Effects of Salt Stimulation on Lunasin Accumulation and Activity during Soybean Germination
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Soybean Germination
2.3. Protein Extraction and Purification
2.4. UPLC-MS/MS Analysis
2.5. Western Blot and ELISA Detection
2.6. Antioxidant-Activity Determination
2.7. Anti-Inflammatory-Activity Assay
2.8. Anticancer-Activity Assay
2.9. Statistical Analysis
3. Results
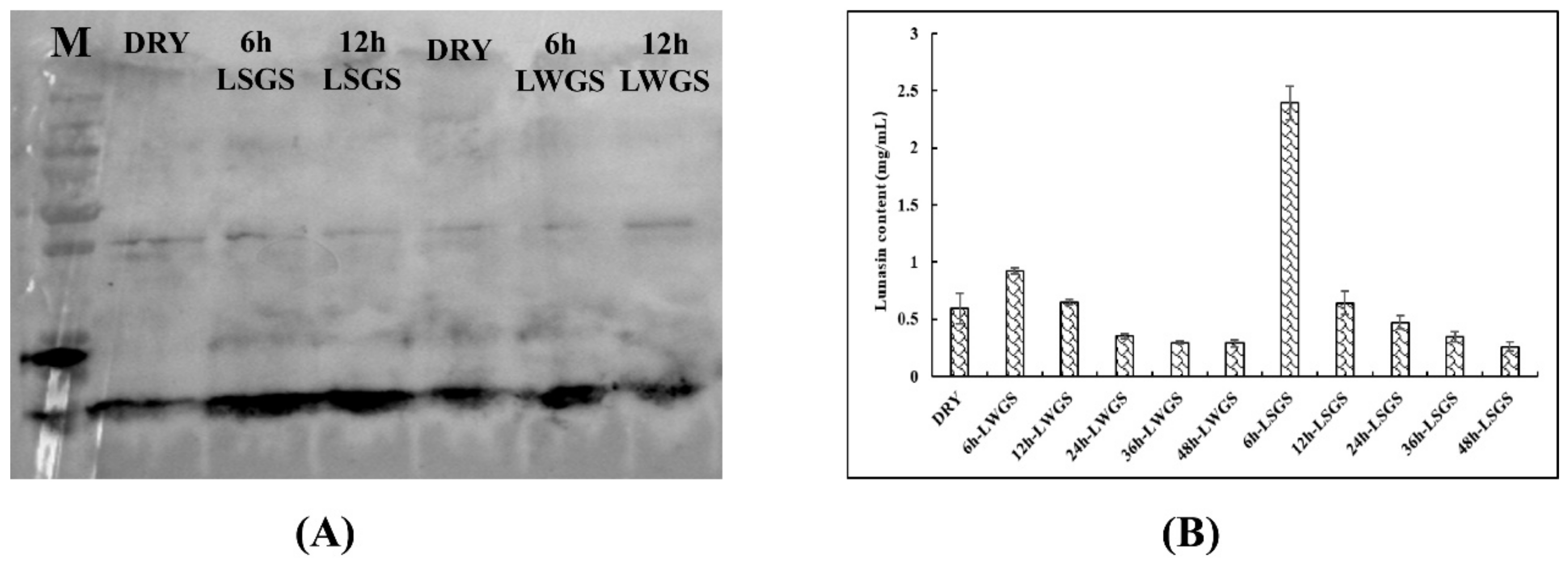
3.1. Lunasin-Content Detection
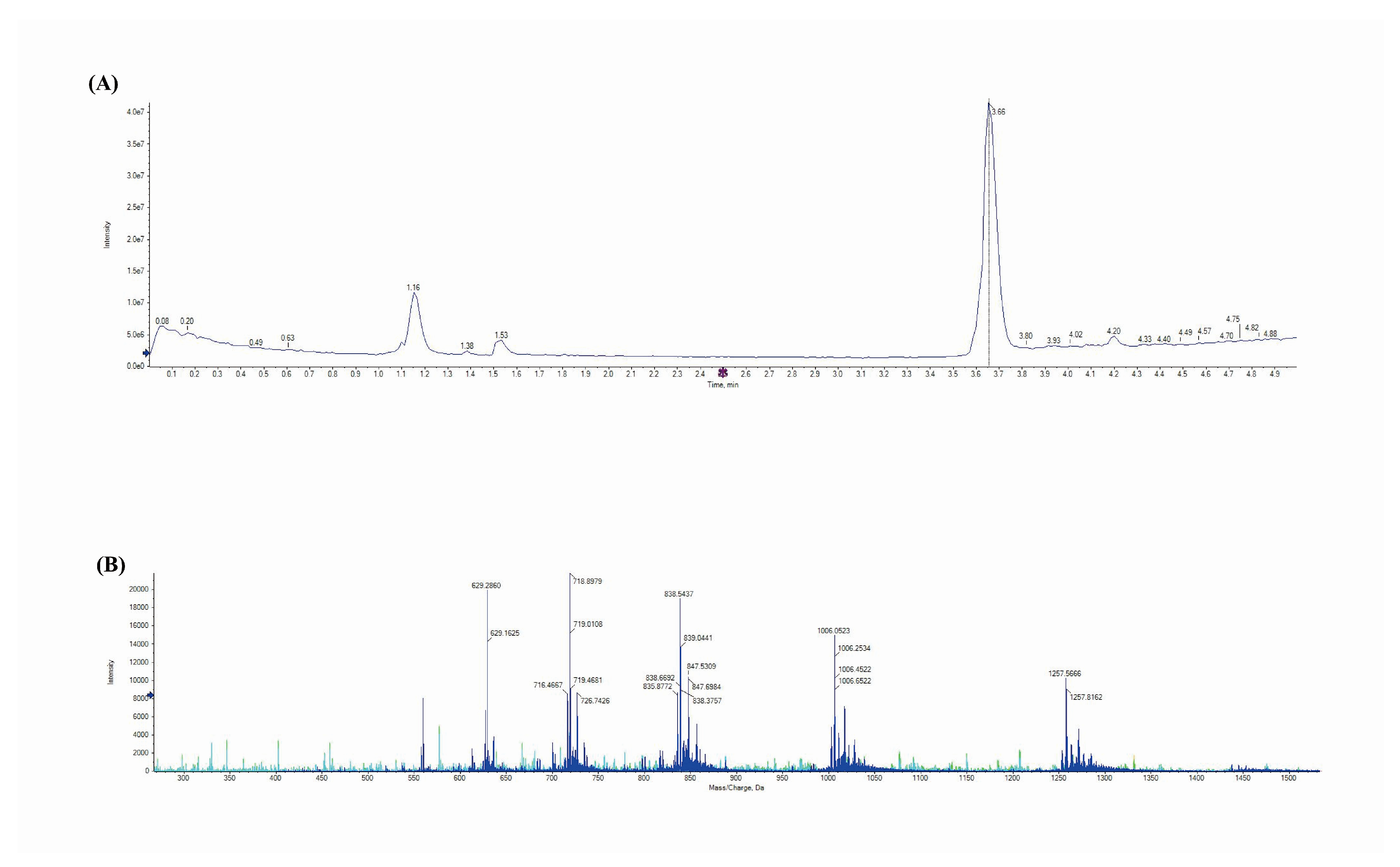
3.2. Mass Spectrometry Analysis
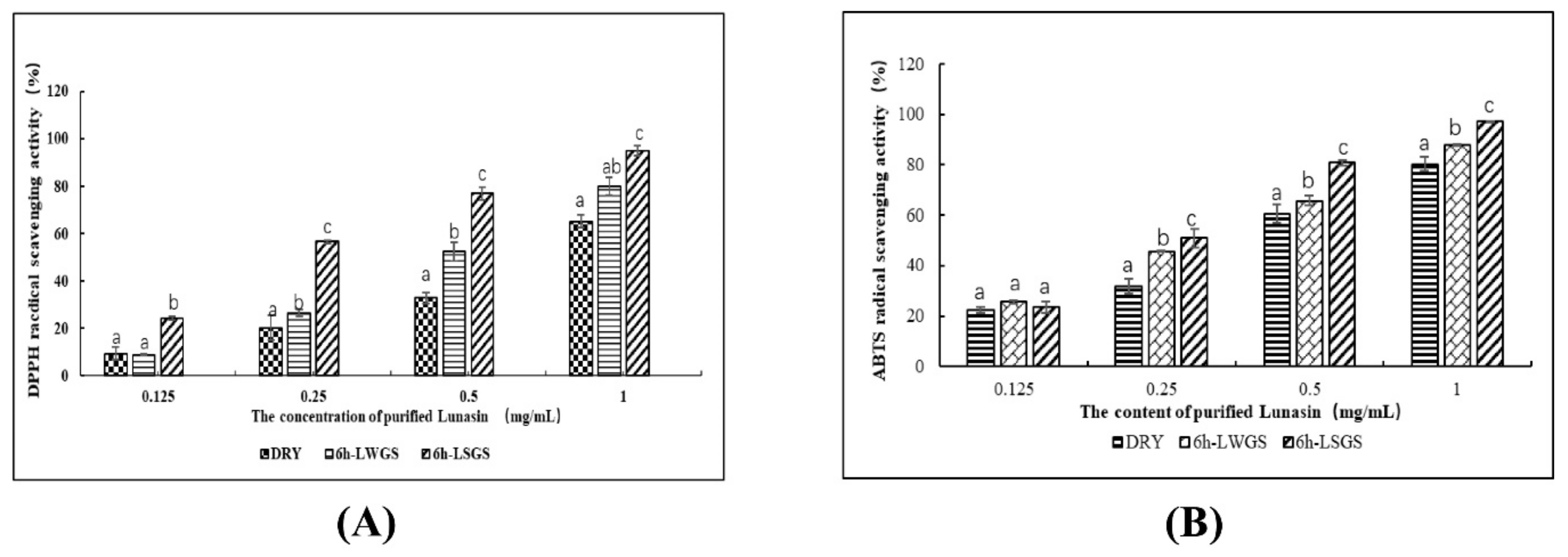
3.3. Antioxidant Activity Assay
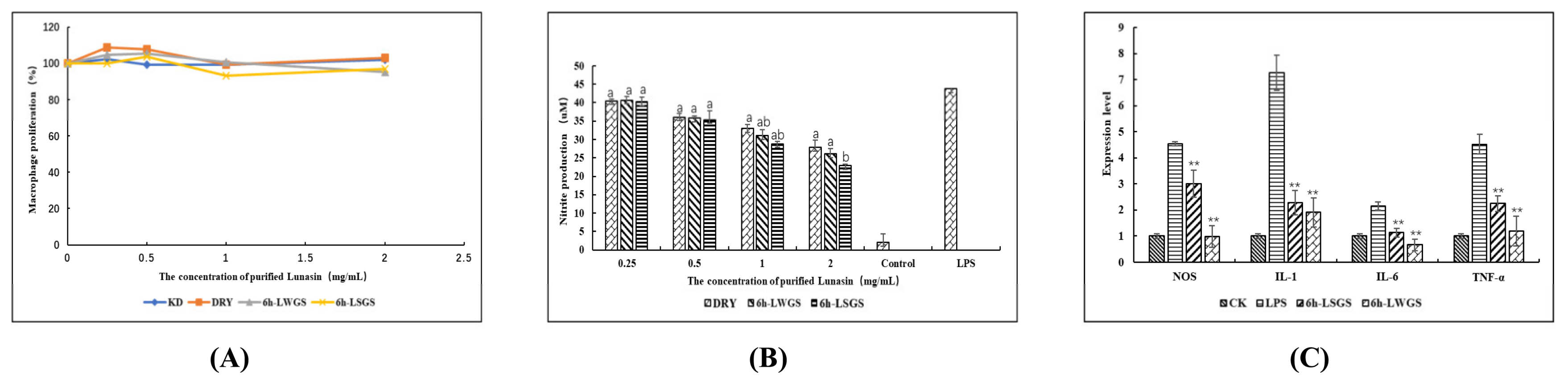
3.4. Anti-Inflammatory-Activity Assay
3.5. Anti-MDA-MB-231 Activity Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jiang, Q.; Pan, Y.; Cheng, Y. Lunasin suppresses the migration and invasion of breast cancer cells by inhibiting matrix metalloproteinase-2/-9 via the FAK/Akt/ERK and NF-κB signaling pathways. Oncol. Rep. 2016, 36, 253–262. [Google Scholar] [CrossRef]
- Ren, G.; Zhu, Y.; Shi, Z. Detection of lunasin in quinoa (Chenopodium quinoa Willd.) and the in vitro evaluation of its antioxidant and anti-inflammatory activities. J. Sci. Food Agric. 2017, 97, 4110–4116. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jeong, H.J.; De Lumen, B.O. Contents and bioactivities of lunasin, bowman-birk inhibitor, and isoflavones in soybean seed. J. Agric. Food Chem. 2005, 53, 7686–7690. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Park, J.H.; Lam, Y. Characterization of Lunasin Isolated from Soybean. J. Agric. Food Chem. 2003, 51, 7901–7906. [Google Scholar] [CrossRef]
- Jeong, H.J.; Lam, Y.; De Lumen, B.O. Barley lunasin suppresses\r, ras\r, -induced colony formation and inhibits core histone acetylation in mammalian cells. J. Agric. Food Chem. 2002, 50, 5903–5908. [Google Scholar] [CrossRef]
- Kim, S.L.; Lee, J.E.; Kwon, Y.U. Introduction and nutritional evaluation of germinated soy germ. Food Chem. 2013, 136, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, M.; Mewis, I.; Huyskens-Keil, S. UV-B-Induced Secondary Plant Metabolites-Potential Benefits for Plant and Human Health. Crit. Rev. Plant Sci. 2012, 31, 229–240. [Google Scholar] [CrossRef]
- Hurkman, W.J.; Tao, H.P.; Tanaka, C.K. Germin-Like Polypeptides Increase in Barley Roots during Salt Stress. Plant Physiol. 1991, 97, 366–374. [Google Scholar] [CrossRef]
- Hurkman, W.J.; Tanaka, C.K.; Dupont, F.M. The Effects of Salt Stress on Polypeptides in Membrane Fractions from Barley Roots. Plant Physiol. 1988, 88, 1263–1273. [Google Scholar] [CrossRef]
- Lopez, F.; Vansuyt, G.; Fourcroy, P.; Casse-Delbart, F. Accumulation of a 22-kDa protein and its mRNA in the leaves of Raphanus sativus in response to salt stress or water deficit. Physiol. Plant. 2006, 91, 605–614. [Google Scholar] [CrossRef]
- Nakurte, I.; Kirhnere, I.; Namniece, J. Detection of the lunasin peptide in oats (Avena sativa L). J. Cereal Sci. 2013, 57, 319–324. [Google Scholar] [CrossRef]
- Hirano, S. Western Blot Analysis. Methods Mol. Biol. 2012, 926, 87–97. [Google Scholar] [PubMed]
- Yang, P.C.; Mahmood, T. Western blot: Technique, theory, and trouble shooting. North Am. J. Med Sci. 2014, 6, 160. [Google Scholar] [CrossRef] [PubMed]
- Dia, V.P.; Wang, W.; Oh, V.L.; De Lumen, B.O.; De Mejia, E.G. Isolation, purification and characterisation of lunasin from defatted soybean flour and in vitro evaluation of its anti-inflammatory activity. Food Chem. 2009, 114, 108–115. [Google Scholar] [CrossRef]
- Menghini, L.; Leporini, L.; Vecchiotti, G. Crocus sativus L. stigmas and byproducts: Qualitative fingerprint, antioxidant potentials and enzyme inhibitory activities. Food Res. Int. 2018, 109, 91–98. [Google Scholar] [CrossRef]
- Frantisek, S.; Fridrich, G.; Peter, K. Synthesis and Free Radical Scavenging Activity of New Hydroxybenzylidene Hydrazines. Molecules 2017, 22, 894. [Google Scholar]
- Kotora, P.; Šeršeň, F.; Filo, J. The Scavenging of DPPH, Galvinoxyl and ABTS Radicals by Imine Analogs of Resveratrol. Molecules 2016, 21, 127. [Google Scholar] [CrossRef]
- Forough, K.; Nikolaus, M.; Wilhelm, C. Yacon (Smallanthus sonchifolius Poepp. and Endl.) as a Novel Source of Health Promoting Compounds: Antioxidant Activity, Phytochemicals and Sugar Content in Flesh, Peel, and Whole Tubers of Seven Cultivars. Molecules 2018, 23, 278. [Google Scholar]
- Thaipong, K.; Boonprakob, U.; Crosby, K. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compos. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Kaviani, B.; Pourkhalili, S.T.; Sajedi, R.H. Salt treatment can change composition of glycinin and β-conglycinin proteins in soybean seed. Plant Omics. 2011, 4, 228–235. [Google Scholar]
- Paucar-Menacho, L.M.; Berhow, M.A.; Mandarino, J.M.G. Optimisation of germination time and temperature on the concentration of bioactive compounds in Brazilian soybean cultivar BRS 133 using response surface methodology. Food Chem. 2010, 119, 636–642. [Google Scholar] [CrossRef]
- Ren, G.; Hao, Y.; Zhu, Y.; Shi, Z.; Zhao, G. Expression of Bioactive Lunasin Peptide in Transgenic Rice Grains for the Application in Functional Food. Molecules 2018, 23, 2373. [Google Scholar] [CrossRef] [PubMed]
- García-Nebot, M.J.; Recio, I.; Hernández-Ledesma, B. Antioxidant activity and protective effects of peptide lunasin against oxidative stress in intestinal Caco-2 cells. Food Chem. Toxicol. 2014, 65, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Shuai, H.; Han, N. The Chemopreventive Peptide Lunasin Inhibits d-Galactose-Induced Experimental Cataract in Rats. Protein Pept. Lett. 2016, 23, 619–625. [Google Scholar]
- Wang, R.L.; Hua, C.; Zhou, F. Effects of NaCl stress on photochemical activity and thylakoid membrane polypeptide composition of a salt-tolerant and a salt-sensitive rice cultivar. Photosynthetica 2009, 47, 125–127. [Google Scholar] [CrossRef]
- Falleh, H.; Inès, J.; Ksouri, R. Effect of salt treatment on phenolic compounds and antioxidant activity of two Mesembryanthemum edule provenances. Plant Physiol. Biochem. 2012, 52, 1–8. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; Hsieh, C.C.; Lumen, B.O.D. Antioxidant and anti-inflammatory properties of cancer preventive peptide lunasin in RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2009, 390, 803–808. [Google Scholar] [CrossRef]
- Cam, A.; Mejia, E.G.D. RGD-peptide lunasin inhibits Akt-mediated NF-κB activation in human macrophages through interaction with the αVβ3 integrin. Mol. Nutr. Food Res. 2012, 56, 1569–1581. [Google Scholar] [CrossRef]
- Galvez, A.F.; Lumen, B.O.D. A soybean cDNA encoding a chromatin-binding peptide inhibits mitosis of mammalian cells. Nat. Biotechnol. 1999, 17, 495–500. [Google Scholar] [CrossRef]
- Lam, Y.; Galvez, A.; Lumen, B.O. Lunasin suppresses E1A-mediated transformation of mammalian cells but does not inhibit growth of immortalized and established cancer cell lines. Nutr. Cancer 2003, 47, 88–94. [Google Scholar] [CrossRef]
- Hsieh, C.C.; Hernández-Ledesma, B.; Lumen, B.O.D. Lunasin–Aspirin Combination against NIH/3T3 Cells Transformation Induced by Chemical Carcinogens. Plant Foods Hum. Nutr. 2011, 66, 107–113. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequences |
|---|---|
| Actin-F | CCATCATGAAGTGTGACGTTG |
| Actin-R | ATCTCCTTCTGCATCCTGTCA |
| IL1b-F | ACTGTGAAATGCCACCTTTTG |
| IL1b-R | TTTGAAGCTGGATGCTCTCAT |
| IL6-F | TCAATTCCAGAAACCGCTATG |
| IL6-R | TTGGGAGTGGTATCCTCTGTG |
| Nos2-F | GTCCGAAGCAAACATCACATT |
| Nos2-R | TGAGGGCTCTGTTGAGGTCTA |
| Tnf-F | GGTTCTCTTCAAGGGACAAGG |
| Tnf-R | GGCAGAGAGGAGGTTGACTTT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Hao, Y.; Teng, C.; Fan, X.; Yang, X.; Liu, M.; Ren, G.; Tan, C. Effects of Salt Stimulation on Lunasin Accumulation and Activity during Soybean Germination. Foods 2020, 9, 118. https://doi.org/10.3390/foods9020118
Zhang W, Hao Y, Teng C, Fan X, Yang X, Liu M, Ren G, Tan C. Effects of Salt Stimulation on Lunasin Accumulation and Activity during Soybean Germination. Foods. 2020; 9(2):118. https://doi.org/10.3390/foods9020118
Chicago/Turabian StyleZhang, Weiyi, Yuqiong Hao, Cong Teng, Xin Fan, Xiushi Yang, Mengjie Liu, Guixing Ren, and Congping Tan. 2020. "Effects of Salt Stimulation on Lunasin Accumulation and Activity during Soybean Germination" Foods 9, no. 2: 118. https://doi.org/10.3390/foods9020118
APA StyleZhang, W., Hao, Y., Teng, C., Fan, X., Yang, X., Liu, M., Ren, G., & Tan, C. (2020). Effects of Salt Stimulation on Lunasin Accumulation and Activity during Soybean Germination. Foods, 9(2), 118. https://doi.org/10.3390/foods9020118







