Microbial Ecology of Greek Wheat Sourdoughs, Identified by a Culture-Dependent and a Culture-Independent Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Physicochemical Characterization
2.3. Microbiological Analyses
2.4. Culture-Dependent Assessment of the Sourdough Microecosystem
2.4.1. Classical Identification
2.4.2. Molecular Identification
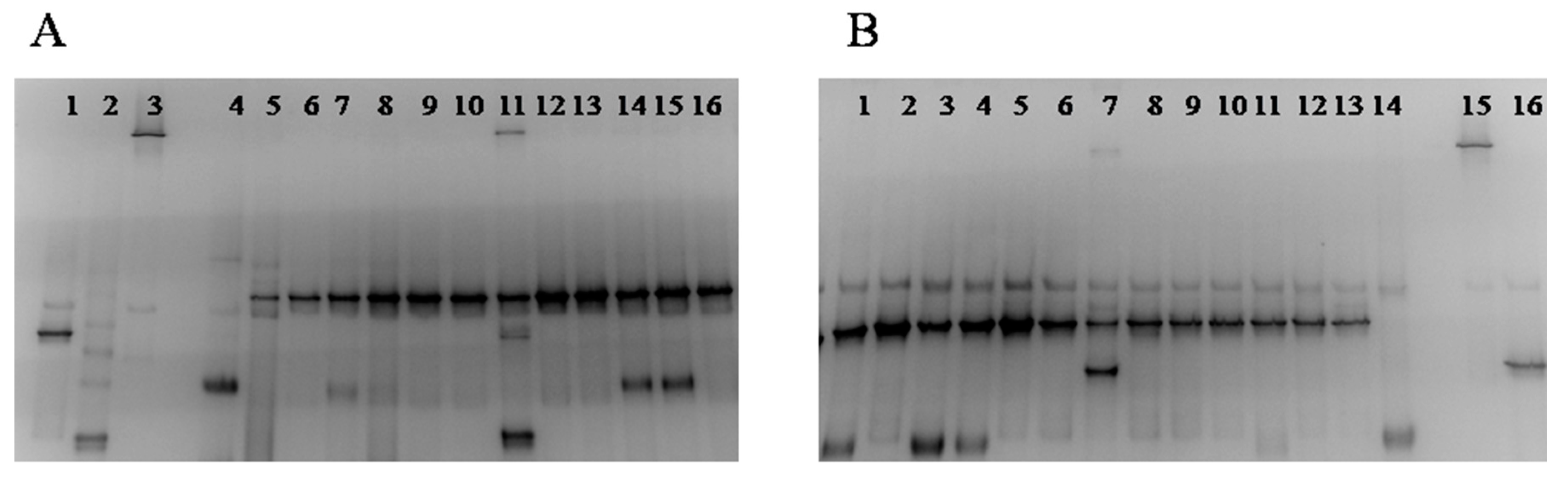
2.5. Culture-Independent Assessment of the Sourdough Microecosystem (PCR-DGGE)
2.6. Statistical Analysis
3. Results
3.1. Physicochemical and Microbiological Characterization
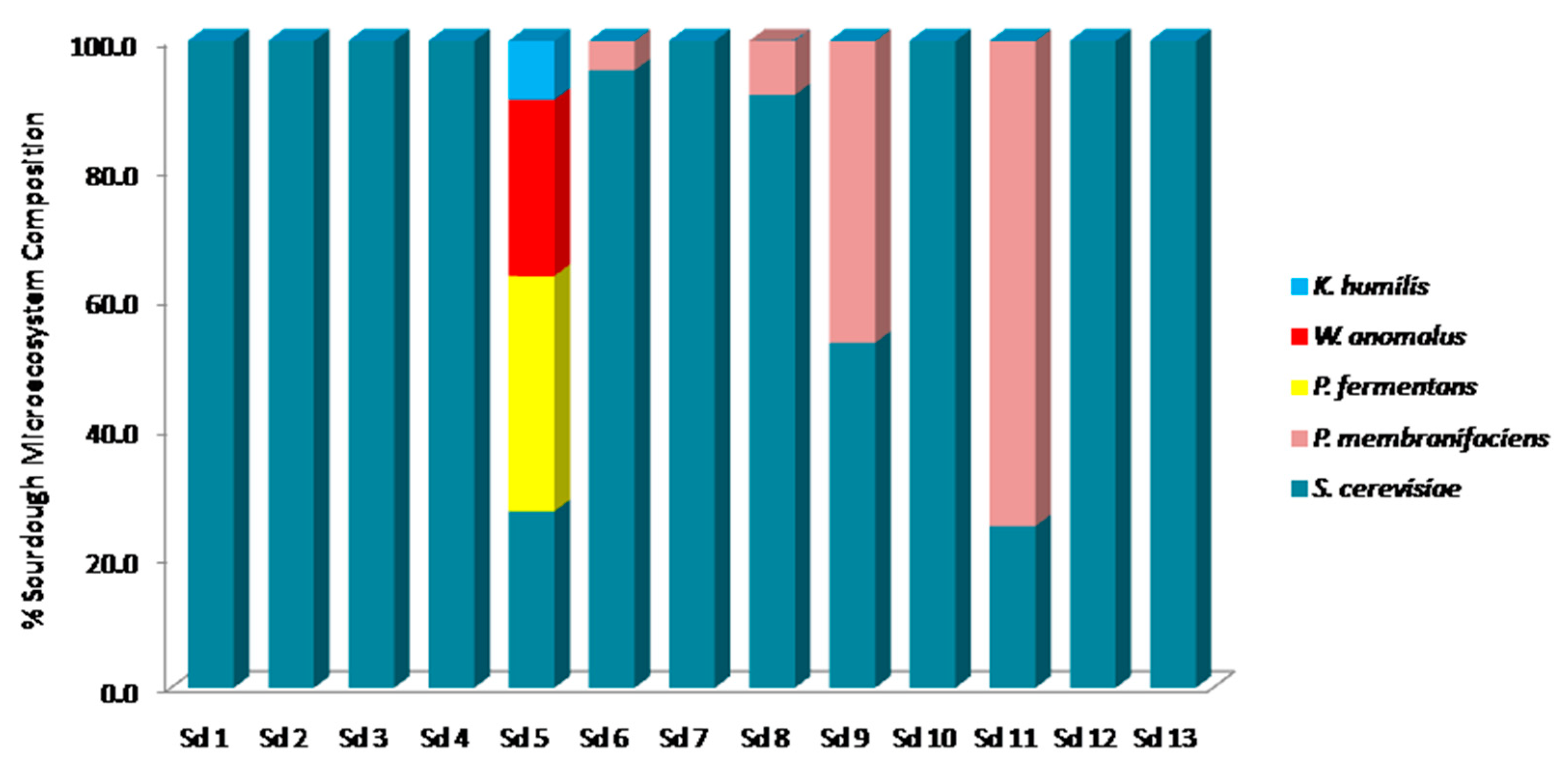
3.2. Culture-Dependent Assessment of Microbiota
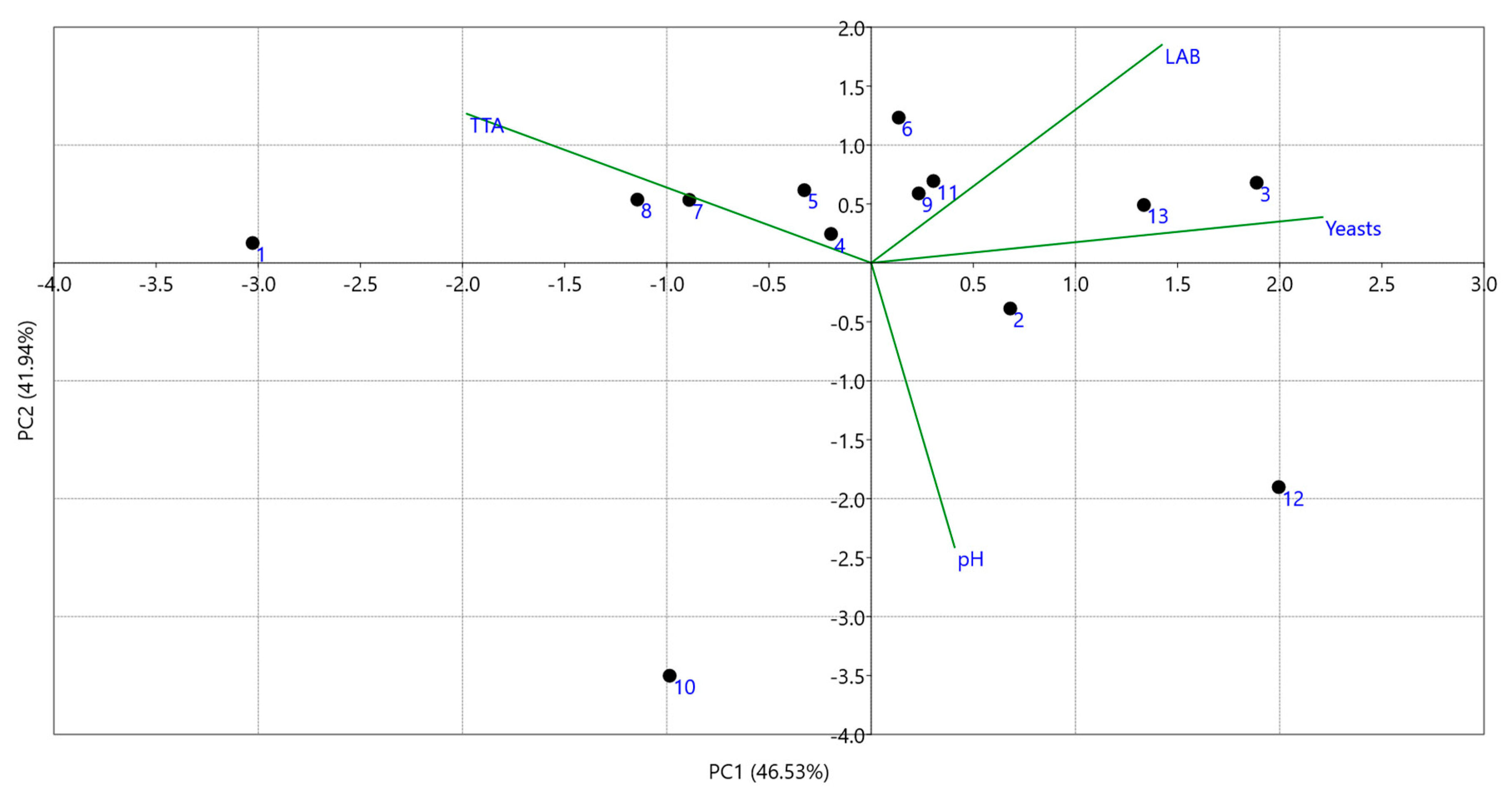
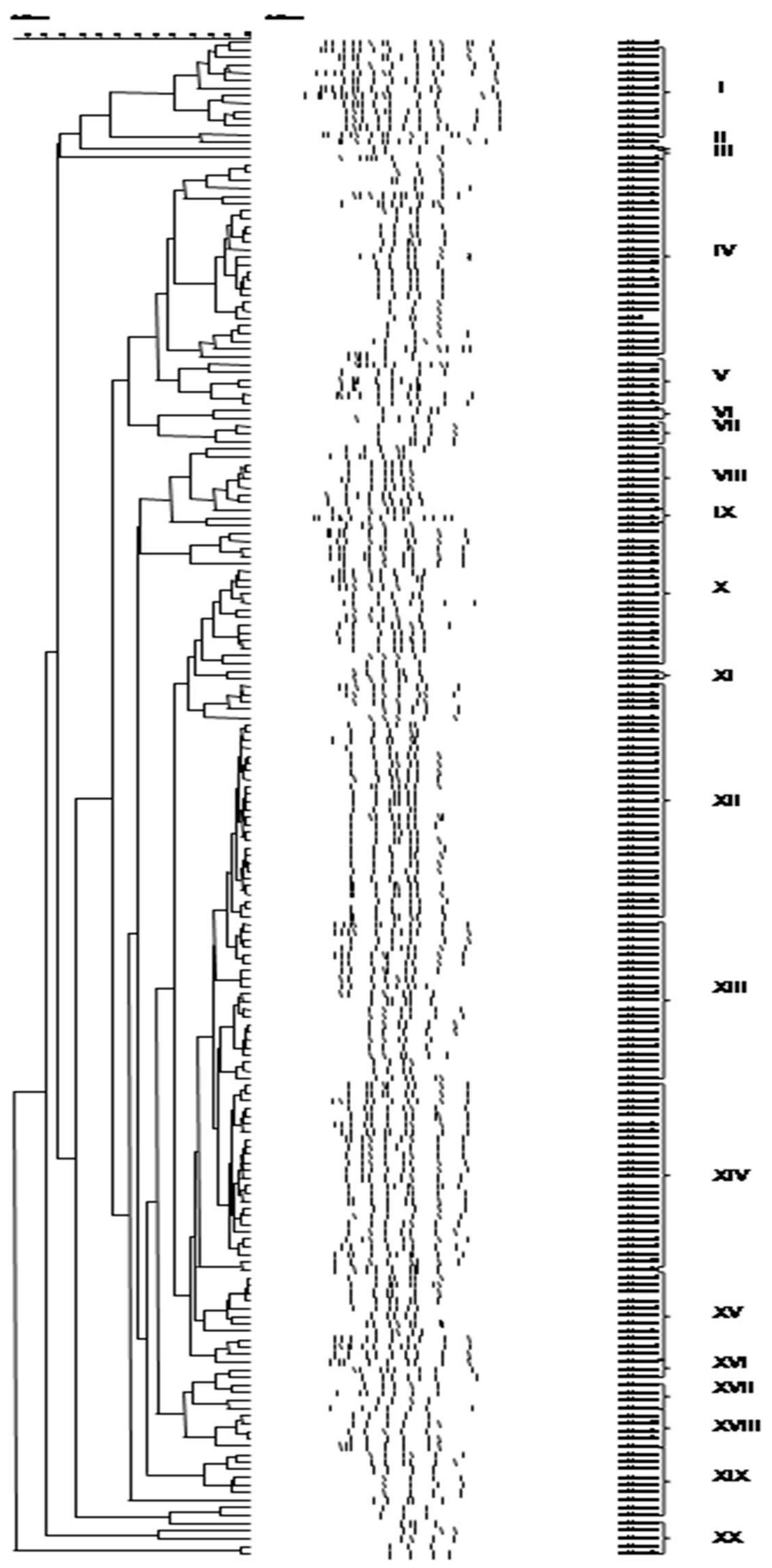
3.3. Culture-Independent Assessment of Microbiota (PCR-DGGE)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gobbetti, M.; Minervini, F.; Pontonio, E.; Di Cagno, R.; De Angelis, M. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. Int. J. Food Microbiol. 2016, 239, 3–18. [Google Scholar] [CrossRef]
- De Angelis, M.; Minervini, F.; Siragusa, S.; Rizzello, C.G.; Gobbetti, M. Wholemeal wheat flours drive the microbiome and functional features of wheat sourdoughs. Int. J. Food Microbiol. 2019, 302, 35–46. [Google Scholar] [CrossRef]
- De Vuyst, L.; Van Kerrebroek, S.; Harth, H.; Huys, G.; Daniel, H.-M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Drosinos, E.H. The sourdough micro-ecosystem: An update. In Fermented Foods, Part II: Technological Interventions; Ray, R.C., Montet, D., Eds.; CRC Science: Cambridge, MA, USA, 2017; pp. 263–283. [Google Scholar]
- Van Kerrebroeck, S.; Maes, D.; De Vuyst, L. Sourdoughs as a function of their species diversity and process conditions, a meta-analysis. Trends Food Sci. Technol. 2017, 68, 152–159. [Google Scholar] [CrossRef]
- Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic acid bacteria isolation from spontaneous sourdough and their characterization including antimicrobial and antifungal properties evaluation. Microorganisms 2020, 8, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boreczek, J.; Litwinek, D.; Żylińska-Urban, J.; Izak, D.; Buksa, K.; Gawor, J.; Gromadka, R.; Bardowski, J.K.; Kowalczyk1, M. Bacterial community dynamics in spontaneous sourdoughs made from wheat, spelt, and rye wholemeal flour. Microbiol. Open 2020, 9, 1009. [Google Scholar] [CrossRef] [PubMed]
- Comasio, A.; Verce, M.; Van Kerrebroeck, S.; De Vuyst, L. Diverse microbial composition of sourdoughs from different origins. Front. Microbiol. 2020, 11, 1212. [Google Scholar] [CrossRef] [PubMed]
- Bockwoldt, J.A.; Stahl, L.; Ehrmann, M.A.; Vogel, R.F.; Jakob, F. Persistence and β-glucan formation of beer-spoiling lactic acid bacteria in wheat and rye sourdoughs. Food Microbiol. 2020, 91, 103539. [Google Scholar] [CrossRef]
- De Vuyst, L.; van Kerrebroeck, S.; Leroy, F. Microbial ecology and process technology of sourdough fermentation. Adv. Appl. Microbiol. 2017, 100, 49–160. [Google Scholar]
- Fraberger, V.; Unger, C.; Kummer, C.; Domig, K.J. Insights into microbial diversity of traditional Austrian sourdough. LWT 2020, 127, 109358. [Google Scholar] [CrossRef]
- Reale, A.; Di Renzo, T.; Boscaino, F.; Nazzaro, F.; Fratianni, F.; Aponte, M. Lactic acid bacteria biota and aroma profile of Italian traditional sourdoughs from the Irpinian area in Italy. Front. Microbiol. 2019, 10, 1621. [Google Scholar] [CrossRef] [Green Version]
- Huys, G.; Daniel, H.-M.; De Vuyst, L. Taxonomy and biodiversity of sourdough yeasts and lactic acid bacteria. In Handbook on Sourdough Biotechnology; Springer: New York, NY, USA, 2013; pp. 105–154. [Google Scholar]
- Minervini, F.; Di Cagno, R.; Lattanzi, A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: Interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 2012, 78, 1251–1264. [Google Scholar] [CrossRef] [Green Version]
- De Vuyst, L.; Harth, H.; van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- De Vuyst, L.; Schrijvers, V.; Paramithiotis, S.; Hoste, B.; Vancanneyt, M.; Swings, J.; Kalantzopoulos, G.; Tsakalidou, E.; Messens, W. The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl. Environ. Microb. 2002, 68, 6059–6069. [Google Scholar] [CrossRef] [Green Version]
- Paramithiotis, S.; Muller, M.R.A.; Ehrmann, M.A.; Tsakalidou, E.; Seiler, H.; Vogel, R.; Kalantzopoulos, G. Polyphasic identification of wild yeast strains isolated from Greek sourdoughs. Syst. Appl. Microbiol. 2000, 23, 156–164. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Tsiasiotou, S.; Drosinos, E.H. Comparative study of spontaneously fermented sourdoughs originating from two regions of Greece: Peloponnesus and Thessaly. Eur. Food Res. Technol. 2010, 231, 883–890. [Google Scholar] [CrossRef]
- Blaiotta, G.; Murru, N.; Di Cerbo, A.; Romano, R.; Aponte, M. Production of probiotic bovine salami using Lactobacillus plantarum 299v as adjunct. J. Sci. Food Agric. 2018, 98, 2285–2294. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-dependent and culture-independent characterization of the olive xylem microbiota: Effect of Sap extraction methods. Front. Plant Sci. 2020, 10, 1708. [Google Scholar] [CrossRef] [Green Version]
- Vogelmann, S.A.; Seitter, M.; Singer, U.; Brandt, M.J.; Hertel, C. Adaptability of lactic acid bacteria and yeasts to sourdoughs prepared from cereals, pseudocereals and cassava and use of competitive strains as starters. Int. J. Food Microbiol. 2009, 130, 205–212. [Google Scholar] [CrossRef]
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Pinto, D.; Siragusa, S.; Rizzello, C.G.; Gobbetti, M. Robustness of Lactobacillus plantarum starters during daily propagation of wheat flour sourdough type I. Food Microbiol. 2010, 27, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Aponte, M.; Boscaino, F.; Sorrentino, A.; Coppola, R.; Masi, P.; Romano, A. Volatile compounds and bacterial community dynamics of chestnut-flour-based sourdoughs. Food Chem. 2013, 141, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Rodríguez, L.; Vera Pingitore, E.; Rollan, G.; Cocconcelli, P.S.; Fontana, C.; Saavedra, L.; Vignolo, G.; Hebert, E.M. Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented quinoa sourdoughs. J. Appl. Microbiol. 2016, 120, 1289–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrigan, W.F.; McCance, M.E. Laboratory Methods in Food and Dairy Microbiology; Academic Press: London, UK, 1976; pp. 47–49. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts-a Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Doulgeraki, A.I.; Paramithiotis, S.; Kagkli, D.M.; Nychas, G.-J.E. Lactic acid bacteria population dynamics during minced beef storage under aerobic or modified atmosphere packaging conditions. Food Microbiol. 2010, 27, 1028–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadjilouka, A.; Andritsos, N.D.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef]
- Berthier, F.; Ehrlich, S.D. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 2006, 161, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paramithiotis, S.; Doulgeraki, A.I.; Karahasani, A.; Drosinos, E.H. Microbial population dynamics during spontaneous fermentation of Asparagus officinalis L. young sprouts. Eur. Food Res. Technol. 2014, 239, 297–304. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Doulgeraki, A.I.; Vrelli, A.; Nychas, G.-J.E.; Drosinos, E.H. Evolution of the microbial community during traditional fermentation of globe artichoke immature inflorescence. Int. J. Clin. Med. Microbiol. 2016, 1, 117. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Gänzle, M.G.; Zheng, J. Lifestyles of sourdough lactobacilli—do they matter for microbial ecology and bread quality? Int. J. Food Microbiol. 2019, 302, 15–23. [Google Scholar] [CrossRef]
- Robert, H.; Gabriel, V.; Fontagné-Faucher, C. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 2009, 135, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Lhomme, E.; Lattanzi, A.; Dousset, X.; Minervini, F.; De Angelis, M.; Lacaze, G.; Onno, B.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of sixteen French traditional sourdoughs. Int. J. Food Microbiol. 2015, 215, 161–170. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; Dinardo, F.R.; De Angelis, M.; Gobbetti, M. Wheat endophytic lactobacilli drive the microbial and biochemical features of sourdoughs. Food Microbiol. 2018, 70, 162–171. [Google Scholar]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Large genetic intraspecific diversity of autochthonous lactic acid bacteria and yeasts isolated from PDO Tuscan bread sourdough. Appl. Sci. 2020, 10, 1043. [Google Scholar] [CrossRef] [Green Version]
- Üçok, G.; Sert, G. Growth kinetics and biomass characteristics of Lactobacillus plantarum L14 isolated from sourdough: Effect of fermentation time on dough machinability. LWT 2020, 129, 109516. [Google Scholar] [CrossRef]
- Caro, I.; Quinto, E.J.; Fuentes, L.; Alessandria, V.; Cocolin, L.S.; Redondo-del-Río, M.P.; Mayo, B.; Flórez, A.B.; Mateo, J. Characterization of Lactococcus strains isolated from artisanal Oaxaca cheese. LWT 2020, 122, 109041. [Google Scholar] [CrossRef]
- Zago, M.; Bardelli, T.; Rossetti, L.; Nazzicari, N.; Carminati, D.; Galli, A.; Giraffa, G. Evaluation of bacterial communities of Grana Padano cheese by DNA metabarcoding and DNA fingerprinting analysis. Food Microbiol. 2021, 93, 103613. [Google Scholar] [CrossRef]
- Raimondi, S.; Nappi, M.R.; Sirangelo, T.M.; Leonardi, A.; Amaretti, A.; Ulrici, A.; Magnani, R.; Montanari, C.; Tabanelli, G.; Gardini, F.; et al. Bacterial community of industrial raw sausage packaged in modified atmosphere throughout the shelf life. Int. J. Food Microbiol. 2018, 280, 78–86. [Google Scholar] [CrossRef]
- Settanni, L.; Barbaccia, P.; Bonanno, A.; Ponte, M.; Di Gerlando, R.; Franciosi, E.; Di Grigoli, A.; Gaglio, R. Evolution of indigenous starter microorganisms and physicochemical parameters in spontaneously fermented beef, horse, wild boar and pork salamis produced under controlled conditions. Food Microbiol. 2020, 87, 103385. [Google Scholar] [CrossRef]
- Diez-Ozaetaa, I.; Amarita, F.; Lavilla, M.; Rainieri, S. Ecology of indigenous lactic acid bacteria from Rioja Alavesa red wines, focusing on biogenic amine production ability. LWT 2019, 116, 108544. [Google Scholar] [CrossRef]
- Dertli, E.; Mercan, E.; Arici, M.; Yilmaz, M.T.; Sağdiç, O. Characterisation of lactic acid bacteria from Turkish sourdough and determination of their exopolysaccharide (EPS) production characteristics. Food Sci. Technol. 2016, 71, 116–124. [Google Scholar] [CrossRef]
- Coda, R.; Kianjam, M.; Pontonio, E.; Verni, M.; Di Cagno, R.; Katina, K.; Rizzello, C.G.; Gobbetti, M. Sourdough-type propagation of faba bean flour: Dynamics of microbial consortia and biochemical implications. Int. J. Food Microbiol. 2017, 248, 10–21. [Google Scholar] [CrossRef] [Green Version]
- Sterr, Y.; Weiss, A.; Schmidt, H. Evaluation of lactic acid bacteria for sourdough fermentation of amaranth. Int. J. Food Microbiol. 2009, 136, 75–82. [Google Scholar] [CrossRef]
- Moroni, A.V.; Arendt, E.K.; Bello, F.D. Biodiversity of lactic acid bacteria and yeasts in spontaneously-fermented buckwheat and teff sourdoughs. Food Microbiol. 2011, 28, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Maidana, S.D.; Ficoseco, C.A.; Bassi, D.; Cocconcelli, P.S.; Puglisi, E.; Savoy, G.; Vignolo, G.; Fontana, C. Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough. Int. J. Food Microbiol. 2020, 316, 108425. [Google Scholar] [CrossRef]
- Boyaci-Gunduz, C.P.; Erten, H. Predominant yeasts in the sourdoughs collected from some parts of Turkey. Yeast 2020, 1–18. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, M.; Jiaxin, C.; Luo, Y.; Ye, F.; Jiao, S.; Lü, X. Bacterial diversity in traditional sourdough from different regions in China. LWT 2018, 96, 251–259. [Google Scholar] [CrossRef]
- Korcari, D.; Ricci, G.; Quattrini, M.; Fortina, M.G. Microbial consortia involved in fermented spelt sourdoughs: Dynamics and characterization of yeasts and lactic acid bacteria. Lett. Appl. Microbiol. 2019, 70, 48–54. [Google Scholar] [CrossRef]
- Cappelli, A.; Ulissi, U.; Valzano, M.; Damiani, C.; Epis, S.; Gabrielli, M.G.; Conti, S.; Polonelli, L.; Bandi, C.; Favia, G.; et al. A Wickerhamomyces anomalus killer strain in the malaria vector Anopheles stephensi. PLoS ONE 2014, 9, 95988. [Google Scholar] [CrossRef]
- Daniel, H.M.; Moons, M.C.; Huret, S.; Vrancken, G.; De Vuyst, L. Wickerhamomyces anomalus in the sourdough microbial ecosystem. Antonie van Leeuwenhoek 2011, 99, 63–73. [Google Scholar] [CrossRef]
- Vrancken, G.; De Vuyst, L.; Van der Meulen, R.; Huys, G.; Vandamme, P.; Daniel, H.M. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 2010, 10, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, M.; Momoda, R.; Tanaka, M.; Zendo, T.; Nakayama, J. Dense tracking of the dynamics of the microbial community and chemicals constituents in spontaneous wheat sourdough during two months of backslopping. J. Biosci. Bioeng. 2019, 128, 170–176. [Google Scholar] [CrossRef]
- Sayevand, H.R.; Bakhtiary, F.; Pointner, A.; Remely, M.; Hippe, B.; Hosseini, H.; Haslberger, A. Bacterial diversity in traditional Doogh in comparison to industrial Doogh. Curr. Microbiol. 2018, 75, 386–393. [Google Scholar] [CrossRef] [Green Version]
- Maoloni, A.; Milanović, V.; Cardinali, F.; Mangia, N.P.; Murgia, M.A.; Garofalo, C.; Clementi, F.; Osimani, A.; Aquilanti, L. Bacterial and fungal communities of Gioddu as revealed by PCR–DGGE analysis. Indian J. Microbiol. 2020, 60, 119–123. [Google Scholar] [CrossRef]
- Ramezani, M.; Hosseini, S.M.; Ferrocino, I.; Amoozegar, M.A.; Cocolin, L. Molecular investigation of bacterial communities during the manufacturing and ripening of semi-hard Iranian Liqvan cheese. Food Microbiol. 2017, 66, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unno, R.; Matsutani, M.; Suzuki, T.; Kodama, K.; Matsushita, H.; Yamasato, K.; Koizumi, Y.; Ishikawa, M. Lactic acid bacterial diversity in Brie cheese focusing on salt concentration and pH of isolation medium and characterisation of halophilic and alkaliphilic lactic acid bacterial isolates. Int. Dairy J. 2020, 109, 104757. [Google Scholar] [CrossRef]
- Cardinali, F.; Milanovi’c, V.; Osimani, A.; Aquilanti, L.; Taccari, M.; Garofalo, C.; Polverigiani, S.; Clementi, F.; Franciosi, E.; Tuohy, K.; et al. Microbial dynamics of model Fabriano-like fermented sausages as affected by starter cultures, nitrates and nitrites. Int. J. Food Microbiol. 2018, 278, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Osimani, A.; Ferrocino, I.; Agnolucci, M.; Cocolin, L.; Giovannetti, M.; Cristani, C.; Palla, M.; Milanović, V.; Roncolini, A.; Sabbatini, R.; et al. Unveiling hαkarl: A study of the microbiota of the traditional Icelandic fermented fish. Food Microbiol. 2019, 82, 560–572. [Google Scholar] [CrossRef] [Green Version]
- Aldrete-Tapia, J.A.; Escalante-Minakata, P.; Martínez-Peniche, R.A.; Tamplin, M.L.; Hernández-Iturriaga, M. Yeast and bacterial diversity, dynamics and fermentative kinetics during small-scale tequila spontaneous fermentation. Food Microbiol. 2020, 86, 103339. [Google Scholar] [CrossRef]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 2017, 250, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Dolci, P.; Zenato, S.; Pramotton, R.; Barmaz, A.; Alessandria, V.; Rantsiou, K.; Cocolin, L. Cheese surface microbiota complexity: RT-PCR-DGGE, a tool for a detailed picture? Int. J. Food Microbiol. 2013, 162, 8–12. [Google Scholar] [CrossRef]
- Garofalo, C.; Bancalari, E.; Milanović, V.; Cardinali, F.; Osimani, A.; Sardaro, M.L.S.; Bottari, B.; Bernini, V.; Aquilanti, L.; Clementi, F. Study of the bacterial diversity of foods: PCR-DGGE versus LHPCR. Int. J. Food Microbiol. 2017, 242, 24–36. [Google Scholar] [CrossRef]
- Iacumin, L.; Cecchini, F.; Manzano, M.; Osualdini, M.; Boscolo, D.; Orlic, S.; Comi, G. Description of the microflora of sourdoughs by culture dependent and culture-independent methods. Food Microbiol. 2009, 26, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Neilson, J.W.; Jordana, F.L.; Maiera, R.M. Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. J. Microbiol. Methods. 2013, 92, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Ercolini, D. PCR-DGGE fingerprinting: Novel strategies for detection of microbes in food. J. Microbiol. Methods. 2004, 56, 297–314. [Google Scholar] [CrossRef]
- Cocolin, L.; Manzano, M.; Cantoni, C.; Comi, G. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 2001, 67, 5113–5121. [Google Scholar] [CrossRef] [Green Version]
- Gafan, G.P.; Spratt, D.A. Denaturing gradient gel electrophoresis gel expansion (DGGEGE)—An attempt to resolve the limitations of co-migration in the DGGE of complex polymicrobial communities. FEMS Microbiol. Lett. 2005, 253, 303–307. [Google Scholar] [CrossRef]
- Scheirlinck, I.; Van der Meulen, R.; Van Schoor, A.; Vancanneyt, M.; De Vuyst, L.; Vandamme, P.; Huys, G. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl. Environ. Microbiol. 2008, 74, 2414–2423. [Google Scholar] [CrossRef] [Green Version]
- Kanagawa, T. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 2003, 96, 317–323. [Google Scholar] [CrossRef]



| Sample No. | Origin | Ingredients a |
|---|---|---|
| 1 | Aetolia-Acarnania | Basil |
| 2 | Aetolia-Acarnania | Basil |
| 3 | Aetolia-Acarnania | Basil |
| 4 | Aetolia-Acarnania | Basil |
| 5 | Arkadia | Basil |
| 6 | Aetolia-Acarnania | Basil |
| 7 | Aetolia-Acarnania | Basil |
| 8 | Thessaloniki | Milk |
| 9 | Thessaloniki | Basil |
| 10 | Thessaloniki | No details available |
| 11 | Thessaloniki | Yoghurt |
| 12 | Viotia | Basil |
| 13 | Salamis island | Basil |
| Sample No | pH | TTA a | Yeasts b | LAB b |
|---|---|---|---|---|
| 1 | 3.76 (0.01) | 1.59 (0.01) | 4.60 | 7.00 |
| 2 | 3.91 (0.13) | 0.79 (0.13) | 6.20 | 7.57 |
| 3 | 3.91 (0.07) | 0.70 (0.07) | 6.32 | 9.20 |
| 4 | 3.72 (0.01) | 0.85 (0.01) | 5.23 | 8.20 |
| 5 | 3.64 (0.07) | 0.99 (0.07) | 5.36 | 8.26 |
| 6 | 3.65 (0.01) | 0.98 (0.01) | 5.30 | 9.18 |
| 7 | 3.85 (0.01) | 1.23 (0.01) | 5.28 | 8.18 |
| 8 | 3.76 (0.04) | 1.21 (0.04) | 5.08 | 8.08 |
| 9 | 3.75 (0.01) | 1.03 (0.01) | 5.94 | 8.23 |
| 10 | 5.05 (0.01) | 0.65 (0.01) | 4.78 | 6.28 |
| 11 | 3.80 (0.06) | 1.10 (0.06) | 6.08 | 8.32 |
| 12 | 4.96 (0.03) | 0.50 (0.03) | 6.30 | 8.20 |
| 13 | 3.64 (0.04) | 0.70 (0.04) | 6.30 | 8.36 |
| Strain Number | Closest Relative | Accession Number | Identity (%) |
|---|---|---|---|
| LQC 2322 | Cb. paralimentarius | KX247775.1 | 100 |
| LQC 2323 | Cb. paralimentarius | MF540546.1 | 100 |
| LQC 2338 | Lb. brevis | MN166306.1 | 100 |
| LQC 2339 | Lb. brevis | LC199964.1 | 100 |
| LQC 2362 | Lb. brevis | MN720522.1 | 100 |
| LQC 2381 | Cb. paralimentarius | MH544805.1 | 100 |
| LQC 2389 | Cb. paralimentarius | MH544805.1 | 100 |
| LQC 2391 | Cb. paralimentarius | MH544805.1 | 100 |
| LQC 2394 | Lb. zymae | KT757254.1 | 100 |
| LQC 2395 | Cb. paralimentarius | MF942368.1 | 100 |
| LQC 2398 | Lb. brevis | CP031174.1 | 100 |
| LQC 2404 | Cb. paralimentarius | KY435699.1 | 100 |
| LQC 2408 | Fb. sanfranciscensis | MH704126.1 | 100 |
| LQC 2410 | Cb. paralimentarius | KC755102.1 | 100 |
| LQC 2412 | Lb. brevis | MN431348.1 | 100 |
| LQC 2428 | Fb. sanfranciscensis | LC483557.1 | 100 |
| LQC 2430 | Lb. brevis | MN049503.1 | 100 |
| LQC 2440 | Lb. brevis | MG646821.1 | 100 |
| LQC 2456 | Lb. sakei | MF428782.1 | 100 |
| LQC 2458 | Lb. brevis | MN720508.1 | 99 |
| LQC 2473 | Lb. sakei | MG462120.1 | 100 |
| LQC 2475 | Lb. curvatus | MN720519.1 | 100 |
| LQC 2494 | Lb. brevis | KX649032.1 | 100 |
| LQC 2508 | Ln. citreum | MG754627.1 | 100 |
| LQC 2510 | Lc. lactis | MN368062.1 | 100 |
| LQC 2511 | Lb. brevis | MH681603.1 | 100 |
| LQC 2512 | Ln. mesenteroides | MG825699.1 | 100 |
| LQC 2517 | Cb. paralimentarius | MH544773.1 | 100 |
| LQC 2537 | Cb. paralimentarius | MH704124.1 | 100 |
| Strain Number | Closest Relative | Accession Number | Identity (%) |
|---|---|---|---|
| LQC 10300 | S. cerevisiae | JQ771733.1 | 100 |
| LQC 10306 | S. cerevisiae | JQ771733.1 | 100 |
| LQC 10308 | S. cerevisiae | CP025108.1 | 100 |
| LQC 10313 | S. cerevisiae | MK397410.1 | 99 |
| LQC 10341 | S. cerevisiae | MN462945.1 | 100 |
| LQC 10345 | K. humilis | MK262977.1 | 100 |
| LQC 10347 | P. fermentans | KJ413162.1 | 98 |
| LQC 10350 | P. fermentans | KM589485.1 | 99 |
| LQC 10351 | S. cerevisiae | JQ771733.1 | 100 |
| LQC 10353 | W. anomalus | MH479120.1 | 99 |
| LQC 10355 | P. fermentans | KY296092.1 | 99 |
| LQC 10361 | W. anomalus | LC178747.1 | 99 |
| LQC 10366 | S. cerevisiae | MK358167.1 | 100 |
| LQC 10369 | S. cerevisiae | MG017585.1 | 100 |
| LQC 10373 | S. cerevisiae | MG017587.1 | 100 |
| LQC 10388 | S. cerevisiae | MG017572.1 | 100 |
| LQC 10389 | S. cerevisiae | MK358167.1 | 99 |
| LQC 10391 | S. cerevisiae | MK027355.1 | 99 |
| LQC 10399 | S. cerevisiae | HM191654.1 | 100 |
| LQC 10403 | S. cerevisiae | MF521985.1 | 100 |
| LQC 10406 | S. cerevisiae | MG017572.1 | 100 |
| LQC 10408 | S. cerevisiae | MF979228.1 | 100 |
| LQC 10412 | S. cerevisiae | MH844381.1 | 100 |
| LQC 10455 | S. cerevisiae | MG386438.1 | 99 |
| LQC 10459 | S. cerevisiae | MG386438.1 | 99 |
| LQC 10460 | S. cerevisiae | MG017586.1 | 99 |
| LQC 10466 | S. cerevisiae | GU080045.1 | 99 |
| LQC 10419 | S. cerevisiae | MG386438.1 | 100 |
| LQC 10420 | S. cerevisiae | KF141642.1 | 100 |
| LQC 10423 | P. membranifaciens | KF141642.1 | 100 |
| LQC 10432 | S. cerevisiae | MF979228.1 | 100 |
| LQC 10441 | P. membranifaciens | KF141642.1 | 100 |
| LQC 10447 | P. membranifaciens | MK358179.1 | 99 |
| LQC 10469 | S. cerevisiae | MF521980.1 | 100 |
| LQC 10472 | S. cerevisiae | MN462933.1 | 100 |
| LQC 10475 | S. cerevisiae | MF979228.1 | 100 |
| LQC 10476 | S. cerevisiae | MK358167.1 | 100 |
| LQC 10482 | S. cerevisiae | MG017585.1 | 99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syrokou, M.K.; Themeli, C.; Paramithiotis, S.; Mataragas, M.; Bosnea, L.; Argyri, A.A.; Chorianopoulos, N.G.; Skandamis, P.N.; Drosinos, E.H. Microbial Ecology of Greek Wheat Sourdoughs, Identified by a Culture-Dependent and a Culture-Independent Approach. Foods 2020, 9, 1603. https://doi.org/10.3390/foods9111603
Syrokou MK, Themeli C, Paramithiotis S, Mataragas M, Bosnea L, Argyri AA, Chorianopoulos NG, Skandamis PN, Drosinos EH. Microbial Ecology of Greek Wheat Sourdoughs, Identified by a Culture-Dependent and a Culture-Independent Approach. Foods. 2020; 9(11):1603. https://doi.org/10.3390/foods9111603
Chicago/Turabian StyleSyrokou, Maria K., Christina Themeli, Spiros Paramithiotis, Marios Mataragas, Loulouda Bosnea, Anthoula A. Argyri, Nikos G. Chorianopoulos, Panagiotis N. Skandamis, and Eleftherios H. Drosinos. 2020. "Microbial Ecology of Greek Wheat Sourdoughs, Identified by a Culture-Dependent and a Culture-Independent Approach" Foods 9, no. 11: 1603. https://doi.org/10.3390/foods9111603





