Swordfish or Shark Slice? A Rapid Response by COIBar–RFLP
Abstract
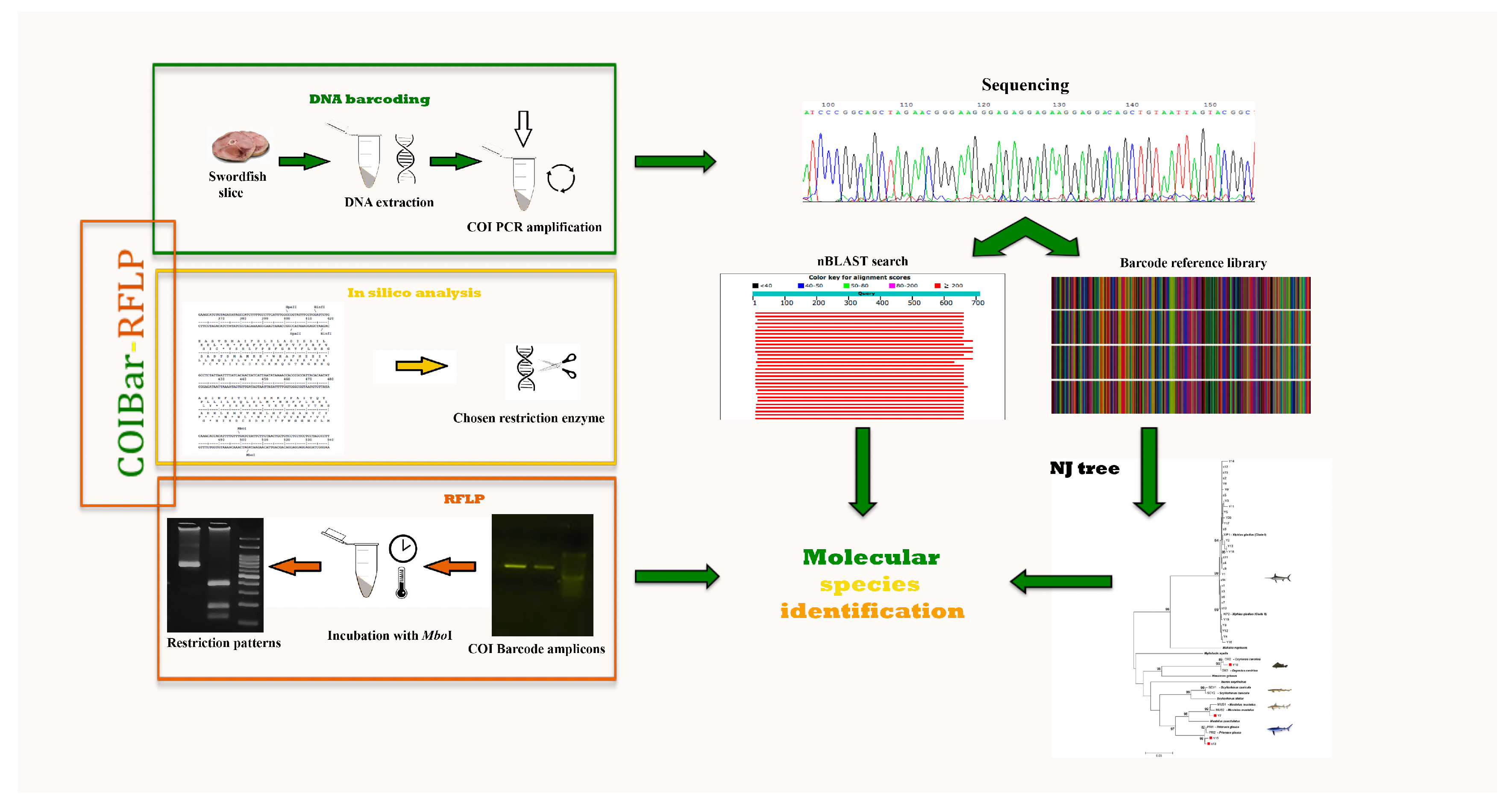
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Barcoding
2.3. COIBar–RFLP
3. Results
3.1. DNA Barcoding
3.2. COIBar–RFLP
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- SCRS Report. Report of the Standing Committee on Research and Statistics (SCRS) International Commission for the Conservation of Atlantic Tunas-Biennial Period 2016–2017; SCRS: Madrid, Spain, 2017. [Google Scholar]
- ISMEA Osservatorio consumi extra-domestici. Analisi & Ricerche n. 3 2005. Available online: http://www.ismea.it (accessed on 25 June 2019).
- Cosmina, M.; Demartini, E.; Gaviglio, A.; Mauracher, C.; Prestamburgo, S.; Trevisan, G. Italian Consumers’ Attitudes Towards Small Palegic Fish. New Medit. 2012, 1, 52–57. [Google Scholar]
- Reilly, A. Overview of Food Fraud in the Fisheries Sector; FAO In Fisheries and Aquaculture Circular; FIAM: Rome, Italy, 2018. [Google Scholar]
- European Union. Regulation (EC) No. 178/2002 of the European parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European food safety authority and laying down procedures in matters of food safety. Off. J. Eur. Union 2002, 31, 1–24. [Google Scholar]
- European Union. Regulation (EC) No. 1169/2011 of the European parliament and of the council of 25 October 2011 on the provision of food information to consumers. Off. J. Eur. Union 2011, 304, 18–63. [Google Scholar]
- European Union. Regulation (EC) No. 1379/2013 of the European parliament and of the council of 11 December 2013 on the common organisation of the markets in fishery and aquaculture products. Off. J. Eur. Union 2013, 354, 1–21. [Google Scholar]
- Altalex IT. Art 440. Codice Penale Italiano, Libro II Titolo VI Capo II. 2017. Available online: http://www.altalex.com/documents/news/2014/06/03/dei-delitti-contro-l-incolumita-pubblica (accessed on 10 April 2019).
- Decreto Legislativo 27 Gennaio 1992, n. 109 Attuazione delle Direttive n. 89/395/CEE e n. 89/396/CEE Concernenti L’etichettatura, la Presentazione e la Pubblicità Dei prodotti Alimentari. Available online: http://www.comune.jesi.an.it/MV/leggi/dlvo109-92.htm (accessed on 27 January 1992).
- Decreto Legislativo 6 settembre 2005, n. 206-codice del consumo, a norma dell’art. 7 della legge 29 luglio 2003, n. 229. Available online: https://www.camera.it/parlam/leggi/deleghe/testi/05206dl.htm (accessed on 6 September 2005).
- Benard-Capelle, J.; Guillonneau, V.; Nouvian, C.; Fournier, N.; Le Loët, K.; Dettai, A. Fish mislabeling in France: Substitution rates and retail types. PeerJ 2015, 2, e714. [Google Scholar] [CrossRef]
- Christiansen, H.; Dettai, A.; Heindler, F.M.; Collins, M.A.; Duhamel, G.; Hautecoeur, M.; Steinke, D.; Volckaert, A.M.; Van de Putte, A.P. Diversity of mesopelagic fishes in the southern ocean—A phylogeographic perspective using DNA barcoding. Front. Ecol. Evol. 2018, 6, 120. [Google Scholar] [CrossRef]
- Do, T.D.; Choi, T.J.; Kim, J.; An, H.E.; Park, Y.J.; Karagozlu, M.Z.; Kim, C.B. Assessment of marine fish mislabelling in South Korea’s markets by DNA barcoding. Food Control 2019, 100, 53–57. [Google Scholar] [CrossRef]
- Garcia-Vazquez, E.; Perez, J.; Martinez, J.L.; Pardiñas, A.F.; Lopez, B.; Karaiskou, N.; Triantafyllidis, A. High level of mislabelling in Spanish and Greek hake markets suggests the fraudulent introduction of African species. J. Agric. Food Chem. 2011, 59, 475–480. [Google Scholar] [CrossRef]
- Von Der Heyden, S.; Barendse, J.; Seebregts, A.J.; Matthee, C.A. Misleading the masses: Detection of mislabelled and substituted frozen fish products in south Africa. ICES J. Mar. Sci. 2010, 67, 176–185. [Google Scholar] [CrossRef]
- Quinteiro, J.; Vidal, R.; Izquierdo, M.; Sotelo, C.G.; Chapela, M.J.; Pérez-Martín, R.I.; Rehbein, H.; Hold, G.L.; Russell, V.J.; Pryde, S.E.; et al. Identification of hake species (Merluccius genus) using sequencing and PCR-RFLP analysis of mitochondrial DNA control region sequences. J. Agric. Food Chem. 2001, 49, 5108–5114. [Google Scholar] [CrossRef]
- Kumar, G.; Kocour, M.; Kunal, S.P. Mitochondrial DNA variation and phylogenetic relationships among five tune species based on sequencing of D-loop region. Mitochondr. DNA Part A 2016, 27, 1976–1980. [Google Scholar]
- Rocco, L.; Ferrito, V.; Costagliola, D.; Marsilio, A.; Pappalardo, A.M.; Stingo, V.; Tigano, C. Genetic divergence among and within four Italian populations of Aphanius fasciatus (Teleostei, Cyprinodontiformes). Ital. J. Zool. 2007, 74, 371–379. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Ferrito, V.; Messina, A.; Patarnello, T.; De Pinto, V.; Guarino, F.; Tigano, C. Genetic structure of the killifish Aphanius fasciatus Nardo 1827 (Teleostei, Cyprinodontidae), results of mitochondrial DNA analysis. J. Fish Biol. 2008, 72, 1154–1173. [Google Scholar] [CrossRef]
- Ferrito, V.; Pappalardo, A.M.; Canapa, A.; Barucca, M.; Doadrio, I.; Olmo, E.; Tigano, C. Mitochondrial phylogeography of the killifish Aphanius fasciatus (Teleostei, Cyprinodontidae) reveals highly divergent Mediterranean populations. Mar. Biol. 2013, 160, 3193–3208. [Google Scholar] [CrossRef]
- Cuttitta, A.; Patti, B.; Maggio, T.; Quinci, E.M.; Pappalardo, A.M.; Ferrito, V.; De Pinto, V.; Torri, M.; Falco, F.; Nicosia, A.; et al. Larval population structure of Engraulis encrasicolus in the strait of Sicily as revealed by morphometric and genetic analyses. Fish Ocean 2015, 24, 135–149. [Google Scholar] [CrossRef]
- Pedrosa-Gerasmio, I.R.; Agmata, A.B.; Santos, M.D. Genetic diversity, population genetic structure, and demographic history of Auxis thazard (Perciformes), Selar crumenophthalmus (Perciformes), Rastrelliger kanagurta (Perciformes) and Sardinella lemuru (Clupeiformes) in Sulu-Celebes sea inferred by mitochondrial DNA sequences. Fish Res. 2015, 162, 64–74. [Google Scholar]
- Machado, V.N.; Willis, S.C.; Teixeira, A.S.; Hrbek, T.; Farias, I.P. Population genetic structure of the Amazonian black flannelmouth characin (Characiformes, Prochilodontidae: Prochilodus nigricans Spix & Agassiz, 1829): Contemporary and historical gene flow of a migratory and abundant fishery species. Environ. Biol. Fishes 2017, 100, 1–16. [Google Scholar]
- Duong, T.; Uy, S.; Chheng, P.; So, N.; Thi Tran, T.; Nguyen, N.T.; Pomeroy, R.; Egna, H. Genetic diversity and structure of striped snakehead (Channa striata) in the lower Mekong basin: Implications for aquaculture and fisheries management. Fish Res. 2019, 218, 166–173. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergence, among closely related species. Proc. R. Soc. Lond. B 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Hajibabaei, M.; Smith, M.A.; Janzen, D.H.; Rodriguez, J.J.; Whitfield, J.B.; Hebert, P.D.N. A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes 2006, 6, 959–964. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA barcoding: How it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Holmes, B.H.; White, W.T.; Last, P.R. DNA barcoding Australasian chondrichthyans: Results and potential uses in conservation. Mar. Fresh Res. 2008, 59, 57–71. [Google Scholar] [CrossRef]
- Vitale, D.G.M.; Viscuso, R.; D’Urso, V.; Gibilras, S.; Sardella, A.; Marletta, A.; Pappalardo, A.M. Morphostructural analysis of the male reproductive system and DNA barcoding in Balclutha brevis Lindberg 1954 (Homoptera, Cicadellidae). Micron 2015, 79, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Lombardo, D.; La Porta, N.; Pappalardo, A.M.; Ferrito, V.; Lombardo, F.; Saccone, S. Rapid molecular identification of necrophagous Diptera by means of variable-length of intron sequences in the wingless gene. J. Forensic Legal Med. 2018, 56, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Conti, E.; Mulder, C.; Pappalardo, A.M.; Ferrito, V.; Costa, G. How soil granulometry, temperature and water predict genetic differentiation in namibian Ariadna spiders and explain their behavior. Ecol. Evol. 2019, 9, 4382–4391. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Steinke, D.; Zemlak, T.S.; Hebert, P.D.N. Barcoding Nemo: DNA-based identifications for the ornamental fish trade. PLoS ONE 2009, 4, e6300. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Guarino, F.; Reina, S.; Messina, A.; De Pinto, V. Swordfish COI-DNA barcoding as a suitable tool for species and stock identification. FEBS J. 2011, 278, 440. [Google Scholar]
- Pappalardo, A.M.; Guarino, F.; Reina, S.; Messina, A.; De Pinto, V. Geographically widespread swordfish barcode stock identification: A case study of its application. PLoS ONE 2011, 6, e25516. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, A.M.; Ferrito, V. DNA barcoding species identification unveils mislabeling of processed flatfish products in southern Italy markets. Fish Res. 2015, 164, 153–158. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Cuttitta, A.; Sardella, A.; Musco, M.; Maggio, T.; Patti, B.; Mazzola, S.; Ferrito, V. DNA barcoding and COI sequence variation in Mediterranean lanternfishes larvae. Hydrobiologia 2015, 745, 155–167. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Federico, C.; Sabella, G.; Saccone, S.; Ferrito, V. A COI nonsynonymous mutation as diagnostic tool for intraspecific discrimination in the European anchovy Engraulis encrasicolus (Linnaeus). PLoS ONE 2015, 10, e0143297. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcode of life data system. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Dawnay, N.; Ogden, R.; McEwing, R.; Carvalho, G.R.; Thorpe, R.S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci. Int. 2007, 173, 1–6. [Google Scholar] [CrossRef]
- Espiñeira, M.; González-Lavín, N.; Vieites, J.M.; Santaclara, F.J. Development of a method for the genetic identification of flatfish species on the basis of mitochondrial DNA sequences. J. Agric. Food Chem. 2008, 56, 8954–8961. [Google Scholar] [CrossRef]
- Cawthorn, D.M.; Steinman, H.A.; Corli Witthuhn, R. Establishment of a mitochondrial DNA sequence database for the identification of fish species commercially available in south Africa. Mol. Ecol. Res. 2011, 11, 979–991. [Google Scholar] [CrossRef]
- Armani, A.; Castigliego, L.; Guidi, A. Fish frauds: The DNA challenge. CAB reviews 071. Anim. Sci. Rev. 2012, 7, 227–238. [Google Scholar]
- Armani, A.; Guardone, L.; La Castellana, R.; Gianfaldoni, D.; Guidi, A.; Castigliego, L. DNA barcoding reveals commercial and health issues in ethnic seafood sold on the Italian market. Food Control 2015, 55, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Khaksar, R.; Carlson, T.; Schaffner, D.W.; Ghorashi, M.; Best, D.; Jandhyala, S.; Traverso, J.; Amini, S. Unmasking seafood mislabeling in U.S. markets: DNA barcoding as a unique technology for food authentication and quality control. Food Control 2015, 56, 71–76. [Google Scholar] [CrossRef]
- Ferrito, V.; Bertolino, V.; Pappalardo, A.M. White fish authentication by COIBar-RFLP: Toward a common strategy for the rapid identification of species in convenience seafood. Food Control 2016, 70, 130–137. [Google Scholar] [CrossRef]
- Nagalakshmi, K.; Annam, P.K.; Venkateshwarlu, G.; Pathakota, G.B.; Lakra, W.S. Mislabelling in Indian seafood: An investigation using DNA barcoding. Food Control 2016, 59, 196–200. [Google Scholar] [CrossRef]
- Ferrito, V.; Pappalardo, A.M. Seafood species identification by DNA barcoding, a molecular tool for food traceability. Biodivers. J. 2017, 8, 65–72. [Google Scholar]
- Pappalardo, A.M.; Copat, C.; Ferrito, V.; Grasso, A.; Ferrante, M. Heavy metal content and molecular species identification in canned tuna: Insights into human food safety. Mol. Med. Rep. 2017, 15, 3430–3437. [Google Scholar] [CrossRef] [Green Version]
- Pappalardo, A.M.; Federico, C.; Saccone, S.; Ferrito, V. Differential flatfish species detection by COIBar-RFLP in processed seafood products. Eur. Food Res. Technol. 2018, 244, 2191–2201. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Petraccioli, A.; Capriglione, T.; Ferrito, V. From fish eggs to fish name: Caviar species discrimination by COIBar-RFLP, an efficient molecular approach to detect fraud in the caviar trade. Molecules 2019, 24, 2468. [Google Scholar] [CrossRef]
- Cocolin, L.; D’Agaro, E.; Manzano, M.; Lanari, D.; Comi, G. Rapid PCR-RFLP method for the identification of marine fish fillets (seabass, seabream, umbrine, and dentex). J. Food Sci. 2000, 65, 1315–1317. [Google Scholar] [CrossRef]
- Hold, G.L.; Russell, V.J.; Pryde, S.E.; Rehbein, H.; Quinteiro, J.; Rey-Mendez, M.; Sotelo, C.G.; Pérez-Martin, R.I.; Santos, A.T.; Rosa, C. Validation of a PCR-RFLP based method for the identification of salmon species in food products. Eur. Food Res. Technol. 2001, 212, 385–389. [Google Scholar] [CrossRef]
- Hsieh, H.S.; Chai, T.; Hwang, D.F. Using the PCR-RFLP method to identify the species of different processed products of billfish meats. Food Control 2007, 18, 369–374. [Google Scholar] [CrossRef]
- Pappalardo, A.M.; Ferrito, V. A COIBar-RFLP strategy for the rapid detection of Engraulis encrasicolus in processed anchovy products. Food Control 2015, 57, 385–392. [Google Scholar] [CrossRef]
- Dent, F.; Clarke, S. State of the Global Market for Shark Products; Food and Agriculture Organization FAO: Rome, Italy, 2015. [Google Scholar]
- Blanco, M.; Pérez-Martín, R.I.; Sotelo, C.G. Identification of shark species in seafood products by forensically informative nucleotide sequencing (FINS). J. Agric. Food Chem. 2008, 56, 9868–9874. [Google Scholar] [CrossRef] [PubMed]
- Barbuto, M.; Galimberti, A.; Ferri, E.; Labra, M.; Malandra, R.; Galli, P.; Casiraghi, M. DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of palombo (Mustelus spp.). Food Res. Int. 2010, 43, 376–381. [Google Scholar] [CrossRef]
- Filonzi, L.; Chiesa, S.; Vaghi, M.; Nonnis Marzano, F. Molecular barcoding reveals mislabelling of commercial fish products in Italy. Food Res. Int. 2010, 43, 1383–1388. [Google Scholar] [CrossRef]
- Herrero, B.; Lago, F.C.; Vieites, J.M.; Espiñeira, M. Authentication of swordfish (Xiphias gladius) by RT-PCR and FINS methodologies. Eur. Food Res. Technol. 2011, 233, 195–202. [Google Scholar] [CrossRef]
- Tortonese, E. Fauna d’Italia, ‘Osteichtyes’: Pesci ossei. Calderini Bologna 1975, 11, 133–139. [Google Scholar]
- Whitehead, P.J.P.; Bauchot, M.L.; Hureau, J.C.; Nielsen, J.; Tortonese, E. Fishes of the North-Eastern Atlantic and the Mediterranean; UNESCO: Paris, France, 1986. [Google Scholar]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- GENECHRON. Available online: http://www.genechron.it/index.php/sanger-sequencing (accessed on 17 November 2018).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- TRANSEQ. Available online: http://www.ebi.ac.uk/Tools/st/emboss_transeq (accessed on 26 July 2019).
- GENBANK. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 12 September 2019).
- Zhang, D.X.; Hewitt, G.M. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol. Evol. 1996, 11, 247–251. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- REMAP EMBOSS. Available online: http://emboss.bioinformatics.nl (accessed on 26 July 2019).
- Alvarado-Bremer, J.R.; Baker, A.J.; Mejuto, J. Mitochondrial DNA control region sequences indicate extensive mixing of swordfish (Xiphias gladius) populations in the Atlantic ocean. Can. J. Fish Aquat. Sci. 1995, 52, 1720–1732. [Google Scholar] [CrossRef]
- Alvarado-Bremer, J.R.; Mejuto, J.; Greig, T.W.; Ely, B. Global population structure of the swordfish (Xiphias gladius L.) as revealed by analysis of the mitochondrial DNA control region. J. Exp. Mar. Biol. Ecol. 1996, 197, 295–310. [Google Scholar] [CrossRef]
- Bradman, H.; Grewe, P.; Appleton, B. Direct comparison of mitochondrial markers for the analysis of swordfish population structure. Fish Res. 2011, 109, 95–99. [Google Scholar] [CrossRef]
- Griffiths, A.M.; Sotelo, C.G.; Mendes, R.; Perez Martin, R.I.; Schröder, U.; Shorten, M.; Silva, H.A.; Verrez-Bagnis, V.; Mariani, S. Current methods for seafood authenticity testing in Europe: Is there a need for harmonisation? Food Control 2014, 45, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Verrez-Bagnis, V.; Sotelo, C.G.; Mendes, R.; Silva, H.; Kappel, K.; Schröder, U. Methods for seafood authenticity testing in Europe. In Bioactive Molecules in Food, Reference Series in Phytochemistry; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer: Berlin, Germany, 2018; pp. 1–55. [Google Scholar]
- EUR-Lex Council Regulation (EC) No. 1224/2009 of 20 November. Off. J. Eur. Union 2009, 343, 50.
- Kappel, K.; Schröder, U. Substitution of high-priced fish with low-priced species: Adulteration of common sole in German restaurants. Food Control 2016, 59, 478–486. [Google Scholar] [CrossRef]



| Code | DNA Sample Voucher | Declared Species | GenBank Access N° | Species Matched by BLAST | Matched Accession From BLAST | % Identity With 100% Coverage |
|---|---|---|---|---|---|---|
| XIP1 | DBGES18-125 | Xiphias gladius * | JN083390 | Xiphias gladius | JN083390 | 100 |
| XIP2 | DBGES18-129 | Xiphias gladius * | MN447670 | Xiphias gladius | JN049558 | 99.71 |
| MUS1 | DBGES18-133 | Mustelus mustelus * | MN447688 | Mustelus mustelus | JN641215 | 99.39 |
| MUS2 | DBGES18-134 | Mustelus mustelus * | MN447689 | Mustelus mustelus | JN641214 | 99.24 |
| OXY1 | DBGES18-111 | Oxynotus centrina * | MN447691 | Oxynotus centrina | JF834320 | 98.96 |
| OXY2 | DBGES18-130 | Oxynotus centrina * | MN447692 | Oxynotus centrina | JF834320 | 98.66 |
| PRI1 | DBGES18-121 | Prionace glauca * | MN447694 | Prionace glauca | KJ146044 | 99.85 |
| PRI2 | DBGES18-138 | Prionace glauca * | MN447695 | Prionace glauca | MH719984 | 99.83 |
| SCY1 | DBGES18-144 | Scyliorhinus canicula * | MN457949 | Scyliorhinus canicula | KJ205311 | 99.70 |
| SCY2 | DBGES18-146 | Scyliorhinus canicula * | MN457950 | Scyliorhinus canicula | KJ205311 | 99.85 |
| X1 | DBGES10-045 | Xiphias gladius | JN083389 | Xiphias gladius | JN083389 | 100 |
| X2 | DBGES10-048 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X3 | DBGES10-049 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X4 | DBGES10-054 | Xiphias gladius | JN083397 | Xiphias gladius | JN083397 | 100 |
| X5 | DBGES10-055 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X6 | DBGES10-058 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X7 | DBGES10-059 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X8 | DBGES10-060 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X9 | DBGES10-068 | Xiphias gladius | JN083393 | Xiphias gladius | JN083393 | 100 |
| X10 | DBGES10-074 | Xiphias gladius | JN049559 | Xiphias gladius | JN049559 | 100 |
| X11 | DBGES10-077 | Xiphias gladius | JN083393 | Xiphias gladius | JN083393 | 100 |
| X12 | DBGES10-080 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| X13 | DBGES10-083 | Xiphias gladius | MN447696 | Prionace glauca | MH719984 | 99.83 |
| X14 | DBGES10-089 | Xiphias gladius | JN083386 | Xiphias gladius | JN083386 | 100 |
| X15 | DBGES10-095 | Xiphias gladius | JN083387 | Xiphias gladius | JN083387 | 100 |
| Y1 | DBGES18-132 | Xiphias gladius | MN447671 | Xiphias gladius | JN049558 | 99.71 |
| Y2 | DBGES18-135 | Xiphias gladius | MN447672 | Xiphias gladius | JN083390 | 99.71 |
| Y3 | DBGES18-136 | Xiphias gladius | MN447673 | Xiphias gladius | JN083387 | 99.71 |
| Y4 | DBGES18-137 | Xiphias gladius | MN447674 | Xiphias gladius | JN049558 | 99.71 |
| Y5 | DBGES18-139 | Xiphias gladius | MN447675 | Xiphias gladius | JN083387 | 99.85 |
| Y6 | DBGES18-141 | Xiphias gladius | MN447676 | Xiphias gladius | JN083387 | 99.71 |
| Y7 | DBGES18-143 | Xiphias gladius | MN447690 | Mustelus mustelus | JN641214 | 99.39 |
| Y8 | DBGES18-144 | Xiphias gladius | MN447677 | Xiphias gladius | JN049558 | 99.85 |
| Y9 | DBGES18-151 | Xiphias gladius | MN447678 | Xiphias gladius | JN083387 | 99.71 |
| Y10 | DBGES18-152 | Xiphias gladius | MN447693 | Oxynotus centrina | JF834320 | 98.07 |
| Y11 | DBGES18-153 | Xiphias gladius | MN447679 | Xiphias gladius | JN083387 | 99.12 |
| Y12 | DBGES18-154 | Xiphias gladius | MN447680 | Xiphias gladius | JN049558 | 99.85 |
| Y13 | DBGES18-161 | Xiphias gladius | MN447681 | Xiphias gladius | JN083390 | 99.41 |
| Y14 | DBGES18-162 | Xiphias gladius | MN447682 | Xiphias gladius | JN083387 | 98.97 |
| Y15 | DBGES18-172 | Xiphias gladius | MN447697 | Prionace glauca | MH194484 | 99.39 |
| Y16 | DBGES18-173 | Xiphias gladius | MN447683 | Xiphias gladius | JN049558 | 99.27 |
| Y17 | DBGES18-174 | Xiphias gladius | MN447684 | Xiphias gladius | JN083387 | 99.85 |
| Y18 | DBGES18-175 | Xiphias gladius | MN447685 | Xiphias gladius | JN083390 | 99.27 |
| Y19 | DBGES18-182 | Xiphias gladius | MN447686 | Xiphias gladius | JN049558 | 99.71 |
| Y20 | DBGES18-187 | Xiphias gladius | MN447687 | Xiphias gladius | JN083387 | 99.56 |
| Species | Sequence Number | Genbank Accession Number | Sequence Size (bases) | Restriction Fragment Size (base pair) |
|---|---|---|---|---|
| Xiphias gladius | 10 | JN083387 | 682 | ≈ 145 - 270 - 220 |
| JN083389 | 682 | ≈ 145 - 270 - 220 | ||
| JN049558 | 682 | ≈ 145 - 270 - 220 | ||
| JF952886 | 652 | ≈ 145 - 265 - 220 | ||
| HQ024928 | 652 | ≈ 145 - 265 - 220 | ||
| HQ024927 | 652 | ≈ 145 - 265 - 220 | ||
| KR086931 | 652 | ≈ 145 - 265 - 220 | ||
| GU324195 | 652 | ≈ 145 - 265 - 220 | ||
| DQ107625 | 655 | ≈ 145 - 265 - 200 | ||
| DQ107623 | 655 | ≈ 145 - 265 - 220 | ||
| Mustelus mustelus | 10 | JN641215 | 676 | ≈ 80 - 390 - 171 |
| JN641214 | 679 | ≈ 80 - 390 - 169 | ||
| JN641213 | 672 | ≈ 80 - 390 - 167 | ||
| JN641212 | 666 | ≈ 80 - 390 - 156 | ||
| JN641211 | 681 | ≈ 80 - 390 - 170 | ||
| KJ768265 | 652 | ≈ 80 - 390 - 142 | ||
| KJ768266 | 652 | ≈ 80 - 390 - 142 | ||
| JN641208 | 656 | ≈ 75 - 390 - 156 | ||
| JN641209 | 669 | ≈ 80 - 390 - 160 | ||
| JN641210 | 664 | ≈ 80 - 390 - 160 | ||
| Oxynotus centrina | 9 | KT307360 | 648 | ≈ 510 - 95 |
| KT307361 | 620 | ≈ 480 - 95 | ||
| KT307362 | 648 | ≈ 510 - 95 | ||
| KT307363 | 648 | ≈ 510 - 95 | ||
| KT307364 | 648 | ≈ 510 - 95 | ||
| JF834320 | 672 | ≈ 505 - 100 | ||
| KY176547 | 642 | ≈ 495 - 105 | ||
| GU805137 | 637 | ≈ 500 - 95 | ||
| GU805138 | 648 | ≈ 510 - 95 | ||
| Prionace glauca | 10 | JN312505 | 652 | ≈ 85 - 405 - 70 |
| JN312504 | 652 | ≈ 85 - 405 - 70 | ||
| JN312503 | 652 | ≈ 85 - 405 - 70 | ||
| KP193446 | 652 | ≈ 85 - 405 - 70 | ||
| KP193455 | 652 | ≈ 85 - 405 - 70 | ||
| KP193350 | 652 | ≈ 85 - 405 - 70 | ||
| KP193339 | 652 | ≈ 85 - 405 - 70 | ||
| KP193159 | 652 | ≈ 85 - 405 - 70 | ||
| KC015834 | 652 | ≈ 85 - 405 - 70 | ||
| KC015833 | 652 | ≈ 85 - 405 - 70 | ||
| Scyliorhinus canicula | 10 | JN641243 | 675 | ≈ 85 - 400 - 100 |
| JN641242 | 676 | ≈ 85 - 400 - 100 | ||
| JN641241 | 652 | ≈ 70 - 400 - 100 | ||
| JN641240 | 680 | ≈ 85 - 400 - 100 | ||
| JN641239 | 675 | ≈ 85 - 400 - 100 | ||
| JN641238 | 671 | ≈ 85 - 400 - 100 | ||
| JN641237 | 680 | ≈ 85 - 400 - 100 | ||
| JN641236 | 674 | ≈ 85 - 400 - 100 | ||
| JN641234 | 672 | ≈ 85 - 400 - 100 | ||
| JN641233 | 680 | ≈ 85 - 400 - 100 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrito, V.; Raffa, A.; Rossitto, L.; Federico, C.; Saccone, S.; Pappalardo, A.M. Swordfish or Shark Slice? A Rapid Response by COIBar–RFLP. Foods 2019, 8, 537. https://doi.org/10.3390/foods8110537
Ferrito V, Raffa A, Rossitto L, Federico C, Saccone S, Pappalardo AM. Swordfish or Shark Slice? A Rapid Response by COIBar–RFLP. Foods. 2019; 8(11):537. https://doi.org/10.3390/foods8110537
Chicago/Turabian StyleFerrito, Venera, Alessandra Raffa, Luana Rossitto, Concetta Federico, Salvatore Saccone, and Anna Maria Pappalardo. 2019. "Swordfish or Shark Slice? A Rapid Response by COIBar–RFLP" Foods 8, no. 11: 537. https://doi.org/10.3390/foods8110537
APA StyleFerrito, V., Raffa, A., Rossitto, L., Federico, C., Saccone, S., & Pappalardo, A. M. (2019). Swordfish or Shark Slice? A Rapid Response by COIBar–RFLP. Foods, 8(11), 537. https://doi.org/10.3390/foods8110537








