenate was supplemented with Fraser half-selective supplements and incubated for 24 h at 30 °C. An aliquot of primary enrichment culture was transferred into Fraser broth and incubated for 48 h at 30 °C for secondary enrichment. Subsequently, both primary and secondary broth cultures (a loopful) were streaked onto ALOA and Oxford Agar and examined for growth after 24 and 48 h at 37 °C. | Bulgaria | Gyurova et al. [288] | |
| 130 dairy milk products (ice cream, butter, and cheese) and meat products (chicken, frankfurter, smoked chicken frankfurter, chicken sandwiches, and chicken lyoner) | 0 | Primary enrichment of 25 g of sample in University of Vermont (UVM) broth (225 mL, i.e., 1:10 dilution) and stomached for close to 2 min. This was incubated at 28–32 °C for 20–26 h. A total of 100 µL of UVM was dispensed into 10 mL of Fraser broth for further incubation performed at 33–37 °C for 18–24 h. Inoculum was plated on chromogenic selective agar, ALOA, and incubated at 33–37 °C for 2 days. ALOA agar plates were assessed for typical L. monocytogenes colonies (blue-green colonies with a halo). | Peninsular Malaysia | Marina et al. [287] | |
| 132 RTE delicatesses (vegetables with mayonnaise sauce, starts without mayonnaise, pasta/rice-based, meat-based and fruit-based courses.) | 17.4 | 25 g of product was diluted with 225 mL of ½ Fraser broth, homogenised for 1 min in a stomacher and incubated at 30 °C for 48 h. Later, 100 µL of the homogenate was plated on Rapid L’monoagar and incubated at 37 °C for 24–48 h according to AFNOR BRD07-0405-09/98 method. In parallel, 1 mL was equally added to 9 mL of whole Fraser broth and incubated at 37 °C for 24 h. Afterwards, 100 µL of the broth was freshly streaked onto Rapid L’monoagar and incubated under the same conditions. One or two presumptive colonies were selected and subsequently identified by Microbact TM Listeria 12L kit. | Northern Italy | Tirloni et al. [292] | |
| 881 RTE meat, fish, and seafood products and RTE milk products | 8.4 | Standard microbiological and enzyme-linked fluorescent immunoassay analysed the presence of L. monocytogenes. A fixed portion (25 g) of test sample was enriched in ½ Fraser broth and incubated for 23 or 26 h. Secondary enrichment was performed in full-strength Fraser broth and incubated at 22 or 26 h. Inoculum of Fraser and ½ Fraser broths were streaked on selective agar and incubated for a maximum of 72 h. Incubated plates were refrigerated for a maximum of 2 days before reading. Confirmation was conducted through immunoassay carried out by automated mini VIDAS system using VIDASR LMX test kit. | Serbia | Branka et al. [305] | |
| 133 retail RTE food, including dairy, meat-based, poultry-based, vegetable-based, fruit-based, and fish-based products. RTE foods were unpackaged while others were packaged and non-frozen. All samples except the vegetable and fruit-based products were heat treated | 12.8 | Two-stage enrichment was carried out for the detection of L. monocytogenes according to the International Organisation for Standardisation standard (ISO) protocol. Each sample (25 g) was added to ½ Fraser broth (225 mL) and homogenised in a stomacher for 3 min. Homogenate was incubated at 29 or 30 °C for 22 or 24 h (primary enrichment). A total of 100 µL of the primary enrichment culture was dispensed into 10 mL of full-strength Fraser broth. The mixture was incubated at 37 °C for 40 or 48 h. A loopful of both the ½ and Fraser broth were streaked on both ALOA and PALCAM agar. Plates were incubated at 37 °C for 24–48 h. Five suspected colonies from both cultured media were picked for identification of the organism via Gram staining, catalase, oxidase, and sugar utilisation tests, CAMP test, motility at 20 °C -25 °C. Confirmation of L. monocytogenes isolates was performed by PCR. | Ankara city (Turkey) | Sentürk et al. [223] | |
| RTE Foods as vehicles | Year of listeriosis outbreak | Human cases involved | Deaths | Countries | References |
| Bologna-style sausage (polony) | 2018 | 937 | 216 | South Africa | Thomas et al. [272]. |
| Turkey meat products | 2012–2016 | 26 | 3 | Czech Republic | Gelbícová et al. [306]. |
| Rillettes (pâte-like meat product) Jellied pork tongue | 1999 2000 | 10 32 | 3 5 | France | De Valk et al. [307]. |
| Cheeses, a sour milk curd termed Quargel | June 2009–Jan. 2010 Dec 2009–Feb. 2010 | 14 20 | 5 3 | Austria, Germany, and Czech Republic | Fretz et al. [308]. |
| Chocolate milk | 1994 | 45 | 0 | USA (Illinois) | Dalton et al. [309]. |
| Butter from pasteurized milk | 1999 | 25 | 6 | Finland | Lyytikäinen et al. [310]. |
| Milkshakes from ice cream | 2015 | 10 | 0 | USA | Pouillot et al. [233]. |
| Unpasteurised milk Pasteurized milk | 2007–2008 | 449 174 | 5 24 | Canada and USA | Sebastianski et al. [311]. |
| Pasteurised milk | 1983 | 49 | 14 | USA (Massachusettes) | Fleming et al. [312]; James et al. [313]. |
| Cheese | 2007 | 5 | 3 | USA (California) | Centre for Disease Control [314]. |
| Unpasteurised chocolate milk | 2014 | 2 | 1 | USA (California and Florida) | Nichols et al. [315]. |
| Cheese from pasteurised milk | 2006–2007 | 189 | 0 | Germany | Koch et al. [316]. |
| Pasteurised ice cream | 2014 | 2 | 1 | USA (Washington) | Rietberg et al. [153]. |
| Cheese dairy | 2018–2020 | 79 | 10 | Switzerland | Nüesch-Inderbinen et al. [317]. |
| Pasteurised chocolate milk | 2015–2016 | 34 | 4 | Canada (Onatrio) | Hanson et al. [318]. |
| Deli meats | 2024 | 61 | 10 | USA (Illinois, Virginia, New York, New Jersey, South Carolina, Florida, New Mexico, and Tennessee | CDC [319]. |
| Fish products (RTE cold-smoked salmon, cream cod, mackerel, herrings, fish-meat product, and fish salad) | 2012–2024 | 73 | 14 | Czehia, Germany, Finland, Italy, the Netherlands, United Kingdom, and Belgium | ECDC, EFSA [320]. |
| Packaged leafy green salads | 2015–2016 | 33 | 5 | USA and Canada | Self [321]. |
4.2.3. Some Specific Food Case Studies Involving L. monocytogenes
- Improvement of the diagnostic methods and increased surveillance in public health.
- The generalisation of food preservation methods, including refrigeration, which permit the growth of L. monocytogenes.
- Industrial development in food generation and the resulting risk associated with large distribution of contaminated food.
- An increase in the population of susceptible individuals, including the elderly and the immunosuppressed.
- The rising consumption level of preservative-free RTE foods.
- The use of antacids and medications that suppress the secretion of gastric acid.
4.3. Transmission of L. monocytogenes (Contamination)
4.3.1. Environmental Sources
4.3.2. Food-Processing Facility/Environment
- The food
- Level of L. monocytogenes contamination in the RTE at retail
- Prevalence of L. monocytogenes in RTE food at retail
- Size of the vulnerable/susceptible population
- Conditions of storage after retail
- Level of consumption
- Virulence of the infecting L. monocytogenes strain
- National surveillance system
4.3.3. Humans
5. Prevention and Control of Listeria Monocytogenes
5.1. Emerging Control Technologies
5.1.1. Phage Therapy
5.1.2. High-Pressure Processing (HPP)
5.1.3. Antimicrobial Active Packaging
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holch, A.; Webb, K.; Lukjancenko, O.; Ussery, D.; Rosenthal, B.M.; Gram, L. Genome Sequencing Identifies Two Nearly Unchanged Strains of Persistent Listeria monocytogenes Isolated at Two Different Fish Processing Plants Sampled 6 Years Apart. Appl. Environ. Microbiol. 2013, 79, 2944–2951. [Google Scholar] [PubMed]
- Castrica, M.; Andoni, E.; Intraina, I.; Curone, G.; Copelotti, E.; Massacci, F.R.; Terio, V.; Colombo, S.; Balzaretti, C.M. Prevalence of Listeria monocytogenes and Salmonella spp. in Different Ready to Eat Foods from Large Retailers and Canteens over a 2-Year Period in Northern Italy. Int. J. Environ. Res. Public Health 2021, 18, 10568. [Google Scholar] [CrossRef]
- Hafner, L.; Pichon, M.; Burucoa, C.; Nusser, S.H.A.; Moura, A.; Garcia-Garcera, M.; Lecuit, M. Listeria monocytogenes faecal carriage is common and depends on the gut microbiota. Nat. Commun. 2021, 12, 6826. [Google Scholar] [CrossRef]
- Manyi-Loh, C.E.; Okoh, A.I.; Lues, R. Occurrence and multidrug resistance in strains of Listeria monocytogenes recovered from the anaerobic Co-digestion sludge contained in a single stage steel biodigester: Implications for antimicrobial stewardship. Microorganisms 2023, 11, 725. [Google Scholar] [CrossRef] [PubMed]
- Linke, K.; Rücker, I.; Brugger, K.; Karpiskova, R.; Walland, J.; Muri-Klinger, S.; Tichy, A.; Wagner, M.; Stessl, B. Reservoirs of Listeria Species in three environmental ecosystems. Appl. Environ. Microbiol. 2014, 80, 5583–5592. [Google Scholar]
- Dufailu, O.A.; Yaqub, M.O.; Owusu-Kwarteng, J.; Addy, F. Prevalence and characteristics of Listeria species from selected African countries. Trop. Dis. Travel. Med. Vaccines 2021, 7, 26. [Google Scholar] [CrossRef]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef] [PubMed]
- Allerberger, F. Listeria: Growth, phenotypic differentiation and molecular microbiology. FEMS Immunol. Med. Microbiol. 2003, 35, 183–189. [Google Scholar]
- Jamali, H.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015, 15, 144. [Google Scholar]
- Project FS101057; Development of An Initial Report—Reducing the Risk of Vulnerable Groups Contracting Listeriosis. Food Standards Agency: London, UK, 2016.
- Luber, P. The codex Alimentarius guidelines on the application of general principles of food hygiene to the control of Listeria monocytogenes in ready-to-eat foods. Food Control 2011, 22, 1482–1483. [Google Scholar]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.R.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319. [Google Scholar] [CrossRef]
- Bierne, H.; Cossart, P. Listeria monocytogenes surface proteins: From genome predictions to function. Microbiol. Mol. Biol. Rev. 2007, 71, 377–397. [Google Scholar] [CrossRef] [PubMed]
- Janez, N.; Skrlj, B.; Sternisa, M.; Klancnik, A.; Sabotic, J. The role of the Listeria monocytogenes surfactome in biofilm formation. Microb. Biotechnol. 2021, 14, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Chang, Y.; Gu, W.; Fischer, N.; McLandsborough, L. Identification of genes involved in Listeria monocytogenes biofilm formation by mariner-based transposon mutagenesis. Appl. Microbiol. Biotechnol. 2012, 93, 2051–2062. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670. [Google Scholar] [CrossRef]
- Soumya, S.; Sudip, K.S.; Smaranika, P.; Sangeeta, R. Review on bacterial biofilm: An universal cause of contamination. Biocatal. Agric. Biotechnol. 2016, 7, 56–66. [Google Scholar]
- Cho, J.-A.; Roh, Y.J.; Son, H.R.; Choi, H.; Lee, J.-W.; Kim, S.J.; Lee, C.-H. Assessment of the biofilm-forming ability on solid surfaces of periprosthetic infection-associated pathogens. Sci. Rep. 2022, 12, 18669. [Google Scholar] [CrossRef]
- Zetzmann, M.; Bucur, F.I.; Crauwels, P.; Borda, D.; Nicolau, A.I.; Grigore-Gurgu, L.; Seibold, G.M.; Riedel, C.U. Characterization of the biofilm phenotype of a Listeria monocytogenes mutant deficient in agr peptide sensing. Microbiologyopen 2019, 8, e00826. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.M.; Martinez-Garcia, E.; Xavier, J.; Durham, V.M.; Kolter, R.; Kim, W.; Foster, K.R. Correction: Biofilm as a response to ecological competition. PLOS Biol. 2015, 13, e1002232. [Google Scholar] [CrossRef] [PubMed]
- Banda, R.; Nduko, J.; Matofari, J. Bacterial Biofilm Formation in Milking Equipments in Lilongwe, Malawi. J. Food Qual. Hazards Control 2020, 7, 142–148. [Google Scholar] [CrossRef]
- Fan, Y.; Qiao, J.; Lu, Z.; Fen, Z.; Tao, Y.; Lv, F.; Zhao, H.; Zhang, C.; Bie, X. Influence of different factors on biofilm formation of Listeria monocytogenes and the regulation of cheY gene. Food Res. Int. 2020, 137, 109405. [Google Scholar] [CrossRef]
- Van Houdt, R.; Michiels, C.W. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef]
- Bhagwat, G.; O’Connor, W.; Grainge, I.; Palanisami, T. Understanding the fundamental basis for biofilm formation on plastic surfaces: Role of conditioning films. Front. Microbiol. 2021, 12, 687118. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, X.; Niu, B.; Yang, J.; Chen, Q. Differences in biofilm formation of Listeria monocytogenes and their effects on virulence and drug resistance of different strains. Foods 2024, 13, 1076. [Google Scholar] [CrossRef]
- Galiè, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, X.; Wang, Q.; Yang, J.; Zhong, Q. The mixed biofilm formed by Listeria monocytogenes and other bacteria: Formation, interaction and control strategies. Crit. Rev. Food Sci. Nutr. 2024, 64, 8570–8586. [Google Scholar] [PubMed]
- Haddad, S.; Elliot, M.; Savard, T.; Deschênes, L.; Smith, T.; Ells, T. Variations in biofilms harbouring Listeria monocytogenes in dual and triplex cultures with pseudomonas fluorescens and lactobacillus plantarum produced under a model system of simulated meat processing conditions, and their resistance to benzalkonium chloride. Food Control 2021, 123, 107720. [Google Scholar] [CrossRef]
- Da Silva Fernandes, M.; Kabuki, D.Y.; Kuaye, A.Y. Behavior of Listeria monocytogenes in a multi-species biofilm with Enterococcus faecalis and Enterococcus faecium and control through sanitation procedures. Int. J. Food Microbiol. 2015, 200, 5–12. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Burmolle, M.; Heyndrickx, M.; Flint, S.; Lu, W.; Chen, W.; Zhao, J.; Zhang, H. Community-wide changes reflecting bacterial interspecific interactions in multispecies biofilms. Crit. Rev. Microbiol. 2021, 47, 338–358. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Fang, K.; Medina, D.; Wan, J.; Lee, J.; Hong, S.H. The probiotic, leuconostoc mesenteroides, inhibits Listeria monocytogenes biofilm formation. J. Food Saf. 2020, 40, e12750. [Google Scholar] [CrossRef]
- de Grandi, A.Z.; Pinto, U.M.; Destro, M.T. Dual-species biofilm of Listeria monocytogenes and Escherichia coli on stainless steel surface. World J. Microbiol. Biotechnol. 2018, 34, 61. [Google Scholar] [CrossRef]
- Alonso, V.P.P.; Kabuki, D.Y. Formation and dispersal of biofilms in dairy substrates. Int. J. Dairy Technol. 2019, 72, 472–478. [Google Scholar] [CrossRef]
- Lee, B.-H.; Cole, S.; Badel-Berchoux, S.; Guillier, L.; Felix, B.; Krezdorn, N.; Hébraud, M.; Bernardi, T.; Sultan, I.; Piveteau, P. Biofilm Formation of Listeria monocytogenes strains under food processing environments and pan-genome-wide association Study. Front. Microbiol. 2019, 10, 2698. [Google Scholar] [CrossRef]
- Blackman, L.D.; Qu, Y.; Cass, P.; Locock, K.E.S. Approaches for the inhibition and elimination of microbial biofilms using macromolecular agents. Chem. Soc. Rev. 2021, 50, 1587–1616. [Google Scholar]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm Formation Ability of Listeria monocytogenes Isolates from Raw Ready-to-Eat Seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef]
- Borucki, M.K.; Gay, C.C.; Reynolds, J.; McElwain, K.L.; Kim, S.H.; Call, D.R.; Knowles, D.P. Genetic diversity of Listeria monocytogenes strains from a high-prevalence dairy farm. Appl. Environ. Microbiol. 2005, 71, 5893–5899. [Google Scholar]
- Carpentier, B.; Chassaing, D. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 2004, 97, 111–122. [Google Scholar]
- Doijad, S.P.; Barbuddhe, S.; Garg, S.; Poharkar, K.V.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Chakraborty, T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE 2015, 10, e0137046. [Google Scholar]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria monocytogenes—How This Pathogen Survives in Food-Production Environments? Front. Microbiol. 2022, 13, 866462. [Google Scholar] [CrossRef]
- Keeney, K.; Trmcic, A.; Zhu, Z.; Delaquis, P.; Wang, S. Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 2018, 67, 530–536. [Google Scholar] [PubMed]
- Ripolles-Avila, C.; Hascoët, A.S.; Guerrero-Navarro, A.E.; Rodríguez-Jerez, J.J. Establishment of incubation conditions to optimize the in vitro formation of mature Listeria monocytogenes biofilms on food-contact surfaces. Food Control 2018, 92, 240–248. [Google Scholar]
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes biofilms in the food industry: Is the current hygiene program sufficient to combat the persistence of the pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef]
- Moretro, T.; Langsrud, S. Listeria monocytogenes: Biofilm formation and persistence in food-processing environments. Biofilms 2004, 1, 107–121. [Google Scholar]
- Chmielewski, R.A.N.; Frank, J.F. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar]
- Mustapha, A.; Liewen, M.B. Destruction of Listeria monocytogenes by sodium hypochlorite and quaternary ammonium sanitizers. J. Food Prot. 1989, 52, 306–311. [Google Scholar]
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordóñez, A. New weapons to fight old enemies: Novel strategies for the (bio)control of bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 1641. [Google Scholar]
- Soares, S.J.A.; Guimarães, F.d.F.; Rossi, G.A.M.; Guerra, S.T.; Dalanezi, F.M.; Lopes, B.C.; Ribeiro Mioni, M.d.S.; Yamakawa, A.C.; da Silva, E.C.; de Moraes, G.N.; et al. Virulence potential, biofilm formation, and disinfectants control of Escherichia coli from Raw Milk Bulk Tanks in the Southeast of Brazil. Dairy 2023, 4, 541–553. [Google Scholar] [CrossRef]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: Relationship among Candida spp. Front. Microbiol. 2015, 6, 205. [Google Scholar] [CrossRef]
- Nalbone, L.; Sorrentino, G.; Giarratana, F.; Schiopu-Mariean, A.; Ziino, G.; Giuffrida, A. Effects of osmotic stress on Listeria monocytogenes ATCC 7644: Persistent cells and heat resistance. Italian J. Food Saf. 2023, 12, 10880. [Google Scholar]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar]
- Saklani-Jusforgues, H.; Fontan, E.; Goossens, P.L. Effect of acid-adaptation in Listeria monocytogenes survival and translocation in a murine intragastric infection model. FEMS Microbiol. Lett. 2000, 193, 155–159. [Google Scholar]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar]
- Lakicevic, B.Z.; Den Besten, H.M.W.; De Biase, D. Landscape of stress response and virulence genes among Listeria monocytogenes strains. Front. Microbiol. 2022, 12, 738470. [Google Scholar] [CrossRef]
- Al-Nabulsi, A.A.; Osaili, T.M.; Awad, A.A.; Olaimat, A.N.; Shaker, R.R.; Holley, R.A. Occurrence and antibiotic susceptibility of Listeria monocytogenes isolated from raw and processed meat products in Amman, Jordan. CyTA-J. Food 2015, 13, 346–352. [Google Scholar] [CrossRef]
- Wiktorczyk-Kapischke, N.; Skowron, K.; Wałecka-Zacharska, E.; Grudlewska-Buda, K.; Wnuk, K.; Buszko, K.; Gospodarek-Komkowska, E. Assessment of the influence of selected stress factors on the growth and survival of Listeria monocytogenes. BMC Microbiol. 2023, 23, 27. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef]
- Rolon, M.L.; Voloshchuk, O.; Bartlett, K.V.; LaBorde, L.F.; Kovac, J. Multi-species biofilms of environmental microbiota isolated from fruit packing facilities promoted tolerance of Listeria monocytogenes to benzalkonium chloride. Biofilm 2024, 7, 100177. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, R.; Saba, T.; Khaliq, H.; Mitchell, J.; Do, L.; Chambi, L.; Diaz, K.; Kennedy, T.; Alkassab, K.; Huynh, T.; et al. Biofilms: Formation, drug resistance and alternatives to conventional approaches. AIMS Microbiol. 2022, 8, 239–277. [Google Scholar] [CrossRef] [PubMed]
- Mpondo, L.; Ebomah, K.E.; Okoh, A.I. Multidrug-resistant listeria Species Shows Abundance in Environmental Waters of a Key District Municipality in South Africa. Ijerph 2021, 18, 481. [Google Scholar] [CrossRef]
- Morvan, A.; Moubareck, C.; Leclercq, A.; Herv_e-Bazin, M.; Bremont, S.; Lecuit, M.; Courvalin, P.; Le Monnier, A. Antimicrobialresistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 2010, 54, 2728–2731. [Google Scholar] [CrossRef]
- Pagliano, P.; Arslan, F.; Ascione, T. Epidemiology and treatment of the commonest form of listeriosis: Meningitis and bacteraemia. Infez. Med. 2017, 25, 2010–2216. [Google Scholar]
- Shamloo, E.; Abdimoghadam, Z.; Nazari, K.; Hosseini, S.M.; Hosseini, H.; Alebouyeh, M. Long term survival of Listeria monocytogenes in stress conditions: High pH and salt concentrations. J. Res. Med. Dental Sci. 2018, 6, 96–100. [Google Scholar]
- Andriyanov, P.A.; Zhurilov, P.A.; Liskova, E.A.; Karpova, T.I.; Sokolova, E.V.; Yushina, Y.K.; Zaiko, E.V.; Bataeva, D.S.; Voronina, O.L.; Psareva, E.K.; et al. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from Humans, Animals, and Food Products in Russia in 1950–1980, 2000–2005, and 2018–2021. Antibiotics 2021, 10, 1206. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Cortés-Cortés, G.; Arenas-Hernández, M.M.P.; Ballesteros-Monrreal, M.G.; Rocha-Gracia, R.C.; Barrios-Villa, E. Editorial: Epidemiology of antimicrobial resistance and virulence factors of emerging and re-emerging bacteria. Front. Cell Infect. Microbiol. 2024, 14, 1387087. [Google Scholar] [CrossRef]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The complex relationship between virulence and antibiotic resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef]
- Ruiz-Bolivar, Z.; Neuque-Rico, M.C.; Poutou-Pinales, R.A.; Carrascal-Camacho, A.K.; Mattar, S. Antimicrobial susceptibility of Listeria monocytogenes food isolates from different cities in Colombia. Foodborne Pathog. Dis. 2011, 8, 913–919. [Google Scholar] [PubMed]
- Moura, A.; Leclercq, A.; Vales, G.; Tessaud-Rita, N.; Bracq-Dieye, H.; Thouvenot, P.; Madec, Y.; Charlier, C.; Lecuit, M. Phenotypic and genotypic antimicrobial resistance of Listeria monocytogenes: An observational study in France. Lancet 2024, 37, 100800. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.F.; Lanza, V.; Duval, M.; Coque, T.M. Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef]
- Yazdanpour, Z.; Tadjrobehkar, O.; Shahkhah, M. Significant association between gene encoding virulence factors with antibiotic resistance and phylogenetic groups in community acquired uropathogenic Escherichia coli isolates. BMC Microbiol. 2020, 20, 241. [Google Scholar] [CrossRef]
- Hanes, R.M.; Huang, Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Health 2022, 19, 5506. [Google Scholar] [CrossRef]
- Borena, B.M.; Dilgasa, L.; Gebremedhin, E.Z.; Sarba, E.Z.; Marami, L.M.; Kelbesa, K.A.; Tadese, N.D. Listeria Species occurrence and associated risk factors and antibiogram of Listeria monocytogenes in milk and milk products in Ambo, Holeta, and Bako Towns, Oromia Regional State, Ethiopia. Hindawi Vet. Med. Int. 2022, 2022, 5643478. [Google Scholar] [CrossRef]
- Moreno, L.Z.; Paixao, R.; Gobbi, D.S.; Raimundo, D.C.; Ferreira, T.P.; Moreno, A.M.; Hofer, E.; Reis, C.M.F.; Matte’, G.R.; Matte, M.H. Characterization of antibiotic resistance in Listeria spp. isolated from slaughterhouse environments, pork and human infections. J. Infect. Dev. Ctries. 2014, 8, 416–423. [Google Scholar] [CrossRef]
- Axelsson, C.; Nilson, B.; Rehnstam-Holm, A.-S. Efficient absorbance-based assay for rapid antibiotic susceptibility testing of Enterobacteriaceae. Antibiotics 2024, 13, 852. [Google Scholar]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Ghimpeteanu, O.M.; Pogurschi, E.N.; Popa, D.C.; Dragomir, N.; Dragotoiu, T.; Mihai, O.D.; Petcu, C.D. Antibiotic use in livestock and residues in food—A public health threat: A review. Foods 2022, 11, 1430. [Google Scholar] [CrossRef]
- Manyi-Loh, C.E.; Lues, R. A South African Perspective on the Microbiological and Chemical Quality of Meat: Plausible Public Health Implications. Microorganisms 2023, 11, 2484. [Google Scholar] [CrossRef]
- Zurawik, A.; Kasperski, T.; Olechowska-Jarzab, A.; Szczesiul-Paszkiewicz, P.; Zak, I.; Wójcicki, M.; Mackiw, E.; Chmielarczyk, A. Genetic Diversity, Virulence Factors and Antibiotic Resistance of Listeria monocytogenes from Food and Clinical Samples in Southern Poland. Pathogens 2024, 13, 725. [Google Scholar] [CrossRef] [PubMed]
- Panera-Martínez, S.; Rodríguez-Melcon, C.; Serrano-Galan, V.; Alonso-Calleja, C.; Rosa Capita, R. Prevalence, quantification and antibiotic resistance of Listeria monocytogenes in poultry preparations. Food Control 2022, 135, 108608. [Google Scholar] [CrossRef]
- Garedew, L.; Taddese, A.; Biru, T.; Nigatu, S.; Kebede, E.; Ejo, M.; Fikru, A.; Birhanu, T. Prevalence and antimicrobial susceptibility profile of Listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 2015, 15, 100. [Google Scholar]
- Centorame, P.; D’Angelo, A.R.; Di Simone, F.; Salini, R.; Cornacchia, A.; Marrone, R.; Anastasio, A.; Pomilio, F. Listeria monocytogenes biofilm production on food packaging materials submitted to physical treatment. Ital. Food Saf. 2017, 6, 6654. [Google Scholar] [CrossRef][Green Version]
- Cepas, V.; L’opez, Y.; Muñoz, E.E.; Rolo, D.; Ardanuy, C.; Marti, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in gram negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef]
- Şanlıbaba, P.; Tezel, B.U.; Çakmak, G.A. Prevalence and Antibiotic Resistance of Listeria monocytogenes isolated from ready-to-eat foods in Turkey. J. Food Qual. 2018, 2018, 7693782. [Google Scholar] [CrossRef]
- Bustamante, F.; Maury-Sintjago, E.; Leal, F.C.; Acuña, S.; Aguirre, J.; Troncoso, M.; Figueroa, G.; Parra-Flores, J. Presence of Listeria monocytogenes in ready-to-eat artisanal chilean foods. Microorganisms 2020, 8, 1669. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Chen, H.; Chen, J.; Zhang, J.; Zhang, Z.; Yang, Y.; Xu, Z.; Zhan, L.; Mei, L. Prevalence, Genotypic Characteristics and Antibiotic Resistance of Listeria monocytogenes From Retail Foods in Bulk in Zhejiang Province, China. Front. Microbiol. 2019, 10, 1710. [Google Scholar] [CrossRef]
- Su, R.; Wen, Y.; Prabakusuma, S.S.; Tang, X.; Huang, A.; Li, L. Prevalence, antibiotic resistance and virulence feature of Listeria monocytogenes isolated from bovine milk in Yunnan, Southwest China. Int. Dairy J. 2023, 144, 105703. [Google Scholar]
- Ebakota, D.O.; Abiodun, O.A.; Nosa, O.O. Prevalence of antibiotics resistant Listeria monocytogenes strains in Nigerian ready-to-eat foods. Food Saf. 2018, 6, 118–125. [Google Scholar] [CrossRef]
- Mirzaei, R.; Golestan, L.; Zahra, F. Listeria monocytogenes isolated from ready-to-eat food products in Tehran: Prevalence and antimicrobial resistance. Arch. Razi Inst. 2024, 79, 1337–1344. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of multidrug-resistant Listeria monocytogenes in milk and milk product and one Health Perspective. PLoS ONE 2022, 17, e0270993. [Google Scholar] [CrossRef]
- Zakaria, A.I.; Sabala, R.F. Potential public health hazards related to consumption of poultry contaminated with antibiotic resistant Listeria monocytogenes in Egypt. BMC Microbiol. 2024, 24, 41. [Google Scholar] [CrossRef]
- Farhoumand, P.; Hassanzadazar, H.; Soltanpour, M.S.; Aminzare, M.; Abbasi, Z. Prevalence, genotyping and antibiotic resistance of Listeria monocytogenes and Escherichia coli in fresh beef and chicken meats marketed in Zanjan. Iran J. Microbiol. 2020, 12, 537–546. [Google Scholar]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes strains isolated from ready-to-eat foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef]
- Duma, M.N.; Ciupescu, L.M.; Dan, S.D.; Crisan-Reget, O.L.; Tabaran, A. Virulence and antimicrobial resistance of Listeria monocytogenes isolated from ready-to-eat food products in Romania. Microorganisms 2024, 12, 954. [Google Scholar] [CrossRef]
- Rippa, A.; Bilei, S.; Peruzy, M.F.; Marrocco, M.G.; Leggeri, P.; Bossù, T.; Murru, N. Antimicrobial resistance of Listeria monocytogenes strains isolated in food and food-processing environments in Italy. Antibiotics 2024, 13, 525. [Google Scholar] [CrossRef]
- Beshiru, A.; Uwhuba, K.E. Detection and characterization of Listeria monocytogenes from locally processed fermented foods in Ethiope West, Delta State, Nigeria. Ife J. Sci. 2023, 25, 483–493. [Google Scholar]
- Elavarasi, S.; Ramesh, B.; Sathiyamurthy, K. Prevalence and Antimicrobial Resistance Pattern of Listeria Monocytogenes in Ready to Eat Foods in Tamil Nadu, India. Indian J. Sci. Technol. 2023, 16, 501–508. [Google Scholar] [CrossRef]
- Rajaei, M.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A. Antibiotic resistance in the pathogenic foodborne bacteria isolated from raw kebab and hamburger: Phenotypic and genotypic study. BMC Microbiol. 2021, 21, 272. [Google Scholar] [CrossRef] [PubMed]
- Şanlibaba, P.; Tezel, U.; Cakmak, G.A.; Keskin, R.; Akçelik, M. occurrence of Listeria spp. and antibiotic resistance profiles of Listeria monocytogenes from raw meat at retail in turkey. Ital. J. Food Sci. 2020, 32, 234–250. [Google Scholar]
- Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt. Foods 2021, 10, 1381. [Google Scholar] [CrossRef]
- Arslan, S.; Özdemir, F. Prevalence and antimicrobial resistance of Listeria species and molecular characterization of Listeria monocytogenes isolated from retail ready-to-eat foods. FEMS Microbiol. Lett. 2020, 367, fnaa006. [Google Scholar] [CrossRef]
- Gana, J.; Gcebe, N.; Moerane, R.; Ngoshe, Y.; Tshuma, T.; Moabelo, K.; Adesiyun, A. Antimicrobial resistance profiles of Listeria species recovered from retail outlets in Gauteng Province, South Africa. J. Food Prot. 2024, 87, 100322. [Google Scholar] [CrossRef]
- Leong, D.; Alvarez-Ordóñez, A.; Jooste, P.; Jordan, K. Listeria monocytogenes in food: Control by monitoring the food processing environment. Afr. J. Microbiol. Res. 2016, 10, 1–14. [Google Scholar]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Ahmad, S.I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- Singh, A.K.; Kaur, R.; Verma, S.; Singh, S. Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, risk, and control. Front. Environ. Sci. 2022, 10, 830861. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar]
- Serwecińska, L. Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and to public health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; González, I.J.; Dittrich, S. Antimicrobial resistance in Africa: A systematic review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef]
- Alkofide, A.; Alhammad, A.M.; Alruwaili, A.; Aldemerdash, A.; Almangour, T.A.; Alsuwayegh, A.; Almoqbel, D.; Albati, A.; Alsaud, A.; Enani, M. Multidrug-resistant and extensively drug- resistant Enterobacteriaceae: Prevalence, Treatments, and Outcomes—A retrospective cohort study. Infect. Drug Resist. 2020, 13, 4653–4662. [Google Scholar] [CrossRef]
- Wartu, J.R.; Butt, A.Q.; Suleiman, U.; Adeke, M.; Tayaza, F.B.; Musa, B.J.; Baba, J. Multidrug resistance by microorganisms: A review. Sci. World J. 2019, 14, 49–56. [Google Scholar]
- Huttner, A.; Harbarth, S.; Carlet, J.; Cosgrove, S.; Goossens, H.; Holmes, A.; Jarlier, V.; Voss, A.; Pittet, D. Antimicrobial resistance: A global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob. Resist. Infect. Control. 2013, 2, 31. [Google Scholar]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- Mpundu, P.; Mbewe, A.R.; Muma, J.B.; Mwasinga, W.; Mukumbuta, N.; Munyeme, M. A global perspective of antibiotic-resistant Listeria monocytogenes prevalence in assorted ready to eat foods: A systematic review. Vet. World 2021, 14, 2219–2229. [Google Scholar] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitraa, S.; Emranc, T.B.; Dhamad, K.; Ripone, M.K.H.; Gajdácsf, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Basak, S.; Singh, P.; Rajurkar, M. Multidrug resistant and extensively drug-resistant bacteria: A Study. J. Pathog. 2016, 2016, 4065603. [Google Scholar] [CrossRef] [PubMed]
- Ammar, A.M.; Abd El-Hamid, M.I.; El-Malt, R.M.S.; Azab, D.S.; Albogami, S.; Al-Sanea, M.M.; Soliman, W.E.; Ghoneim, M.M.; Bendary, M.M. Molecular Detection of Fluoroquinolone Resistance among Multidrug-, Extensively Drug-, and Pan-Drug-Resistant Campylobacter Species in Egypt. Antibiotics 2021, 10, 1342. [Google Scholar] [CrossRef]
- Bendary, M.M.; Abd El-Hamid, M.I.; Alhomrani, M.; Alamri, A.S.; Elshimy, R.; Mosbah, R.A.; Bahnass, M.M.; Omar, N.N.; Al-Sanea, M.M.; Elmanakhly, A.R. M.; Elmanakhly, A.R. What is behind the correlation analysis of diarrheagenic, E. coli pathotypes? Biology 2022, 11, 1004. [Google Scholar]
- Brink, A. Antibiotic resistance and virulence. Int. J. Infect. Dis. 2014, 21, 64. [Google Scholar] [CrossRef][Green Version]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Sant’Ana, A.; Igarashi, M.C.; Langraf, M.; Destro, M.T.; Franco, B.D.G. Prevalence, populations and pheno- and genotypic characteristics of Listeria monocytogenes isolated from ready-to-eat vegetables marketed in São Paulo, Brazil. Int. J. Food Microbiol. 2012, 155, 1–9. [Google Scholar]
- Pan, Y.; Zeng, J.; Li, L.; Yang, J.; Tang, Z.; Xiong, W.; Li, Y.; Chen, S.; Zeng, Z. Coexistence of antibiotic resistance genes and virulence factors deciphered by large-scale complete genome analysis. mSystems 2020, 5, e00821-19. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole genome-based characterization of Listeria monocytogenes isolates recovered from the Food Chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.; Bernier, S.P.; Kuchma, S.L.; Hammond, J.H.; Hasan, F.; O’Toole, G.A. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 2010, 13, 207–212. [Google Scholar]
- Billings, N.; Birjinuik, A.; Samad, T.S.; Doyle, P.S.; Ribbeck, K. Material properties of biofilm-a review of methods for understanding permeability mechanics. Rep. Prog. Phys. 2015, 78, 036601. [Google Scholar]
- Capita, R.; Castaño-Arriba, A.; Rodríguez-Melcón, C.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C. Diversity, antibiotic resistance, and biofilm-forming ability of enterobacteria isolated from red meat and poultry preparations. Microorganisms 2020, 8, 1226. [Google Scholar] [CrossRef]
- Lambrechts, K.; Diane Rip, D. Listeria monocytogenes in the seafood industry: Exploring contamination sources, outbreaks, antibiotic susceptibility and genetic diversity. Microbiol. Open 2024, 13, e003. [Google Scholar] [CrossRef]
- Takeuchi-Storm, N.; Hansen, L.T.; Nielsen, N.L.; Andersen, J.K. Presence and persistence of Listeria monocytogenes in the Danish ready-to -eat food production environment. Hygiene 2023, 3, 18–32. [Google Scholar] [CrossRef]
- Finn, L.; Onyeaka, H.; O’Neill, S. Listeria monocytogenes biofilms in food-associated environments: A persistent enigma. Foods 2023, 12, 3339. [Google Scholar] [CrossRef] [PubMed]
- Unrath, N.; McCabe, E.; Macori, G.; Fanning, S. Application of whole genome sequencing to aid in deciphering the persistence Potential of Listeria monocytogenes in food production environments. Microorganisms 2021, 9, 1856. [Google Scholar] [CrossRef]
- Nogueira, R.; Cabo, M.L.; García-Sanmartín, L.; Sánchez-Ruiloba, L.; Rodríguez-Herrera, J.J. Risk factor-based clustering of Listeria monocytogenes in food processing environments using principal component analysis. Food Res. Int. 2023, 170, 112989. [Google Scholar] [CrossRef]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Charlotte Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Chang, Z.; Liu, X.; Chen, W.; Yu, Y.; Wang, X.; Dong, Q.; Ye, Y.; Zhang, X. Listeria monocytogenes contamination characteristics in Two ready-to-eat meat plants from 2019 to 2020 in Shanghai. Front. Microbiol. 2021, 12, 729114. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar]
- Meesilp, N.; Mesil, N. Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci. Biotechnol. 2019, 28, 289–296. [Google Scholar] [PubMed]
- Rodríguez-Campos, D.; Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Persistent Listeria monocytogenes isolates from a poultry-processing facility form more biofilm but do not have a greater resistance to disinfectants than sporadic strains. Pathogens 2019, 8, 250. [Google Scholar] [CrossRef] [PubMed]
- Palaiodimou, L.; Fanning, S.; Fox, E.M. Genomic insights into persistence of Listeria species in the food processing environment. J. Appl. Microbiol. 2021, 131, 2082–2094. [Google Scholar]
- Roilides, E.; Simitsopoulou, M.; Katragkou, A.; Walsh, T.J. How biofilms evade host defenses. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Pang, X.; Wong, C.; Chung, H.-J.; Yuk, H.-G. Biofilm formation of Listeria monocytogenes and its resistance to quaternary ammonium compounds in a simulated salmon processing environment. Food Control 2019, 98, 200–208. [Google Scholar]
- Donelli, G.; Vuotto, C. Biofilm-based infections in long-term care facilities. Future Microbiol. 2014, 9, 175–188. [Google Scholar] [CrossRef]
- Trang, P.N.; Ngoc, T.T.A.; Masuda, Y.; Hohjoh, K.; Miyamoto, T. Biofilm Formation From Listeria monocytogenes Isolated From Pangasius Fish-processing Plants. J. Food Protect. 2023, 86, 100044. [Google Scholar]
- Rietberg, K.; Lloyd, U.; Melius, B.; Wyan, P.; Treadwell, R.; Plson, G.; Kang, M.-G.; Duchin, D. Outbreak of Listeria monocytogenes infection linked to a pasteurized ice cream product served to hospitalized patients. Epidemiol. Infect. 2016, 144, 2728–2731. [Google Scholar]
- Holah, J.T.; Bri, C. UK Cleaning and Disinfection Practices in Food Processing; Woodhead Publishing Limited: Cambridge, UK, 2014; Volume 259, ISBN 9780857094292. [Google Scholar]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 2019, 13, e331. [Google Scholar]
- Silvetti, T.; Morandi, S.; Hintersteiner, M.; Brasca, M. Chapter 22—Use of Hen Egg White Lysozyme in the Food Industry; Academic Press: San Diego, CA, USA, 2017; pp. 233–242. [Google Scholar]
- Nguyen, U.T.; Burrows, L.L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 2014, 187, 26–32. [Google Scholar] [PubMed]
- Shen, C.; Islam, M.T.; Masuda, Y.; Ichi Honjoh, K.; Miyamoto, T. Transcriptional changes involved in inhibition of biofilm formation by ε-polylysine in Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 2020, 104, 5427–5436. [Google Scholar]
- Qi, J.; Yang, H.; Wang, X.; Zhu, H.; Wang, Z.; Zhao, C.; Li, B.; Liu, Z. State-of-the-art on animal manure pollution control and resource utilization. J. Environ. Chem. Eng. 2023, 11, 110462. [Google Scholar]
- Gray, J.A.; Chandry, P.S.; Kaur, M.; Kocharunchitt, C.; Bowman, J.P.; Fox, E.M. Novel biocontrol methods for Listeria monocytogenes biofilms in food production facilities. Front. Microbiol. 2018, 9, 605. [Google Scholar] [CrossRef]
- Webb, L.; Ma, L.; Lu, X. Impact of lactic acid bacteria on the control of Listeria monocytogenes in ready-to-eat foods. Food Qual. Saf. 2022, 6, fyac045. [Google Scholar] [CrossRef]
- Hill, D.; Sugrue, I.; Arendt, E.; Hill, C.; Stanton, C. Recent advances in microbial fermentation for dairy and health [version 1; referees:3 approved]. F1000Research 2017, 6, 751. [Google Scholar] [CrossRef]
- Branson, S.R.; Broadbent, J.R.; Carpenter, C.E. Internal pH and acid anion accumulation in Listeria monocytogenes and Escherichia coli exposed to lactic or acetic acids at mildly Acidic pH. Front. Microbiol. 2022, 12, 803271. [Google Scholar] [CrossRef]
- Mani-López, E.; Hugo, G.; Aurelio, L.-M. Organic acids as antimicrobial to control Salmonella in meat and poultry products. Food Res. Int. 2012, 45, 713–721. [Google Scholar] [CrossRef]
- Arcari, T.; Feger, M.-L.; Guerreiro, D.N.; Wu, J.; O’Byrne, C.P. Comparative Review of the Responses of Listeria monocytogenes and Escherichia coli to Low pH Stress. Genes 2020, 11, 1330. [Google Scholar] [CrossRef]
- Wu, J.; Wang, C.; O’Byrne, C. Metabolic reprogramming in the food-borne pathogen Listeria monocytogenes as a critical defence against acid stress. FEMS Microbiol. Lett. 2024, 371, fnae060. [Google Scholar] [CrossRef]
- Cotter, P.D.; Gahan, C.G.M.; Hill, C. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 2000, 60, 137–146. [Google Scholar] [CrossRef]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: The role of stress responses. Food Res. Int. 2015, 67, 75–90. [Google Scholar] [CrossRef]
- Smith, J.L.; Liu, Y.; Paoli, G.C. How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 2013, 59, 141–152. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Vázquez-Sánchez, D.; López Cabo, M. Current knowledge on Listeria monocytogenes biofilms in food-related environments: Incidence, resistance to biocides, ecology and biocontrol. Foods 2018, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Yang, Y.; Dong, Z.; Wang, X.; Fang, C.; Yang, M.; Sun, J.; Xiao, L.; Fang, W.; Song, H. Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughputmicroplate-based fluorometry. Front. Microbiol. 2015, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.P.; Kendall, P.A.; Sofos, J.N. Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7514–7516. [Google Scholar] [CrossRef]
- Karatzas, K.-A.G.; Suur, L.; O’Byrne, C.P. Characterization of the intracellular glutamate decarboxylase system: Analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2012, 78, 3571–3579. [Google Scholar] [CrossRef]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: A review. Pathogens 2000, 9, 528. [Google Scholar] [CrossRef]
- Giotis, E.S.; McDowell, D.A.; Blair, I.S.; Wilkinson, B.J. Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 2007, 73, 997–1001. [Google Scholar]
- Chiancone, E.; Ceci, P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta (BBA) Gen. Subj. 2010, 1800, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Milecka, D.; Samluk, A.; Wasiak, K.; Krawczyk-Balska, A. An essential role of a ferritin-like protein in acid stress tolerance of Listeria monocytogenes. Arch. Microbiol. 2015, 197, 347–351. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Lues, R. Reduction of Bacterial Pathogens in a Single-Stage Steel Biodigester Co-Digesting Saw Dust and Pig Manure at Psychrophilic Temperature. Appl. Sci. 2022, 12, 10071. [Google Scholar] [CrossRef]
- Roberts, B.N.; Chakravarty, D.; Gardner, J.C.; Ricke, S.C.; Donaldson, J.R. Listeria monocytogenes response to anaerobic environments. Pathogens 2020, 9, 210. [Google Scholar] [CrossRef]
- Yu, M.; Jiang, C.; Meng, Y.; Wang, F.; Qian, J.; Fei, F.; Yin, Z.; Zhao, W.; Zhao, Y.; Liu, H. Effect of low temperature on the resistance of Listeria monocytogenes and Escherichia coli O157:H7 to acid electrolyzed water. Food Res. Int. 2023, 168, 112776. [Google Scholar]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Cordero, N.; Maza, F.; Navea-Perez, H.; Aravena, A.; Marquez-Fontt, B.; Navarrete, P.; Figueroa, G.; González, M.; Latorre, M.; Reyes-Jara, A. Different transcriptional responses from slow and fast growth rate strains of Listeria monocytogenes adapted to low temperature. Front. Microbiol. 2016, 7, 229. [Google Scholar] [CrossRef]
- Touche, C.; Hamchaoui, S.; Quilleré, A.; Darsonval, M.; F Dubois-Brissonnet, F. Growth of Listeria monocytogenes is promoted at low temperature when exogenous unsaturated fatty acids are incorporated in its membrane. Food Microbiol. 2023, 110, 104170. [Google Scholar] [CrossRef]
- NicAogáin, K.; O’Byrne, C.P. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the Food Chain. Front. Microbiol. 2016, 7, 1865. [Google Scholar]
- Angelidis, A.S.; Smith, G.M. Three transporters mediate uptake of glycine betaine and carnitine by Listeria monocytogenes in response to hyperosmotic stress. Appl. Environ. Microbiol. 2003, 69, 1013–1022. [Google Scholar] [PubMed]
- Hébraud, M.; Guzzo, J. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 2000, 190, 29–34. [Google Scholar]
- Eshwar, A.K.; Guldimann, C.; Oevermann, A.; Tasara, T. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella-based motility in Listeria monocytogenes. Front. Cell. Infect. Microbiol. 2017, 7, 453. [Google Scholar] [CrossRef]
- Muchaamba, F.; Eshwar, A.; Stevens, M.J.A.; Stephan, R.; Tasara, T. Different shades of Listeria monocytogenes: Strain, serotype and lineage-based variability in virulence and stress tolerance profiles. Food Microbiol. 2022, 12, 792162. [Google Scholar] [CrossRef]
- Arioli, S.; Montanari, C.; Magnani, M.; Tabanelli, G.; Patrignani, F.; Lanciotti, R.; Mora, D.; Gardini, F. Modelling of Listeria monocytogenes Scott A after a mild heat treatment in the presence of thymol and carvacrol: Effects on culturability and viability. J. Food Eng. 2019, 240, 73–82. [Google Scholar] [CrossRef]
- Doyle, M.E.; Mazzotta, A.S.; Wang, T.; Wiseman, D.W.; Scott, V.N. Heat resistance of Listeria monocytogenes. J. Food Prot. 2001, 64, 410–429. [Google Scholar] [CrossRef]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar]
- Schirmer, E.C.; Glover, J.R.; Singer, M.A.; Lindquist, S. HSP100/Clp proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 1996, 21, 289–296. [Google Scholar] [CrossRef]
- Amezaga, M.R.; Davidson, I.; McLaggan, D.; Verheul, A.; Abee, T.; Booth, I.R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology 1995, 141, 41–49. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Bowen, B.; Wiedmann, M.; Boor, K.J. Listeria monocytogenes shows temperature-dependent and independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 2012, 78, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Cotter, P.D.; Sleator, R.D.; Gahan, C.G.M. Bacterial stress response in Listeria monocytogenes: Jumping the hurdles imposed by minimal processing. Int. Dairy J. 2002, 12, 273–283. [Google Scholar] [CrossRef]
- Ballal, A.; Basu, B.; Apte, S.K. The Kdp-ATPase system and its regulation. J. Biosci. 2007, 32, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Sleator, R.D.; Gahan, C.G.; Hill, C. A postgenomic appraisal of osmotolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 1–9. [Google Scholar] [CrossRef]
- Bayles, D.O.; Wilkinson, B.J. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett. Appl. Microbiol. 2000, 30, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, T.; Buys, E.M. Listeria monocytogenes Pathogenesis: The role of stress adaptation. Microorganisms 2022, 10, 1522. [Google Scholar] [CrossRef]
- Disson, O.; Moura, A.; Lecuit, M. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 2021, 29, 811–822. [Google Scholar]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, a1869. [Google Scholar] [CrossRef]
- Churchill, K.J.; Sargeant, J.M.; Farber, J.M.; O’Connor, A.M. Prevalence of Listeria monocytogenes in select ready-to-eat foods—Deli meat, soft cheese, and packaged salad: A systematic review and meta-analysis. J. Food Prot. 2019, 82, 344–357. [Google Scholar] [CrossRef]
- Chen, H.; Neetoo, H.; Ye, M.; Joerger, R.D. Differences in pressure tolerance of Listeria monocytogenes strains are not correlated with other stress tolerances and are not based on differences in CtsR. Food Microbiol. 2009, 26, 404–408. [Google Scholar] [CrossRef]
- Carvalho, F.; Sousa, S.; Cabanes, D. How Listeria monocytogenes organizes its surface for virulence. Front. Cell Infect. Microbiol. 2014, 4, 48. [Google Scholar] [CrossRef]
- Jeyaletchumi, P.; Tunung, R.; Selina, P.M.; Chai, L.C.; Radu, S.; Farinazleen, M.G.; Cheah, Y.K.; Mitsuaki, N.; Yoshitsugu, N.; Pradeep Kumar, M.P. Assessment of Listeria monocytogenes in salad vegetables through kitchen simulation study. Int. Food Res. J. 2012, 17, 1–11. [Google Scholar]
- Quereda, J.J.; Meza-Torres, J.; Cossart, P.; Pizarro-Cerda, J. Listeriolysin S: A bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microbes 2017, 8, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Gahan, C.G.; Hill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Front. Cell Infect. Microbiol. 2014, 4, 9. [Google Scholar] [CrossRef]
- Neves, D.; Job, V.; Dortet, L.; Cossart, P.; Dessen, A. Structure of internalin InlK from the human pathogen Listeria monocytogenes. J. Mol. Biol. 2013, 425, 4520–4529. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Filipello, V.; Dall’Asta, C.; Finazzi, G.; Galaverna, G.; Losio, M.N. A structural study on the Listeria monocytogenes internalin A—Human E-cadherin interaction: A molecular tool to investigate the effects of missense mutations. Toxins 2020, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Ireton, K.; Mortuza, R.; Gyanwali, G.C.; Gianfelice, A.; Hussain, M. Role of internalin proteins in the pathogenesis of Listeria monocytogenes. Mol. Microbiol. 2021, 116, 1407–1419. [Google Scholar] [CrossRef]
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect. Immun. 2010, 78, 5062–5073. [Google Scholar] [CrossRef]
- Drolia, R.; Tenguria, S.; Durkes, A.C.; Turner, J.R.; Bhunia, A.K. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 2018, 23, 470–484.e7. [Google Scholar] [CrossRef]
- Henderson, B.; Nair, S.; Pallas, J.; Williams, M.A. Fibronectin: A multidomain host adhesin targeted by bacterial fibronectin binding proteins. FEMS Microbiol. Rev. 2011, 35, 147–200. [Google Scholar] [CrossRef]
- Hymes, J.P.; Klaenhammer, T.R. Stuck in the middle: Fibronectin-binding proteins in Gram-positive bacteria. Front. Microbiol. 2016, 7, 1504. [Google Scholar] [CrossRef]
- Suárez, M.; González-Zorn, B.; Vega, Y.; Chico-Calero, I.; Vázquez-Boland, J.A. A role for ActA in epithelial cell invasion by Listeria monocytogenes. Cell. Microbiol. 2001, 3, 853–864. [Google Scholar]
- Kühn, S.; Enninga, J. The actin comet guides the way: How Listeria actin subversion has impacted cell biology, infection biology and structural biology. Cell. Microbiol. 2020, 22, e13190. [Google Scholar]
- Hamon, M.A.; Ribet, D.; Stavru, F.; Cossart, P. Listeriolysin O: The Swiss army knife of Listeria. Trends Microbiol. 2012, 20, 360–368. [Google Scholar] [PubMed]
- Phelps, C.C.; Vadia, S.; Arnett, E.; Tan, Y.; Zhang, X.; Pathak-Sharma, S.; Gavrilin, M.A.; Seveau, S. Relative Roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018, 86, e00555-18. [Google Scholar] [PubMed]
- Pizarro-Cerda, J.; Kuhbacher, A.; Cossart, P. Entry of Listeria monocytogenes in mammalian epithelial cells: An updated view. Cold Spring Harb. Perspect. Med. 2012, 2, a010009. [Google Scholar] [CrossRef] [PubMed]
- Coffey, A.; Burg, B.V.D.; Veltman, R.; Abee, T. Characteristics of the biologically active 35-kDa metalloprotease virulence factor from Listeria monocytogenes. J. Appl. Microbiol. 2000, 88, 132–141. [Google Scholar]
- Gründler, T.; Quednau, N.; Stump, C.; Orian-Rousseau, V.; Ishikawa, H.; Wolburg, H.; Schroten, H.; Tenenbaum, T.; Schwerk, C. The surface proteins InlA and InlB are interdependently required for polar basolateral invasion by Listeria monocytogenes in a human model of the blood-cerebrospinal fluid barrier. Microbes Infect. 2013, 15, 291–301. [Google Scholar] [CrossRef]
- Tiensuu, T.; Guerreiro, D.N.; Oliveira, A.H.; O’Byrne, C.; Johansson, J. Flick of a switch: Regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 2019, 165, 819–833. [Google Scholar] [CrossRef]
- Şentürk, E.; Buzrul, S.; Şanlıbaba, P. Prevalence of Listeria monocytogenes in ready-to-eat foods, and growth boundary modeling of the selected strains in broth as a function of temperature, salt and nisin. Int. J. Food Prop. 2022, 25, 2237–2253. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Chrysadakou, M.; Tzakoniati, A.; Bikouli, V.C.; Nychas, G.-J.; Skandamis, P.N. Variability of Listeria monocytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quaternary ammonium compounds. Int. J. Food Microbiol. 2016, 237, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Mateus, T.; Silva, J.; Maia, R.L.; Teixeira, P. Listeriosis during pregnancy: A public health concern. Int. Sch. Res. Notices 2013, 20, 1385–1712. [Google Scholar] [CrossRef] [PubMed]
- Matle, I.; Mbatha, K.R.; Lentsoane, O.; Magwedere, K.; Morey, L.; Madoroba, E. Occurrence, serotypes, and characteristics of Listeria monocytogenes in meat and meat products in South Africa between 2014 and 2016. J. Food Saf. 2019, 39, e12629. [Google Scholar] [CrossRef]
- Bagatella, S.; Tavares-Gomes, L.; Oevermann, A. Listeria monocytogenes at the interface between ruminants and humans: A comparative pathology and pathogenesis review. Vet. Pathol. 2022, 59, 186–210. [Google Scholar] [CrossRef]
- Koopmans, M.M.; Brouwer, M.C.; Vázquez-Boland, J.A.; van de Beek, D. Human Listeriosis. Clin. Microbiol. Rev. 2023, 36. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Monnier, A.; Sylvain Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Zelalem, T.; Dima, C. Epidemiology and antimicrobial resistance of Listeria monocytogenes in food items: A review. Int. J. Adv. Res. Biol. Sci. 2023, 10, 47–58. [Google Scholar] [CrossRef]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef]
- Pouillot, R.; Klontz, K.C.; Chen, Y.; Burall, L.S.; Macarisin, D.; Doyle, M.; Bally, K.M.; Strain, E.; Datta, A.R.; Hammack, T.S.; et al. Infectious dose of Listeria monocytogenes in outbreak linked to ice cream, United States, 2015. Emerg. Infect. Dis. 2016, 22, 2113–2119. [Google Scholar] [CrossRef]
- Heiman, K.E.; Garalde, V.B.; Gronostaj, M.; Jackson, K.A.; Beam, S.; Joseph, L.; Asaupe, A.; Ricotta, E.; Waechter, H.; Wellman, A.; et al. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol. Infect. 2016, 144, 2698–2708. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Trallero, E.; Zigorraga, C.; Artieda, J.; Alkorta, M.; Marimón, J.M. Two outbreaks of Listeria monocytogenes infection, northern Spain. Emerg. Infect. Dis. 2014, 20, 2155–2157. [Google Scholar] [CrossRef]
- Lachmann, R.; Halbedel, S.; Lüth, S.; Holzer, A.; Adler, M.; Pietzka, A.; Al Dahouk, S.; Stark, K.; Flieger, A.; Kleta, S.; et al. Invasive listeriosis outbreaks and salmon products: A genomic, epidemiological study. Emerg. Microbes Infect. 2022, 11, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Khan, J.; Miskeen, S.; Tango, C.N.; Park, J.S.; Oh, D.H. Prevalence and control of Listeria monocytogenes in the food industry—A review. Czech J. Food Sci. 2016, 34, 469–487. [Google Scholar] [CrossRef]
- Bhatia, V.; Vag, R.; Burgess, C.M.; Gaffney, M.; Celayeta, J.M.F.C.; Cummins, E. Microbial risks associated with Ready-To-Eat Fresh Produce (RTEFP)—A focus on temperate climatic conditions. Postharvest Biol. Technol. 2024, 213, 112924. [Google Scholar] [CrossRef]
- Bantawa, K.; Rai, K.; Limbu, D.S.; Khanal, H. Food borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC Res. Notes 2018, 11, 618. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Wu, Q.; Zhang, J.; Zhang, F.; Yang, X.; Wu, H.; Zeng, H.; Chen, M.; Ding, Y.; et al. Staphylococcus aureus isolated from retail meat and meat products in China: Incidence, antibiotic resistance, and genetic diversity. Front. Microbiol. 2018, 9, 2767. [Google Scholar] [CrossRef]
- Osman, I.; Osman, S.; Mokhtar, I.; Setapa, F.; Shukor, S.A.M.; Temyati, Z. Family food consumption: Desire towards convenient food products. Procedia Soc. Behav. Sci. 2014, 121, 223–231. [Google Scholar] [CrossRef]
- Sibanda, T.; Ntuli, V.; Neetoo, S.H.; Habib, I.; Njage, P.M.K.; Kunadu, A.P.-H.; Andoh, A.H.A.; Ranil Coorey, R.; Buys, E.M. Listeria monocytogenes at the food–human interface: A review of risk factors influencing transmission and consumer exposure in Africa. Int. J. Food Sci. Technol. 2023, 58, 4114–4126. [Google Scholar] [CrossRef]
- Zakrzewski, A.J.; Gajewska, J.; Chajęcka-Wierzchowska, W.; Załuski, D.; Zadernowsk, A. Prevalence of Listeria monocytogenes and other Listeria species in fish, fish products and fish processing environment: A systematic review and meta-analysis. Sci. Total Environ. 2024, 907, 167912. [Google Scholar] [CrossRef]
- Ajayeoba, T.A.; Atanda, O.O.; Obadina, A.O.; Bankole, M.O.; Adelowo, O.O. The incidence and distribution of Listeria monocytogenes in ready-to-eat vegetables in South- Western Nigeria. Food Sci. Nutr. 2016, 4, 59–66. [Google Scholar]
- Adeoye, I.B.; Olufunmi, O.O.; Idris, B.A.; Okafor, B.N.; Ajetunmobi, T. Consumers’ preference for common exotic vegetables in Oyo and Kano State, Nigeria. Continent. J. Agric. Sci. 2010, 4, 60–65. [Google Scholar]
- Amoah, P.; Drechsel, P.; Abaidoo, R.C.; Abraham, E.M. Improving food hygiene in Africa where vegetables are irrigated with polluted water. In Proceedings of the Regional Sanitation and Hygiene Symposium, Accra, Ghana, 3–5 November 2009. [Google Scholar]
- Farber, J.M.; Zwietering, M.; Wiedmann, M.; Schaffner, D.; Hedberg, C.W.; Harrison, M.A.; Hartnett, E.; Chapman, B.; Donnelly, C.W.; Goodburn, K.E.; et al. Alternative approaches to the risk management of Listeria monocytogenes in low risk foods. Food Control 2021, 123, 107601. [Google Scholar] [CrossRef]
- Brown, S.R.B.; Kozak, S.M.; D’Amico, D.J. Applications of edible coatings formulated with antimicrobials inhibit Listeria monocytogenes growth on Queso Fresco. Front. Sustain. Food Syst. 2018, 2, 1. [Google Scholar] [CrossRef]
- Adebesin, I.O.; Amoka, S.O.; Olomoko, C.R.; Olubunmi, O.H.; Balogun, A.O. Listeria monocytogenes in Food production and Food Safety: A review. J. Food Sci. Nutr. Ther. 2024, 10, 57–68. [Google Scholar] [CrossRef]
- Malley, T.J.V.; Butts, J.; Wiedmann, M. Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J. Food Prot. 2015, 78, 436–445. [Google Scholar]
- Magdovitz, B.F.; Gummalla, S.; Thippareddi, H.; Harrison, M.A. Evaluating environmental monitoring protocols for Listeria spp. and Listeria monocytogenes in frozen food manufacturing facilities. J. Food Prot. 2020, 83, 172–187. [Google Scholar] [CrossRef]
- Leong, D.; Alvarez-Ordóñez, A.; Jordan, K. Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front. Microbiol. 2014, 5, 436. [Google Scholar] [CrossRef]
- Jordan, K.; McAuliffe, O. Listeria monocytogenes in foods. In Biological Emerging Risks in Foods (Advances in Food and Nutrition Research); Rodriguez-Lazaro, D., Ed.; Elsevier: Cambridge, UK, 2018; Volume 86, pp. 181–213. [Google Scholar] [CrossRef]
- Calvo-Arrieta, K.; Matamoros-Montoya, K.; Arias-Echandi, M.L.; Huete-Soto, A.; Redond-Solano, M. Prsenece of L. monocytogenes in ready-to-eat meat products sold at retail stores in Costa Rica and analysis of contributing factors. J. Food Prot. 2021, 84, 1729–1740. [Google Scholar] [CrossRef]
- PChemat, F.; Hoarau, N. Hazard analysis and critical control point (HACCP) for an ultrasound food processing operation. Ultrason. Sonochemistry 2005, 11, 257–260. [Google Scholar] [CrossRef]
- Herrea, A.G. The hazard analysis and critical control point system in food safety. Methods Mol. Biol. 2004, 268, 235–280. [Google Scholar] [CrossRef]
- Tompkin, R.B. Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 2002, 65, 709–725. [Google Scholar] [CrossRef]
- Salza, S.; Piras, G.; Melillo, R.; Molotzu, M.; Giagnoni, L.; Doneddu, L.; Tondello, A.; Cecchinato, A.; Stevanato, P.; Squartini, A.; et al. Environmental monitoring of Listeria monocytogenes contamination in dairy processing facilities combining culturing techniques and molecular methods. LWT Food Sci. Technol. 2024, 211, 116870. [Google Scholar] [CrossRef]
- Gupta, P.; Adhikari, A. Novel approaches to environmental monitoring and control of Listeria monocytogenes in food production facilities. Foods 2022, 11, 1760. [Google Scholar] [CrossRef] [PubMed]
- FDA. Draft Guidance for Industry: Control of Listeria Monocytogenes in Ready-to-Eat Foods. FDA-2008-D-0096. 2017. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/draft-guidance-industry-control-listeria-monocytogenes-ready-eat-foods (accessed on 27 February 2025).
- Zoellner, C.; Ceres, K.; Ghezzi-Kopel, K.; Wiedmann, M.; Ivanek, R. Design elements of Listeria environmental monitoring programs in food processing facilities: A scoping review of research and guidance materials. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1156–1171. [Google Scholar] [CrossRef]
- Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M.; Sip, A. Effectiveness of phage-based inhibition of Listeria monocytogenes in Food products and food processing environments. Microorganisms 2020, 8, 1764. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. J. Eur. Union. 2005. L338, 9. Available online: https://eurlex.europa.eu (accessed on 26 February 2025).
- Costa, P.V.; Nascimento, J.D.S.; Costa, L.E.D.O.; Ferreira, P.B.D.M.; Brandã, M.L.L. Listeria monocytogenes: Challenges of microbiological control of food in Brazil. Food Sci. Technol. Camp. 2022, 42, e08322. [Google Scholar] [CrossRef]
- Braga, V.; Vázqueza, S.; Vico, V.; Pastorinoa, V.; Mota, M.I.; Legnania, M.; Schelotto, F.; Lancibidada, G.; Varela, G. Prevalence and serotype distribution of Listeria monocytogenes isolated from foods in Montevideo-Uruguay. Braz. J. Microbiol. 2017, 48, 689–694. [Google Scholar] [CrossRef]
- Jibo, G.G.; Raji, Y.E.; Salawudeen, A.; Amin-Nordin, S.; Mansor, R.; Jamaluddin, T.Z.M.T. A systematic review and meta-analysis of the prevalence of Listeria monocytogenes in South-East Asia; a one-health approach of human-animal-food-environment. One Health 2022, 15, 100417. [Google Scholar] [CrossRef]
- Dhama, K.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Barbuddhe, S.; Malik, S.V.S.; Singh, R.K. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: A comprehensive review. Vet. Q. 2015, 35, 11–235. [Google Scholar] [CrossRef]
- Meza-Bone, G.A.; Meza-Bone, J.S.; Cedeño, Á.; Martín, I.; Martín, A.; Maddela, N.R.; Córdoba, J.J. Prevalence of Listeria monocytogenes in RTE meat products of Quevedo (Ecuador). Foods 2023, 12, 2956. [Google Scholar] [CrossRef] [PubMed]
- Kurpas, M.; Wieczorek, K.; Osek, J. Ready-to-eat meat products as a source of Listeria monocytogenes. J. Vet. Res. 2018, 61, 49–55. [Google Scholar] [CrossRef]
- Barrett, C.B.; Reardon, T.; Swinnen, J.; Zilberman, D. Agri-food value chain revolutions in low-and middle-income countries. J. Econ. Lit. 2020, 60, 1316–1377. [Google Scholar]
- Rodriguez, C.; Taminiau, B.; García-Fuentes, E.; Daube, G.; Korsak, N. Listeria monocytogenes dissemination in farming and primary production: Sources, shedding and control measures. Food Control 2021, 120, 107540. [Google Scholar] [CrossRef]
- Thomas, J.; Govender, N.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; Sekwadi, P.; Ntshoe, G.; et al. Outbreak of Listeriosis in South AfricaaAssociated with processed meat. N. Engl. J. Med. 2020, 382, 632–643. [Google Scholar]
- Schutte, C.-M.; Van Der Meyden, C.H.; Kakaza, M.; Lockhat, Z.; Van Der Walt, E. Life-threatening Listeria meningitis: Need for revision of south African acute bacterial meningitis treatment guidelines. S. Afr. Med. J. 2019, 109, 296–298. [Google Scholar]
- Milford, A.B.; Le Mouël, C.; Bodirsky, B.L.; Rolinski, S. Drivers of meat consumption. Appetite 2019, 141, 104313. [Google Scholar]
- de Bruin, S.; Dengerink, J.; van Vliet, J. Urbanisation as driver of food system transformation and opportunities for rural livelihoods. Food Secur. 2021, 13, 781–798. [Google Scholar]
- Reardon, T. The hidden middle: The quiet revolution in the midstream of agrifood value chains in developing countries. Oxf. Rev. Econ. Policy 2015, 31, 45–63. [Google Scholar] [CrossRef]
- Barrett, C.B.; Reardon, T.; Swinnen, J.; Zilberman, D. Structural Transformation and Economic Development: Insights from the Agri-food Value Chain Revolution; Dyson School of Applied Economics and Management, Cornell University: Ithaca, NY, USA, 2019; pp. 1–56. Available online: https://barrett.dyson.cornell.edu/files/papers/BRSZ%2013%20Aug%202019.pdf (accessed on 26 February 2025).
- Steyn, N.P.; Labadarios, D.; Nel, J.H. Factors which influence the consumption of street foods and fast foods in South Africa—A national survey. Nutr. J. 2011, 10, 104. [Google Scholar] [CrossRef]
- Kok, R.; Balkaran, R. Street food vending and hygiene practices and implications for consumers. J. Econ. Behav. Stud. 2014, 6, 188–193. [Google Scholar] [CrossRef]
- Nkosi, N.V.; Tabit, F.T. The food safety knowledge of street food vendors and the sanitary conditions of their street food vending environment in the Zululand District, South Africa. Heliyon 2021, 7, e07640. [Google Scholar] [CrossRef] [PubMed]
- Rane, S. Street vended food in developing world: Hazard analyses. Ind. J. Microbiol. 2011, 51, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Nordic Workshop on Listeria Monocytogenes in Copenhagen 26th and 27th September, 2001; Nordic Committee of Senior Officials for Food Issues: Copenhagen, Denmark, 2001. [Google Scholar]
- Lukinmaa, S.; Miettinen, M.; Nakari, U.-M.; Korkeala, H.; Siitonen, A.A. Listeria monocytogenes isolates from invasive infections: Variation of sero- and genotypes during an 11-year period in Finland. J. Clin. Microbiol. 2003, 41, 1694–1700. [Google Scholar] [CrossRef]
- Centre for Disease Prevention and Control. When Food Bites Back. Protecting People from Deadly Listeria Food Poisoning. 2013. Available online: http://www.cdc.gov (accessed on 26 December 2024).
- Morshdy, A.E.M.A.; Nagaty, E.A.; Abdallah, K.M.E.; Darwish, W.S.; Mahmoud, A.F.A. Prevalence of Listeria monocytogenes in meat products retailed in Egypt and worldwide: A Review. J. Adv. Vet. Res. 2023, 13, 1487–1490. [Google Scholar]
- Tenny, S.; Hoffman, M.R. Prevalence; StatPearls Publishing: Treasure Islands, FL, USA, 2025. [Google Scholar]
- Marina, A.R.; Ismail, M.; Nurilyana, M.T.; Mohd Fharok, Y.; Azzura, A.L. Incidence of Listeria monocytogenes in dairy and food products of animal origin in central region of peninsular Malaysia. Mal. J. Vet. Res. 2019, 10, 106–112. [Google Scholar]
- Gyurova, E.; Krumova-Vulcheva, G.; Hristo Daskalov, H.; Gogov, Y. Prevalence of Listeria monocytogenes in ready-to-eat foods in Bulgaria. J. Hyg. Eng. Des. 2020, 112–118. [Google Scholar]
- Skowron, K.; Wałecka-Zacharska, E.; Wiktorczyk-Kapischke, N.; Skowron, K.J.; Grudlewska-Buda, K.; Bauza-Kaszewska, J.; Bernacia, Z.; Borkowski, M.; Gospodarek-Komkowska, E. Assessment of the Prevalence and Drug Susceptibility of Listeria monocytogenes Strains Isolated from Various Types of Meat. Foods 2020, 9, 1293. [Google Scholar] [CrossRef]
- Shimojima, Y.; Kanai, Y.; Takatoshi Moriyama, T.; Sayoko Arakawa, S.; Tamura, Y.; Yumiko Okada, Y.; Morita, Y. Environmental monitoring of food manufacturing facilities for Listeria: A case study. J. Food Prot. 2023, 86, 100149. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, L.; Wang, Y.; Li, Z.; Wang, Z.; Han, J. Effect of high hydrostatic pressure on solubility and conformation changes of soybean protein isolate glycated with flaxseed gum. Food Chem. 2020, 333, 127530. [Google Scholar] [CrossRef]
- Tirloni, E.; Centorotola, G.; Pomilio, F.; Torresi, M.; Bernardi, C.; Stella, S. Listeria monocytogenes in ready-to-eat (RTE) delicatessen foods: Prevalence, genomic characterization of isolates and growth potential. Int. J. Food Microbiol. 2024, 410, 110515. [Google Scholar] [CrossRef] [PubMed]
- Diriba, K.; Awulachew, E.; Diriba, K.D.K.; Awulachew, E.; Diribsa, K. The prevalence of Listeria species in diferent food items of animal and plant origin in Ethiopia: A systematic review and meta-analysis. Eur. J. Med. Res. 2021, 26, 60. [Google Scholar] [CrossRef]
- Osek, J.; Wieczorek, K. Listeria monocytogenes—How this pathogen uses its virulence mechanisms to infect the hosts. Pathogens 2022, 11, 1491. [Google Scholar] [CrossRef]
- Meloni, D. Presence of Listeria monocytogenes in Mediterranean-Style Dry Fermented Sausages. Foods 2015, 4, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.K.; Roof, S.; Boyle, E.A.; Burson, D.; Thippareddi, H.; Geornaras, I.; Sofos, J.N.; Wiedmann, M.; Nightingale, K. Molecular ecology of Listeria monocytogenes and other Listeria species in small and very small ready-to-eat meat processing plants. J. Food Prot. 2011, 74, 63–77. [Google Scholar]
- El-Demerdash, A.S.; Raslan, M.T. Molecular characterization of Listeria monocytogenes isolated from different animal-origin food items from urban and rural areas. Adv. Anim. Vet. Sci. 2019, 7, 51–56. [Google Scholar] [CrossRef]
- Deka, A.; Hazarika, R.A.; Barua, A.G.; Saikia, G.K.; Borah, P.; Shakuntala, I.; Roychoudhury, P.; Bora, D.P. Prevalence of Listeria monocytogenes in Foods of animal origin: Study from Assam, A North-Eastern State of India. Eur. J. Vet. Med. 2022, 2, 20–25. [Google Scholar]
- Koskar, J.; Kramarenko, T.; Meremäe, K.; Kuningas, M.; Sõgel, J.; Mäesaar, M.; Anton, D.; Lillenberg, M.; Roasto, M. Prevalence and Numbers of Listeria monocytogenes in various ready-to-eat foods over a 5-year period in Estonia. J. Food Prot. 2019, 82, 597–604. [Google Scholar]
- Martinez-Laorden, A.; Arraiz-Fernandez, C.; Cantalejo, M.J.; Gonzalez-Fandos, E. Prevalence, identification and antimicrobial resistance of Listeria monocytogenes and Listeria spp. isolated from poultry and pork meat. Int. J. Food Sci. Technol. 2024, 59, 2667–2675. [Google Scholar] [CrossRef]
- Amajoud, N.; Leclercq, A.; Soriano, J.M.; Bracq-Dieye, H.; Maadoudi, M.; Senhaji, N.S.; Kounnoun, A.; Moura, A.; Lecuit, M.; El Abrini, J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control 2018, 84, 436–441. [Google Scholar] [CrossRef]
- Seyoum, E.T.; Woldetsadik, D.A.; Mekonen, T.K.M.; Gezahegn, H.A.; Gebreyes, W.A. Prevalence of Listeria monocytogenes in raw bovine milk and milk products from central highlands of Ethiopia. J. Infect. Dev. Ctries. 2015, 9, 1204–1209. [Google Scholar] [CrossRef]
- Moabelo, K.C.; Gcebe, N.; Gana, J.; Ngoshe, Y.B.; Adesiyun, A.A. Contamination of beef and beef products by Listeria spp. and molecular characterization of L. monocytogenes in Mpumalanga, South Africa. J. Food Saf. 2023, 43, e13055. [Google Scholar] [CrossRef]
- Chen, M.; Wu, Q.; Zhang, J.; Wu, S.; Guo, W. Prevalence enumeration, and pheno- and genotypic characteristics of Listeria monocytogenes isolated from raw foods in South China. Front. Microbiol. 2015, 6, 1026. [Google Scholar] [CrossRef]
- Branka, B.; Tatjana, B.; Brankica, L.; Vesna, J.; Radmila, M.; Jelena, J.; Slobodan, L. Prevalence of Listeria monocytogenes in ready-to-eat food of animal origin. Tehnologija 2014, 55, 117–122. [Google Scholar]
- Gelbíčová, T.; Zobaníková, M.; Tomáštíková, Z.; Van Walle, I.; Ruppitsch, W.; Karpíšková, R. An outbreak of listeriosis linked to Turkey meat products in the Czech Republic, 2012–2016. Epidemiol. Infect. 2018, 146, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- De Valk, H.; Vaillant, V.; Jacquet, C.; Rocourt, J.; Le Querrec, F.; Stainer, F.; Quelquejeu, N.; Pierre, O.; Pierre, V.; Desenclos, J.-C.; et al. Two consecutive nationwide outbreaks of listeriosis in France, October 1999–February 2000. Am. J. Epidemiol. 2001, 154, 944–950. [Google Scholar]
- Fretz, R.; Sagel, U.; Ruppitsch, W.; Pietzka, A.; Stoger, A.; Huhulescu, S.; Heuberger, S.; Pichler, J.; Much, P.; Pfaff, G.; et al. Listeriosis outbreak caused by acid curd cheese Quargel, Austria and Germany 2009. Euro. Surveill. 2010, 15, 19477. [Google Scholar]
- Dalton, C.B.; Austin, C.C.; Sobel, J.; Hayes, P.S.; Bibb, W.F.; Graves, L.M.; Swaminathan, B.; Proctor, M.E.; Griffin, P.M. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 1997, 336, 100–105. [Google Scholar]
- Lyytikainen, O.; Autio, T.; Maijala, R.; Ruutu, P.; Honkanen-Buzalski, T.; Miettinen, M.; Hatakka, M.; Mikkola, J.; Anttila, V.-J.; Johansson, T.; et al. An Outbreak of Listeria monocytogenes Serotype 3a Infections from Butter in Finland. J. Infect. Dis. 2000, 181, 1838–1841. [Google Scholar]
- Sebastianski, M.; Bridger, N.A.; Featherstone, R.M.; Robinson, J.L. Disease outbreaks linked to pasteurized and unpasteurized dairy products in Canada and the United States: A systematic review. Can. J. Public Health 2022, 113, 569–578. [Google Scholar] [CrossRef]
- Fleming, D.W.; Cochi, S.L.; MacDonald, K.L.; Brondum, J.; Hayes, P.S.; Plikaytis, B.D.; Holmes, M.B.; Audurier, A.; Broome, C.V.; Reingold, A.L. Pasteurised milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 1985, 312, 404–407. [Google Scholar] [CrossRef]
- James, S.M.; Fannin, S.L.; Agee, B.A.; Hall, B.; Parker, E.; Vogt, J.; Run, G.; Williams, J.; Lieb, L.; Salminen, C.; et al. Listeriosis outbreak associated with Mexicanstyle cheese—California. Morb. Mort. Weekly Rep. 1985, 34, 357–359. [Google Scholar]
- Centers for Disease Control and Prevention. Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy–Massachusetts, 2007. MMWR 2008, 57, 1097–1100. [Google Scholar]
- Nichols, M.; Conrad, A.; Whitlock, L.; Stroika, S.; Strain, E.; Weltman, A.; Johnson, L.; DeMent, J.; Reporter, R.; Williams, I. Short communication: Multistate outbreak of Listeria monocytogenes infections retrospectively linked to unpasteurized milk using whole-genome sequencing. J. Dairy Sci. 2020, 103, 176–178. [Google Scholar] [CrossRef]
- Koch, J.; Dworak, R.; Prager, R.; Becker, B.; Brockmann, S.; Wicke, A.; Wichmann-Schauer, H.; Hof, H.; Werber, D.; Stark, K. Large listeriosis outbreak linked to cheese made from pasteurized milk, Germany, 2006–2007. Foodborne Pathog. Dis. 2010, 7, 1581–1584. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Bloemberg, G.V.; Müller, A.; Stevens, M.J.A.; Cernela, N.; Kollöffel, B.; Stephan, R. Listeriosis Caused by Persistence of Listeria monocytogenes Serotype 4b Sequence Type 6 in Cheese Production Environment. Emerg. Infect. Dis. 2021, 27, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Hanson, H.; Whitfield, Y.; Lee, C.; Badiani, T.; Minielly, C.; Fenik, J.; Makrostergios, T.; Kopko, C.; Majury, A.; Hillyer, E.; et al. Listeria monocytogenes Associated with Pasteurized Chocolate Milk, Ontario, Canada. Emerg. Infect. Dis. 2019, 25, 581–584. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention. Listeria Outbreak Linked to Meats Sliced at Delis. 2024. Available online: https://www.cdc.gov/listeria/outbreaks/delimeats-7-24/index.html (accessed on 16 February 2025).
- European Centre for Disease Prevention and Control, European Food Safety Authority. Prolonged Multicountry Outbreak of Listeria monocytogenes ST173 Linked to Consumption of Fish Products—19 June 2024. ISBN 978-92-9498-726-6; Catalogue number TQ-09-24-482-EN-N. 2024. Available online: https://www.cdc.gov/listeria/outbreaks/delimeats-7-24/investigation.html (accessed on 12 February 2024).
- Self, J.L.; Conrad, A.; Stroika, S.; Jackson, A.; Whitlock, L.; Jackson, K.A.; Beal, J.; Wellman, A.; Fatica, M.K.; Bidol, S.; et al. Multistate Outbreak of Listeriosis Associated with Packaged Leafy Green Salads, United States and Canada, 2015–2016. Emerg. Infect. Dis. 2019, 25, 1461–1468. [Google Scholar] [CrossRef]
- Lamont, R.F.; Sobel, J.; Mazaki-Tovi, S.; Kusanovic, J.P.; Vaisbuch, E.; Kim, S.K.; Uldbjerg, N.; Romero, R. Listeriosis in human pregnancy: A systematic review. J. Perinat. Med. 2011, 39, 227–236. [Google Scholar] [CrossRef]
- Desai, A.N.; Anyoha, A.; Madoff, C.L.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of proMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Mishra, S.; Tuglo, L.S.; Sarangi, A.K.; Kandi, V.; Al Ibrahim, A.A.; Alsaif, H.A.; Rabaan, A.A.; Zahan, M.K. Recurring food source-based Listeria outbreaks in the United States: An unsolved puzzle of concern? Health Sci. Rep. 2024, 7, e1863. [Google Scholar] [CrossRef]
- Datta, A.; Burall, L. Current trends in foodborne human listeriosis. Food Saf. 2018, 6, 1–6. [Google Scholar] [CrossRef]
- Calahorrano-Moreno, M.B.; Ordoñez-Bailon, J.J.; Baquerizo-Crespo, R.J.; Dueňas-Rivadeneira, A.A.; Montenegro, M.C.B.S.M.; Rodríguez-Díaz, J.M. Contaminants in the cow’s milk we consume? Pasteurization and other technologies in the elimination of contaminants. F1000Research 2022, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Medjahdi, K.; Didouh, N.; Araujo, R. Pasteurized milk: A highlight on potential sources of contamination by aerobic spore-forming bacteria. Food Control 2025, 171, 111134. [Google Scholar] [CrossRef]
- Londoño-Carmona, J.; Blandón-Escobar, S.; Montoya-Zuluaga, J.; Betancourt-Chaves, P.; Castillo-Moreno, S.; Arboleda-Múnera, C.; Vallejo-Timarán, D. Antibiotic residues and microbial contamination in pasteurized whole milk intended for human consumption. Vet. World 2024, 17, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wang, C.; Liu, Y.; Zhang, P.; Wu, Y.; Li, M.; Zhao, J.; Zhang, X.; Ma, X. Pre-packaged cold-chain ready-to-eat food as a source of sporadic listeriosis in Beijing, China. J. Infect. 2024, 89, 106254. [Google Scholar] [CrossRef]
- Burall, L.S.; Chen, Y.; Macarisin, D.; Pouillot, R.; Strain, E.; De Jesus, A.J.; Laasri, A.; Wang, H.; Ali, L.; Tatavarthy, A.; et al. Enumeration and characterization of Listeria monocytogenes in novelty ice cream samples manufactured on a specific production line linked to a listeriosis outbreak. Food Control 2017, 82, 1–7. [Google Scholar] [CrossRef]
- Lundén, J.; Tolvanen, R.; Korkeala, H. Human Listeriosis outbreaks linked to dairy products in Europe. J. Dairy Sci. 2004, 87, E6–E11. [Google Scholar] [CrossRef]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef]
- Edwards, D.; Dunlop, D.J. Food Microbiology. In Food Microbiol, 5th ed.; Mp, D., Diez-Gonzalez, F., Hill, C., Eds.; ASM Press: Washington, DC, USA, 2019; Volume 1093. [Google Scholar]
- Lecuit, M. Listeria monocytogenes, a model in infection biology. Cell. Microbiol. 2020, 22, e13186. [Google Scholar] [CrossRef]
- Harrand, A.S.; Strawn, L.K.; Illas-Ortiz, P.M.; Wiedmann, M.; Weller, D. Listeria monocytogenes prevalence varies more within fields than between fields or over time on conventionally farmed New York produce fields. J. Food Prot. 2020, 83, 1958–1966. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Windham, K.; Martin, K.E.; Yeung, M.; Wiedmann, M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 2005, 71, 8764–8772. [Google Scholar] [PubMed]
- Dalzini, E.; Bernini, V.; Bertasi, B.; Daminelli, P.; Losio, M.-N.; Varisco, G. Survey of prevalence and seasonal variability of Listeria monocytogenes in raw cow milk from Northern Italy. Food Control 2016, 60, 466–470. [Google Scholar]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar]
- Schoder, D.; Guldimann, C.; Märtlbauer, E. Asymptomatic Carriage of Listeria monocytogenes by animals and humans and its impact on the food chain. Foods 2022, 11, 3472. [Google Scholar] [CrossRef] [PubMed]
- Chandler-Khayd, C.; Di Francesco, J.; Baron, J.N.; Ramos, T.D.M.; Aminabadi, P.; Jay-Russell, M.T.; Haghani, V.; Millner, P.D.; Pagliari, P.H.; Hutchinson, M.; et al. Risk factors associated with the prevalence of Listeria monocytogenes in manured soils on certified organic farms in four regions of the United States. Front. Sustain. Food Syst. 2023, 7, 1222192. [Google Scholar]
- Fangueiro, D.; Alvarenga, P.; Fragoso, R. Horticulture and orchards as new markets for manure valorisation with less environmental Impacts. Sustainability 2021, 13, 1436. [Google Scholar] [CrossRef]
- Raheem, D. Outbreaks of listeriosis associated with deli meats and cheese: An overview. AIMS Microbiol. 2016, 23, 230–250. [Google Scholar] [CrossRef]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fern’ández Escamez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Nørrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Alegbeleye, O.; Sant’Ana, A.S. Survival and growth behaviour of L. monocytogenes in ready-to-eat foods. Food Control 2022, 138, 109023. [Google Scholar]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and death Decision Making in a Pathogen. Front. Microbiol. 2020, 11, 1505. [Google Scholar] [CrossRef] [PubMed]
- Ravindhiran, R.; Sivarajan, K.; Sekar, J.N.; Murugesan, R.; Dhandapani, K. Listeria monocytogenes an Emerging Pathogen: A comprehensive overview on listeriosis, virulence determinants, detection, and anti-listerial interventions. Microb. Ecol. 2023, 86, 2231–2251. [Google Scholar] [CrossRef] [PubMed]
- Sotohy, S.A.; Elnaker, Y.F.; Omar, A.M.; Alm Eldin, N.K.; Diab, M.S. Prevalence, antibiogram and molecular characterization of Listeria monocytogenes from ruminants and humans in New Valley and Beheira Governorates, Egypt. BMC Vet. Res. 2024, 20, 297. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, A.; Linke, K.; Wagner, M.; Stessl, B. The saprophytic lifestyle of Listeria monocytogenes and entry into the Food-Processing Environment. Front. Microbiol. 2022, 13, 789801. [Google Scholar] [CrossRef]
- Nkhebenyane, J.S.; Lues, R. The knowledge, attitude, and practices of food handlers in central South African hospices. Food Sci Nutr. 2020, 8, 2598–2607. [Google Scholar] [CrossRef]
- Terentjeva, M.; Šteingolde, Ž.; Meistere, I.; Elferts, D.; Avsejenko, J.; Streikiša, M.; Gradovska, S.; Alksne, L.; Kibilds, J.; Bērzinš, A. Prevalence, genetic diversity and factors fssociated with distribution of Listeria monocytogenes and other Listeria spp. in cattle farms in Latvia. Pathogens 2021, 10, 851. [Google Scholar] [CrossRef]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes—From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, e1236–e1243. [Google Scholar] [CrossRef]
- Rocourt, J.; Jacquet, C.; Reilly, A. Epidemiology of human listeriosis and seafoods. Int. J. Food Microbiol. 2000, 62, 197–209. [Google Scholar] [CrossRef]
- Goulet, V.; de Valk, H.; Pierre, O.; Stainer, F.; Rocourt, J.; Vaillant, V.; Jacquet, C.; Desenclos, J.C. Effect of prevention measures on incidence of human listeriosis, France, 1987-1997. Emerg. Infect. Dis. 2001, 7, 983–989. [Google Scholar] [CrossRef]
- Elsayed, M.E.; Abd El-Hamid, M.I.; El-Gedawy, A.; Bendary, M.M.; ELTarabili, R.M.; Alhomrani, M.; Alamri, A.S.; Alghamdi, S.A.; Arnout, M.; Binjawhar, D.N.; et al. New insight into Listeria monocytogenes antimicrobial resistance, virulence attributes and their prospective correlation. Antibiotics 2022, 11, 1447. [Google Scholar] [CrossRef]
- Graves, L.M.; Hunter, S.B.; Ong, A.R.; Schoonmaker-Bopp, D.; Hise, K.; Kornstein, L.; DeWitt, W.E.; Hayes, P.S.; Dunne, E.; Mead, P.; et al. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 2005, 43, 2350–2355. [Google Scholar]
- Yousefi, M.; Khorshidian, N.; Hosseini, H. Potential application of essential oils for mitigation of Listeria monocytogenes in Meat and Poultry Products. Front. Nutr. 2020, 7, 577287. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T. Outer membrane vesicles (OMVs) of Gram-negative bacteria: A perspective update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Denoya, G.I.; Polenta, G.A.; Apostolo, N.M.; Budde, C.O.; Sancho, A.M.; Vaudagna, S.R. Optimization of high hydrostatic pressure processing for the preservation of minimally processed peach pieces. Innov. Food Sci. Emerg. Technol. 2016, 33, 84–93. [Google Scholar] [CrossRef]
- Jadhav, H.B.; Choudhary, P. Emerging techniques for the processing of food to ensure higher food safety with enhanced food quality: A review. Discover Food. 2024, 4, 20. [Google Scholar] [CrossRef]
- Lajnaf, R. Introductory Chapter: Novel thermal and non-thermal technologies for food processing. In Food Processing and Preservation; IntechOpen: London, UK, 2023. [Google Scholar] [CrossRef]
- Putnik, P.; Kresoja, Ž.; Bosiljkov, T.; Režek Jambrak, A.; Barba, F.J.; Lorenzo, J.M.; Roohinejad, S.; Granato, D.; Žuntar, I.; Kovačević, B. Comparing the effects of thermal and non-thermal technologies on pomegranate juice quality: A review. Food Chem. 2019, 279, 150–161. [Google Scholar] [CrossRef]
- Bahrami, A.; Babolia, Z.M.; Schimmel, K.; Jafarid, S.M.; Williams, L. Efficiency of novel processing technologies for the control of Listeria monocytogenes in food products. Trends Food Sci. Technol. 2020, 96, 61–67. [Google Scholar]
- Russo, G.L.; Langellotti, A.L.; Torrieri1, E.; Masi1, P. Emerging technologies in seafood processing: An overview of innovations reshaping the aquatic food industry. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13281. [Google Scholar] [CrossRef]
- Liao, X.; Liu, D.; Xiang, Q.; Ahn, J.; Chen, S.; Ye, X.; Ding, T. Inactivation mechanisms of non-thermal plasma on microbes: A review. Food Control 2017, 75, 83–91. [Google Scholar] [CrossRef]
- Yin, W.; Xu, S.; Wang, Y.; Zhang, Y.; Cho, S.-H.; Gaiperin, M.Y. Ways to control harmful biofilms:prevention, inhibtion and eradication. Crit. Rev. Microbiol. 2021, 47, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.T.; Siguemoto, É.S.; Funcia, E.S.; Augusto, P.E.; Curet, S.; Boillereaux, L.; Sastry, S.K.; Gut, J.A. Non-thermal effects of microwave and ohmic processing on microbial and enzyme inactivation: A critical review. Curr. Opin. Food Sci. 2020, 35, 36–48. [Google Scholar] [CrossRef]
- Cheng, C.; Jiang, L.; Li, X.; Song, H.; Fang, W. Can natural preservatives serve as a new line of protective technology against bacterial pathogens in meat and meat products? Food Qual. Saf. 2024, 8, fyad049. [Google Scholar] [CrossRef]
- Song, Y.; Peters, T.L.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Characterization of a novel group of Listeria phages that target serotype 4b Listeria monocytogenes. Viruses 2021, 13, 671. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Sorek, R. Contemporary phage biology: From classic models to new insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xia, X.; Liu, Y.; Li, Z.; Shi, T.; Zhang, H.; Dong, Q. Recent advances on the formation, detection, resistance mechanism, and control technology of Listeria monocytogenes biofilm in food industry. Food Res. Int. 2024, 180, 114067. [Google Scholar] [CrossRef]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef]
- Ranveer, S.A.; Dasriya, V.; Ahmad, F.; Dhillon, H.S.; Samtiya, M.; Shama, E.; Anand, T.; Dhewa, T.; Chaudhary, V.; Chaudhary, P.; et al. Positive and negative aspects of bacteriophages and their immense role in the food chain. npj Sci. Food 2024, 8, 1. [Google Scholar] [CrossRef]
- Hagens, S.; Loessner, M.J. Phages of Listeria offer novel tools for diagnostics and biocontrol. Front. Microbiol. 2014, 5, 159. [Google Scholar] [CrossRef]
- Gutierrez, D.; Rodríguez-Rubio, L.; Fernandez, L.; Martínez, B.; Rodríguez, A.; García, P. Applicability of commercial phage-based products against Listeria monocytogenes for improvement of food safety in Spanish dry-cured ham and food contact surfaces. Food Control 2017, 73, 1474–1482. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.R.; Kim, H.-S.; Ha, S.-D. Effective ness of a phage cocktail as a biocontrol agent against L. monocytogenes biofilms. Food Control 2017, 78, 256–263. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Schmuki, M.M.; Erne, D.; Loessner, M.J.; Klumpp, J. Bacteriophage P70: Unique morphology and unrelatedness to other Listeria Bacteriophages. J. Virol. 2012, 86, 13099–13102. [Google Scholar]
- Vongkamjan, K.; Switt, A.M.; Bakker, H.C.D.; Fortes, E.D.; Wiedmann, M. Silage collected from dairy farms harbors an abundance of Listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol. 2012, 78, 8666–8675. [Google Scholar]
- Sumrall, E.T.; Keller, A.P.; Shen, Y.; Loessner, M.J. Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 2020, 113, 627–637. [Google Scholar]
- Huang, Y.; Morvay, A.A.; Shi, X.; Suo, Y.; Shi, C.; Knøchel, S. Comparison of oxidative stress response and biofilm formation of Listeria monocytogenes serotypes 4b and 1/2a. Food Control 2018, 85, 416–422. [Google Scholar] [CrossRef]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef]
- Stone, E.; Lhomet, A.; Neve, H.; Grant, I.R.; Campbell, K.; McAuliffe, O. Isolation and characterization of Listeria monocytogenes PhagvB_LmoH_P61, a Phage with biocontrol potential on different Food Matrices. Front. Sustain. Food Syst. 2020, 4, 521645. [Google Scholar] [CrossRef]
- Ribeiro, A.C.; Almeida, F.A.; Medeiros, M.M.; Miranda, B.R.; Pinto, U.M.; Alves, V.F. Listeria monocytogenes: An inconvenient hurdle for the dairy industry. Dairy 2023, 4, 316–344. [Google Scholar] [CrossRef]
- El-Bakry, M.; Sanchez, A.; Mehta, B.M.; El-Bakry, M.; Sanchez, A.; Mehta, B.M. Microstructure of Dairy Products, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA, 2018; ISBN 978-1-118-96421-7. [Google Scholar]
- Grigore-Gurgu, L.; Bucur, F.I.; Mihalache, O.A.; Nicolau, A.I. Comprehensive review on the biocontrol of Listeria monocytogenes in food products. Foods 2024, 13, 734. [Google Scholar] [CrossRef] [PubMed]
- Cucić, S.; Ells, T.; Guri, A.; Kropinski, A.M.; Khursigara, C.M.; Anany, H. Degradation of Listeria monocytogenes biofilm by phages belonging to the genus Pecentumvirus. Appl. Environ. Microbiol. 2024, 90, e0106223. [Google Scholar] [CrossRef]
- Gómez-Galindo, M.; Truchado, P.; Allende, A.; Gil, M.I. Optimisation of the use of a commercial phage-based product as a control strategy of Listeria monocytogenes in the fresh-cut industry. Foods 2023, 12, 3171. [Google Scholar] [CrossRef]
- Schellekens, M.M.; Woutersi, J.; Hagens, S.; Hugenholtz, J. Bacteriophage P100 application to control Listeria monocytogenes on smeared cheese. Milchwissenschaft 2007, 62, 284–287. [Google Scholar]
- Perera, M.N.; Abuladze, T.; Li, M.; Woolston, J.; Sulakvelidze, A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015, 52, 42–48. [Google Scholar]
- Shannon, R.; Radford, D.R.; Balamurugan, S. Impacts of food matrix on bacteriophage and endolysin antimicrobial efficacy and performance. Crit. Rev. Food Sci. Nutr. 2020, 60, 1631–1640. [Google Scholar]
- Kocot, A.M.; Briers, Y.; Plotka, M. Phages and engineered lysins as an effective tool to combat Gram-negative foodborne pathogens. Compr. Rev. Food Sci. Food Saf. 2023, 22, 2235–2266. [Google Scholar]
- Tsevdou, M.; Gogou, E.; Taoukis, P. High hydrostatic pressure processing of foods. In Green Food Processing Techniques; Chemat, F., Vorobiev, E., Eds.; Elsevier Inc.: Paris, France, 2019; pp. 87–137. [Google Scholar] [CrossRef]
- Raur, B.P.; Rao, P.S. Modelling the combined effect of pressure and mild heat on the inactivation kinetics of Escherichia coli, Listeria innocua and Staphylococcus aureus in black tiger shrimp (Penaeus monodon). Front. Microbiol. 2017, 8, 1311. [Google Scholar]
- Roobab, U.; Chacha, J.S.; Abida, A.; Rashid, S.; Muhammad Madni, G.; Lorenzo, J.M.; Zeng, X.-A.; Aadil, R.M. Emerging Trends for Nonthermal Decontamination of Raw and Processed Meat: Ozonation, High-Hydrostatic Pressure and Cold Plasma. Foods 2022, 11, 2173. [Google Scholar] [CrossRef]
- Georget, E.; Sevenich, R.; Reineke, K.; Mathys, A.; Heinz, V.; Callanan, M.; Rauh, C.; Knorr, D. Inactivation of microorganisms by high isostatic pressure processing in complex matrices: A review. Innov. Food Sci. Emerg. Technol. 2015, 27, 1–14. [Google Scholar] [CrossRef]
- Kulawik, P.; Rathod, N.B.; Ozogul, Y.; Ozogul, F.; Zhang, W. Recent developments in the use of cold plasma, high hydrostatic pressure, and pulsed electric fields on microorganisms and viruses in seafood. Crit Rev. Food Sci. Nutr. 2022, 63, 9716–9730. [Google Scholar] [CrossRef] [PubMed]
- Khouryieh, H.A. Novel and emerging technologies used by the U.S. food processing industry. Innov. Food Sci. Emerg. Technol. 2021, 67, 102559. [Google Scholar] [CrossRef]
- Aganovic, K.; Hertel, C.; Vogel, R.F.; Johne, R.; Schlüter, O.; Schwarzenbolz, U.; Jäger, H.; Holzhauser, T.; Bergmair, J.; Roth, A.; et al. Aspects of high hydrostatic pressure food processing: Perspectives on technology and food safety. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3225–3266. [Google Scholar] [CrossRef]
- Zhao, Y.; Teixeira, J.S.; Saldaňa, M.D.A.; Gänzle, M.G. Antimicrobial activity of bioactive starch packaging films against Listeria monocytogenes and reconstituted meat microbiota on ham. Int. J. Food Microbiol. 2019, 305, 108253. [Google Scholar] [CrossRef]
- Fadiji, T.; Rashvand, M.; Daramola, M.O.; Iwarere, S.A. A Review on antimicrobial packaging for extending the shelf life of food. Processes 2023, 11, 590. [Google Scholar] [CrossRef]
- Fidan, H.; Esatbeyoglu, T.; Simat, V.; Trif, M.; Tabanelli, G.; Kostka, T.; Montanari, C.; Ibrahim, S.A.; Özogul, F. Recent Developments of Lactic Acid Bacteria and Their Metabolites on Foodborne Pathogens and Spoilage Bacteria: Facts and Gaps. Food Biosci. 2022, 47, 101741. [Google Scholar] [CrossRef]
- Espitia, P.J.P.; Pacheco, J.J.R.; De Melo, N.R.; Soares, N.D.F.F. Packaging properties and control of Listeria monocytogenes in bologna by cellulosic films incorporated with pediocin. Braz. J. Food Technol. 2013, 16, 226–235. [Google Scholar] [CrossRef]
- Özel, B.; Sim¸sek, Ö.; Akçelik, M.; Saris, P.E.J. Innovative Approaches to Nisin Production. Appl. Microbiol. Biotechnol. 2018, 102, 6299–6307. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, L.; Dong, P.; Liang, R.; Mao, Y.; Yang, X.; Zhang, Y.; Luo, X. Acid Tolerance Response of Listeria monocytogenes in various external pHs with different concentrations of lactic acid. Foodborne Pathog Dis. 2020, 17, 253–261. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Bahrami, A.; Delshadi, R.; Jafari, S.M.; Williams, L. Nanoencapsulated nisin: An engineered natural antimicrobial system for the food industry. Trends Food Sci. Technol. 2019, 94, 20. [Google Scholar]
- Khaleque, M.A.; Keya, C.A.; Hasan, K.N.; Hoque, M.M.; Inatsu, Y.; Bari, M.L. Use of cloves and cinnamon essential oil to inactivate Listeria monocytogenes in ground beef at freezing and refrigeration temperatures. Food Sci. Technol. 2016, 74, 219–223. [Google Scholar] [CrossRef]
- Sharma, P.; Kaur, J.; Sharma, G.; Kashyap, P. Plant derived antimicrobial peptides: Mechanism of target, isolation techniques, sources and pharmaceutical applications. J. Food Biochem. 2022, 46, e14348. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chen, W.; Pelton, R.; Griffiths, M.W. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in Meat by Using Phages Immobilized on Modified Cellulose Membranes. Appl. Environ. Microbiol. 2011, 77, 6379–6387. [Google Scholar] [CrossRef]
- Radford, D.; Guild, B.; Strange, P.; Ahmed, R.; Lim, L.-T.; Balamurugan, S. Characterization of antimicrobial properties of Salmonella phage Felix O1 and Listeria phage A511 embedded in xanthan coatings on Poly (lactic acid) films. Food Microbiol. 2017, 66, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Barton Behravesh, C.; Jones, T.F.; Vugia, D.J.; Long, C.; Marcus, R.; Smith, K. Deaths associated with bacterial pathogens transmitted commonly through food: Foodborne diseases active surveillance network (FoodNet), 1996–2005. J. Infect. Dis. 2011, 204, 263–267. [Google Scholar] [CrossRef]
- Goutard, F.L.; Bordier, M.; Calba, C.; Erlacher-Vindel, E.; Góchez, D.; de Balogh, K.; Benigno, C.; Kalpravidh, W.; Roger, F.; Vong, S.J.B. Antimicrobial policy interventions in food animal production in South East Asia. BMJ 2017, 358, j3544. [Google Scholar] [CrossRef]
- Tadesse, T.; Zenebe, T.; Kebede, T.; Gizaw, O. Prevalence and antimicrobial susceptibility profile of Listeria monocytogenes in humans, animals, and foods of animal origin in Ethiopia: A systematic review and meta-analysis. Cogent Food Agric. 2024, 10, 2306018. [Google Scholar] [CrossRef]

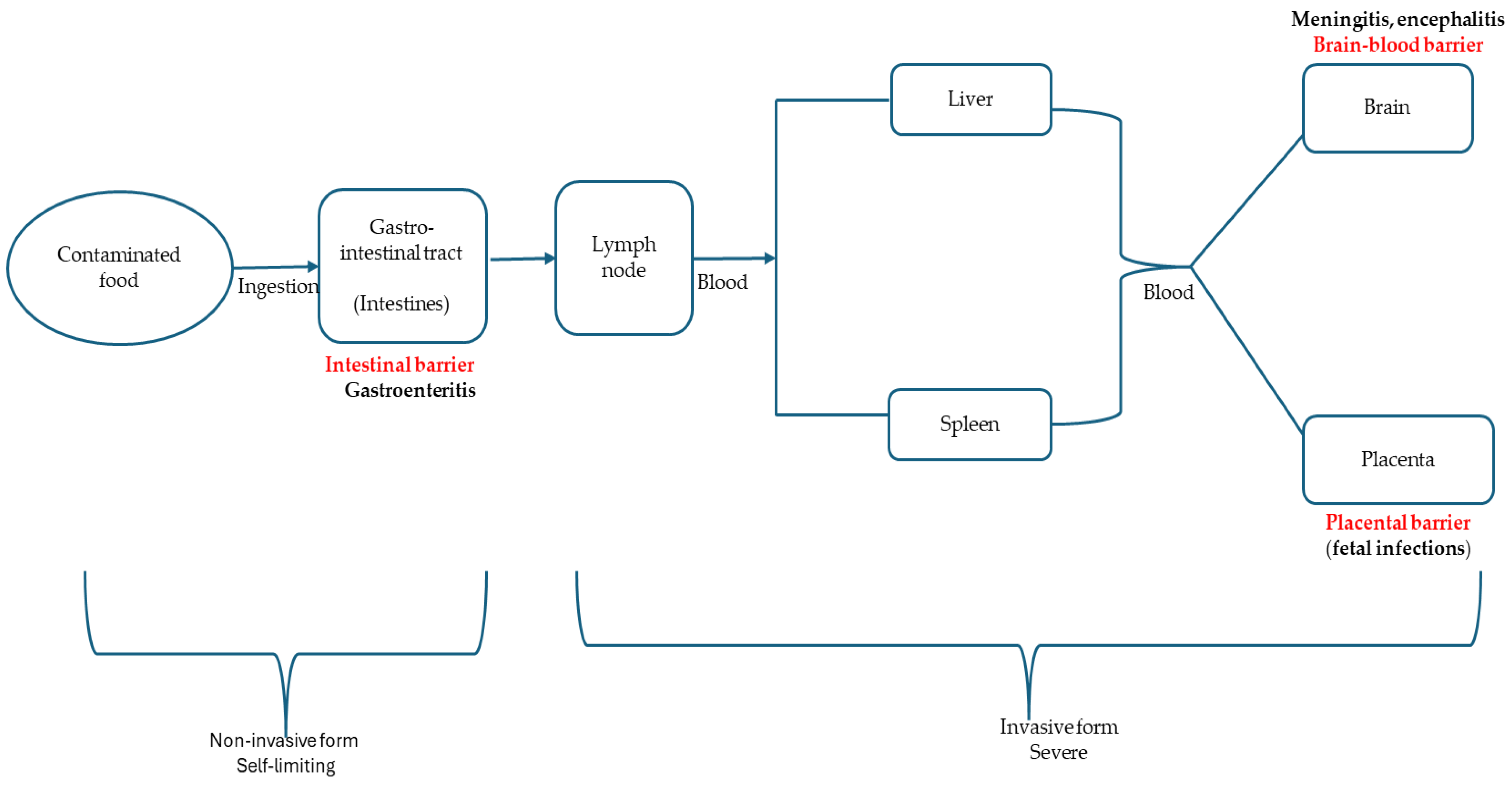
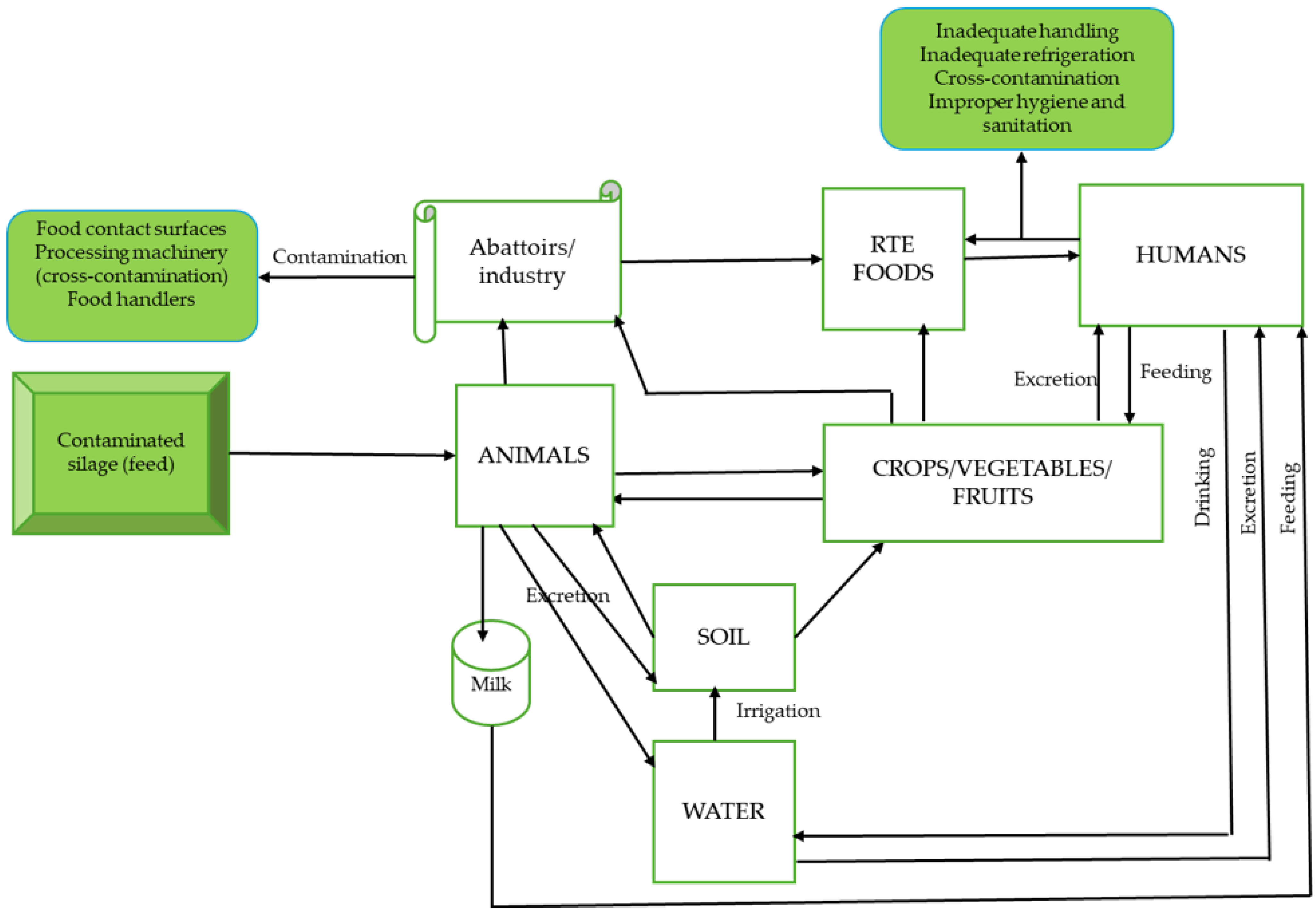
| Lineages | Serotypes | Clonal Complexes | Sources | Listeria Pathogenicity Islands |
|---|---|---|---|---|
| I | 1/2b,3b,4b,4e | CC1, CC2, CC4, CC6 (hypervirulent) | Clinical isolates of humans | LIPI-1, LIPI-3, LIPI-4 |
| II | 1/2a, 1/2c, 3a, 3c | CC7, CC9, CC121 (hypovirulent) | Clinical and food, but more in food | LIPI-1 |
| III | 4b, 1/2a, 4a, 4c | Rarely isolated | Predominantly animal sources | LIPI-1, LIPI-4 |
| IV | 4a, 4c | LIPI-1, LIPI-4 |
| Hosts | Infectious Dose | Symptoms | Disease Outcomes | Examples | References |
|---|---|---|---|---|---|
| Healthy population (immunocompetent) | 107–109 colony-forming units | Nausea, headache, fever, diarrhoea, vomiting, muscle pain, abdominal pain | Febrile gastroenteritis | Healthy individuals. | Pouillot et al. [233]; Bagatella et al. [227]. |
| Susceptible/vulnerable/high-risk population | 104–106 colony-forming units | Encephalitis, meningitis, endocarditis, septicaemia | Invasive listeriosis | Elderly, neonates. HIV/AIDS individuals, patients undergoing chemotherapy. | Heiman et al. [234]; Pérez-Trallero et al. [235]; Lachmann et al. [236]. |
| L. monocytogenes Phages | Descriptions | References |
|---|---|---|
| ListShieldTM (formerly LMP-102) | Commercially available phage product. A cocktail of six (6) distinct lytic phages as follows: LIST-36 (ATCC#PTA-5376) LMSP-25 (ATCC#PTA-8353) LMTA-34 (ATCC#PTA-8354) LMTA-57(ATCC#PTA-8355) LMTA-94 (ATCC#PTA-8356) LMTA-148 (ATCC#PTA-8357) | Sadekuzzaman et al. [377]. |
| PhageGuard ListexTM (formerly ListexTM P100) | Commercially available product. Only one broad host range, phage P100 | Moye et al. [378]. |
| P70 | Broad host range infecting Listeria species serovars: 1/2a, 1/2b,1/2c,4a,4c,4d,4e5,6a, and 6b with similar efficiencies. It has a distinct virion morphology, genomic size, and structure that is unrelated to any Listeria phage so far identified | Schmuki et al. [379]. |
| LP-018 | Assigned to Homburgvirus genus with the distinct trait to infect phage-resistant mutants | Vongkapamin et al. [380]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manyi-Loh, C.E.; Lues, R. Listeria monocytogenes and Listeriosis: The Global Enigma. Foods 2025, 14, 1266. https://doi.org/10.3390/foods14071266
Manyi-Loh CE, Lues R. Listeria monocytogenes and Listeriosis: The Global Enigma. Foods. 2025; 14(7):1266. https://doi.org/10.3390/foods14071266
Chicago/Turabian StyleManyi-Loh, Christy E., and Ryk Lues. 2025. "Listeria monocytogenes and Listeriosis: The Global Enigma" Foods 14, no. 7: 1266. https://doi.org/10.3390/foods14071266
APA StyleManyi-Loh, C. E., & Lues, R. (2025). Listeria monocytogenes and Listeriosis: The Global Enigma. Foods, 14(7), 1266. https://doi.org/10.3390/foods14071266






