Rapid Identification and Visualization of Jowl Meat Adulteration in Pork Using Hyperspectral Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Image Acquisition and Calibration
2.3. Region of Interests (ROI) Identification and Spectral Extraction
2.4. Spectral Pretreatments
2.5. Modeling Method
2.6. Wavelengths Selection Methods
2.7. Models Performance Assessment
2.8. Distribution Maps of the Adulterant
3. Results and Discussion
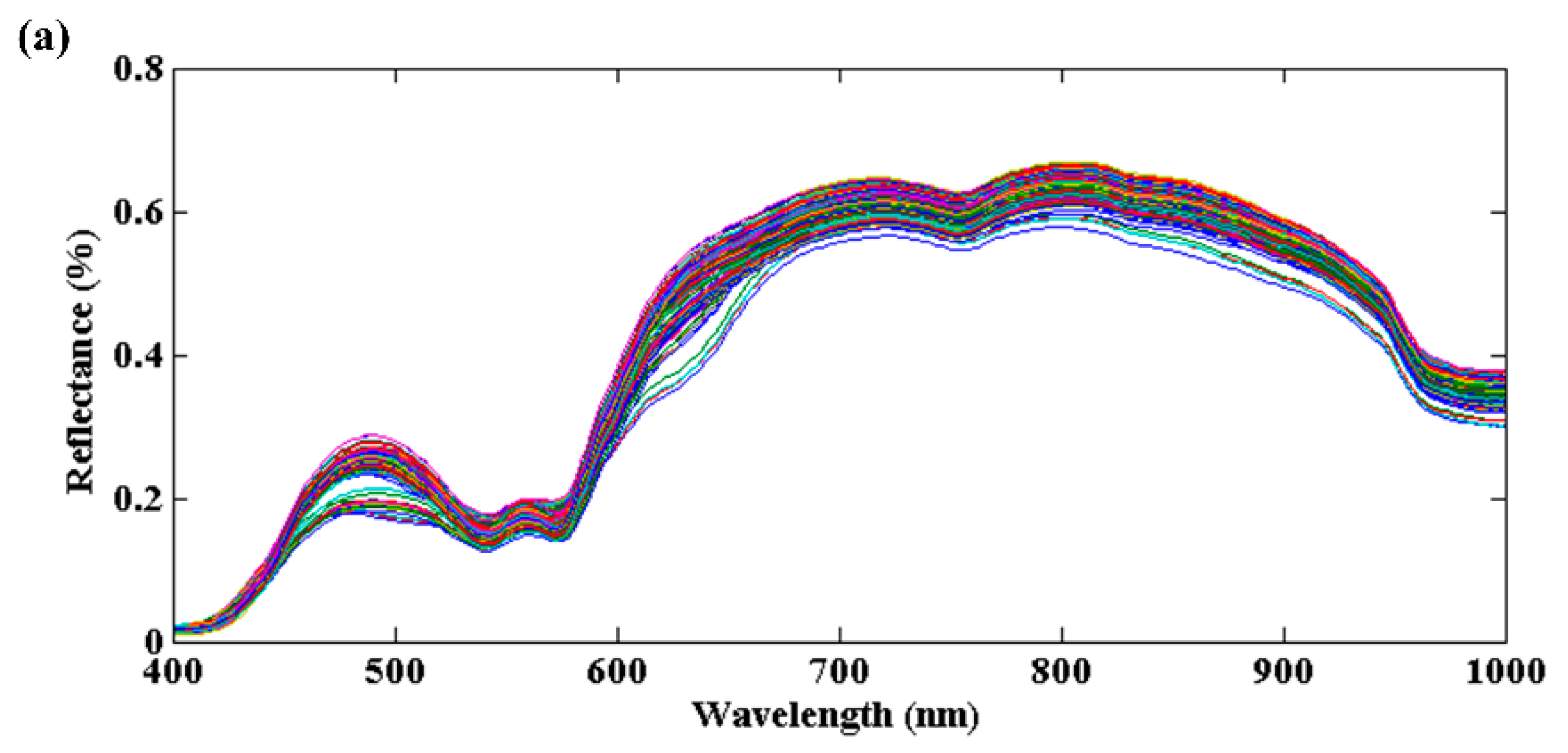
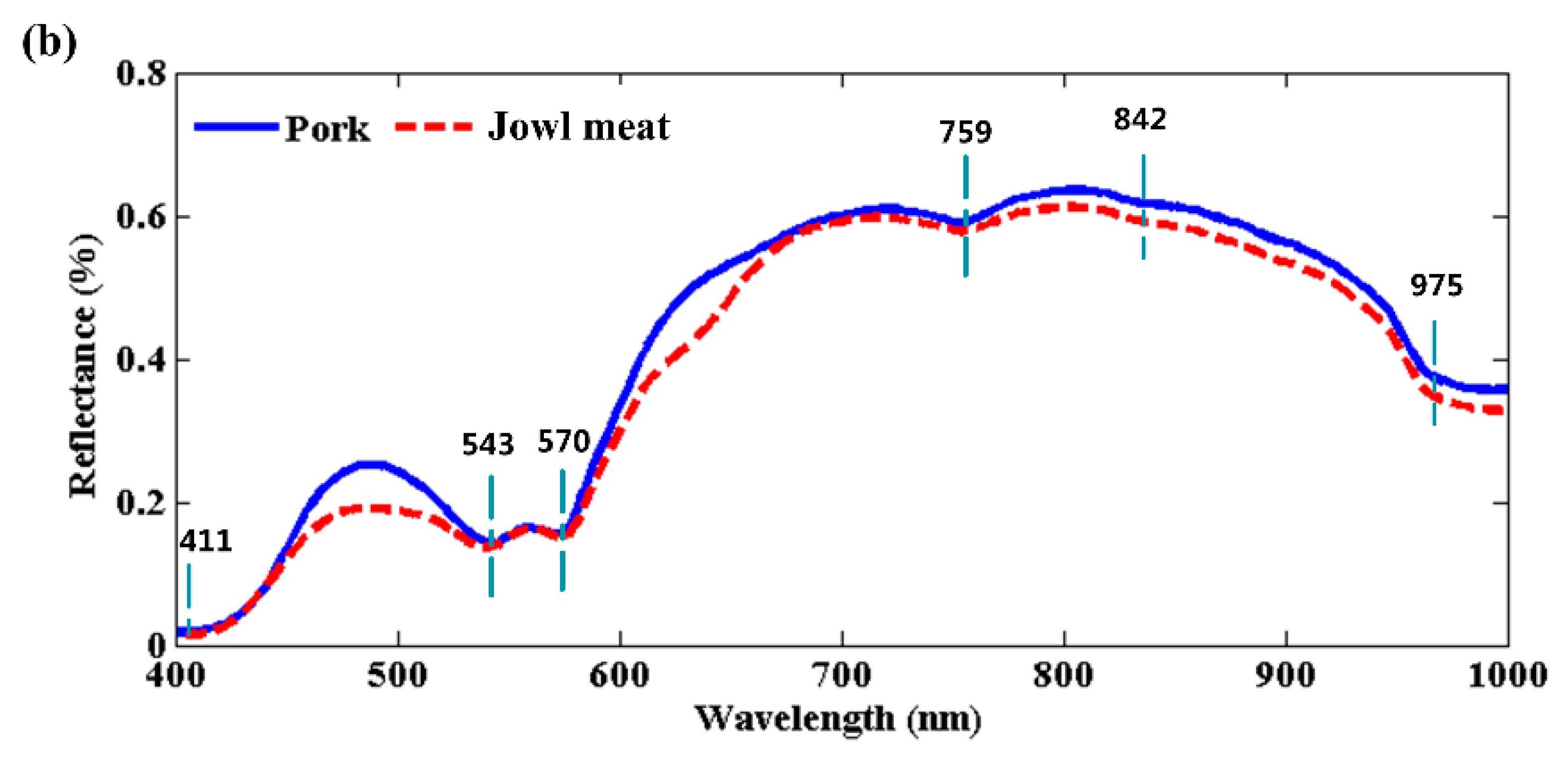
3.1. Spectral Profiles
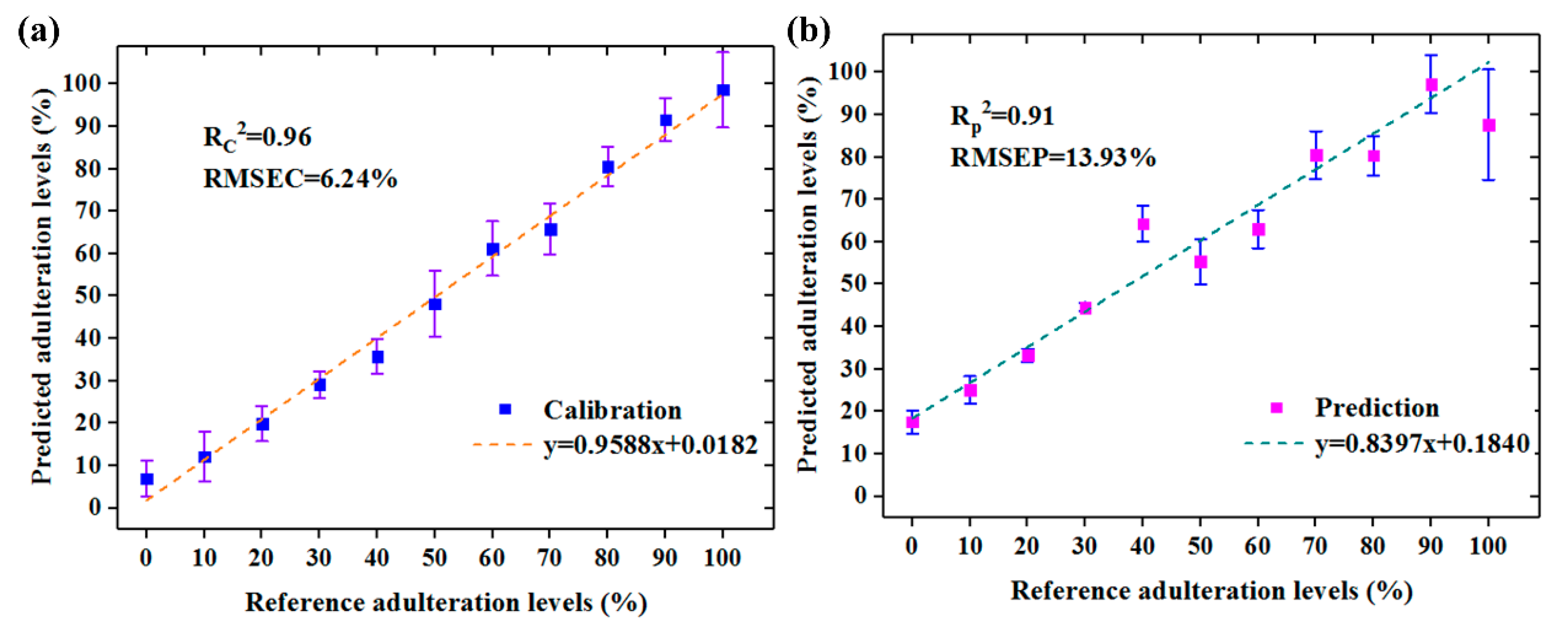
3.2. PLSR Models
3.3. Wavelengths Selection
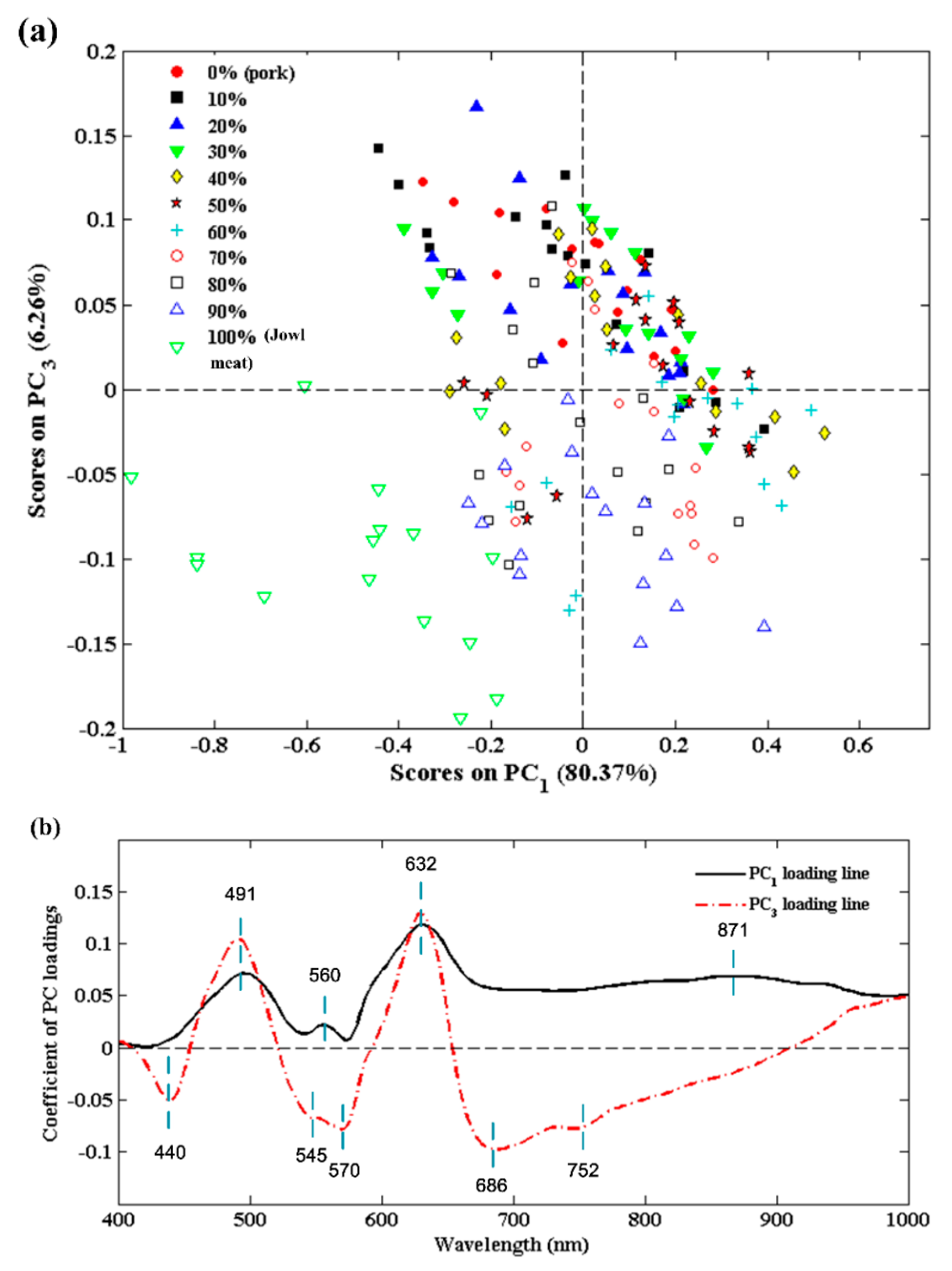
3.3.1. PCA Explanatory Analysis
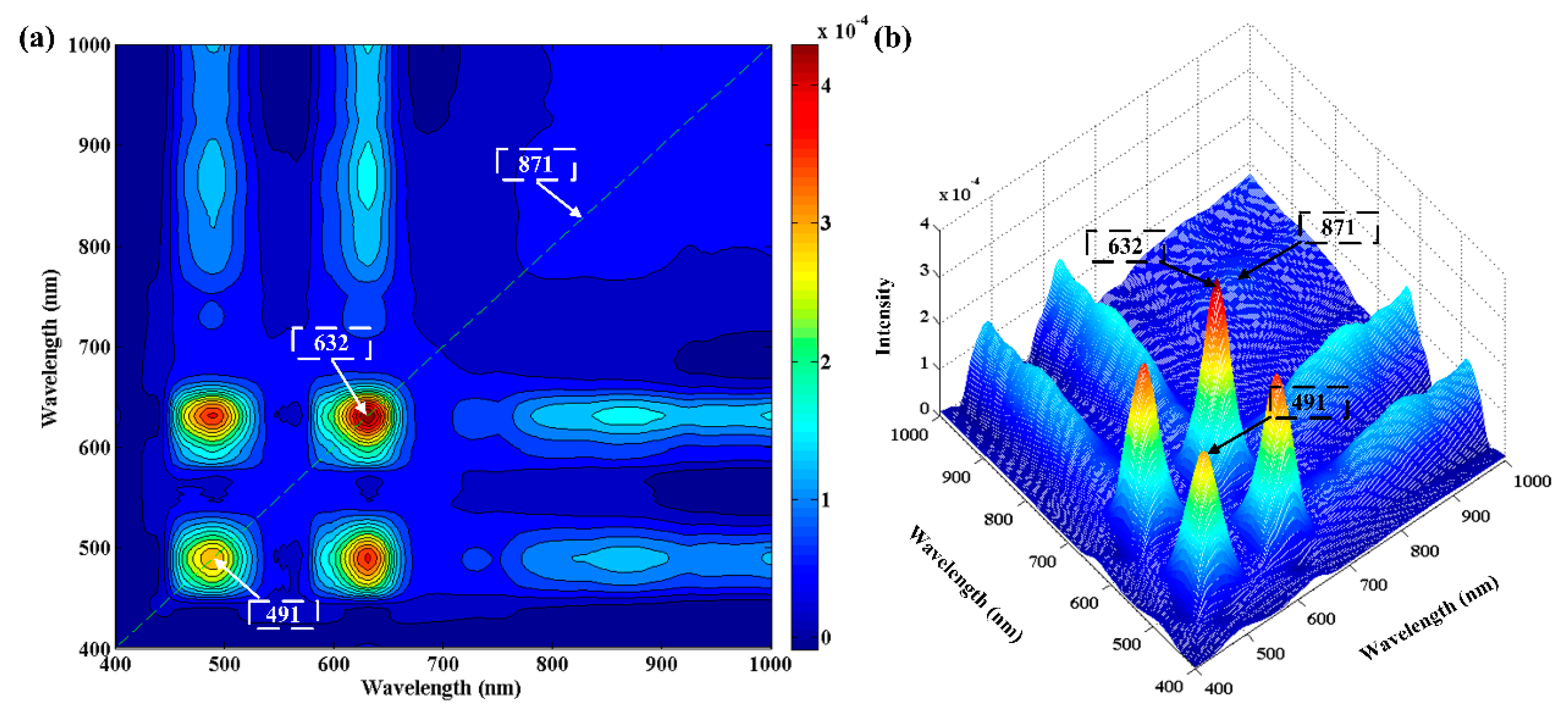
3.3.2. Two-Dimensional Correction Spectroscopy
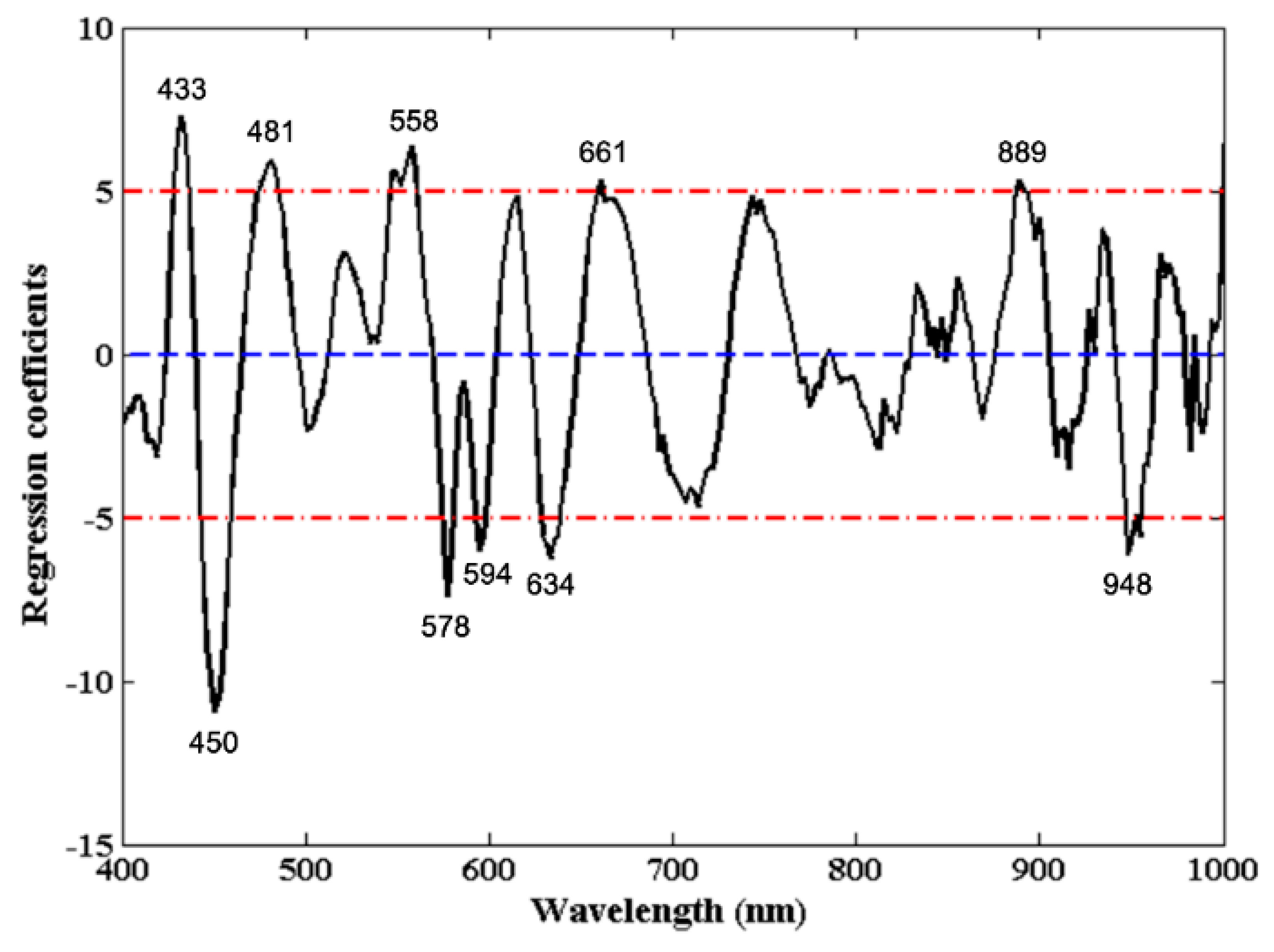
3.3.3. Regression Coefficients
3.4. Multispectral Models Development
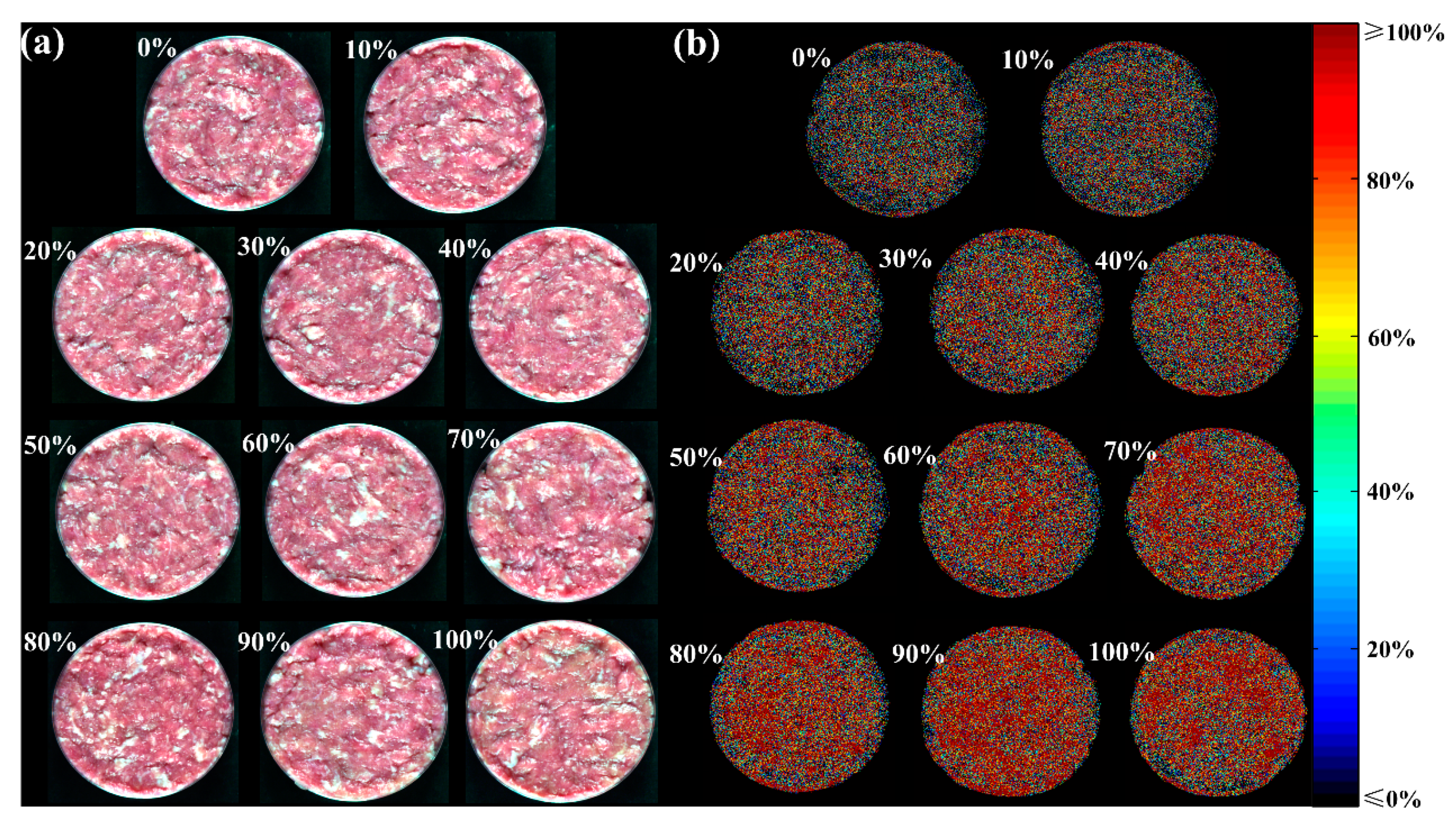
3.5. Visualization of the Adulteration Levels
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Smet, S.; Vossen, E. Meat: The balance between nutrition and health. A review. Meat Sci. 2016, 120, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, S.; Julkapli, N.M.; Yehye, W.A.; Basirun, W.J. Identification of meat origin in food products—A review. Food Control 2016, 68, 379–390. [Google Scholar] [CrossRef]
- Ballin, N.Z. Authentication of meat and meat products. Meat Sci. 2010, 86, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Alamprese, C.; Amigo, J.M.; Casiraghi, E.; Engelsen, S.B. Identification and quantification of turkey meat adulteration in fresh, frozen-thawed and cooked minced beef by FT-NIR spectroscopy and chemometrics. Meat Sci. 2016, 121, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Shao, H.; Yang, Y. Discussion on the edible safety of pork jowl meat. Shandong Food Ferment 2013, 03, 56–58. (In Chinese) [Google Scholar]
- Al-Rashood, K.A.; Abdel-Moety, E.M.; Rauf, A.; Abou-Shaaban, R.R.; Al-Khamis, K.I. Triacylglycerols-profiling by high performance liquid chromatography: A tool for detection of pork fat (lard) in processed foods. J. Liq. Chromatogr. Relat. Technol. 1995, 18, 2661–2673. [Google Scholar] [CrossRef]
- He, H.; Hong, X.; Feng, Y.; Wang, Y.; Ying, J.; Liu, Q.; Qin, Y.; Zhou, X.; Wang, D. Application of quadruple multiplex PCR detection for beef, duck, mutton and pork in mixed meat. J. Food Nutr. Res. 2015, 3, 392–398. [Google Scholar] [CrossRef] [Green Version]
- Black, C.; Chevallier, O.P.; Cooper, K.M.; Haughey, S.A.; Balog, J.; Takats, Z.; Elliott, C.T.; Cavin, C. Rapid detection and specific identification of offals within minced beef samples utilising ambient mass spectrometry. Sci. Rep. 2019, 9, 6295. [Google Scholar] [CrossRef] [Green Version]
- Dahimi, O.; Rahim, A.A.; Abdulkarim, S.M.; Hassan, M.S.; Hashari, S.B.Z.; Mashitoh, A.S.; Saadi, S. Multivariate statistical analysis treatment of DSC thermal properties for animal fat adulteration. Food Chem. 2014, 158, 132–138. [Google Scholar] [CrossRef]
- Macedo-Silva, A.; Barbosa, S.F.C.; Alkmin, M.D.G.A.; Vaz, A.J.; Shimokomaki, M.; Tenuta-Filho, A. Hamburger meat identification by dot-ELISA. Meat Sci. 2000, 56, 189–192. [Google Scholar] [CrossRef]
- Alamprese, C.; Casale, M.; Sinelli, N.; Lanteri, S.; Casiraghi, E. Detection of minced beef adulteration with turkey meat by UV–Vis, NIR and MIR spectroscopy. LWT Food Sci. Technol. 2013, 53, 225–232. [Google Scholar] [CrossRef]
- Pieszczek, L.; Czarnik-Matusewicz, H.; Daszykowski, M. Identification of ground meat species using near-infrared spectroscopy and class modeling techniques—Aspects of optimization and validation using a one-class classification model. Meat Sci. 2018, 139, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Morsy, N.; Sun, D.W. Robust linear and non-linear models of NIR spectroscopy for detection and quantification of adulterants in fresh and frozen-thawed minced beef. Meat Sci. 2013, 93, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Downey, G.; O’Donnell, C.P. Detection of adulteration in fresh and frozen beefburger products by beef offal using mid-infrared ATR spectroscopy and multivariate data analysis. Meat Sci. 2014, 96, 1003–1011. [Google Scholar] [CrossRef]
- Zhao, M.; Downey, G.; O’Donnell, C.P. Dispersive Raman spectroscopy and multivariate data analysis to detect offal adulteration of thawed beefburgers. J. Agric. Food Chem. 2015, 63, 1433–1441. [Google Scholar] [CrossRef]
- Velioglu, H.M.; Sezer, B.; Bilge, G.; Baytur, S.E.; Boyaci, I.H. Identification of offal adulteration in beef by laser induced breakdown spectroscopy (LIBS). Meat Sci. 2018, 138, 28–33. [Google Scholar] [CrossRef]
- Nolasco-Perez, I.M.; Rocco, L.A.; Cruz-Tirado, J.P.; Pollonio, M.A.; Barbon, S.; Barbon, A.P.A.; Barbin, D.F. Comparison of rapid techniques for classification of ground meat. Biosyst. Eng. 2019, 183, 151–159. [Google Scholar] [CrossRef]
- Jiang, X.; Zhao, T.; Liu, X.; Zhou, Y.; Shen, F.; Ju, X.; Liu, X.; Zhou, H. Study on Method for On-Line Identification of Wheat Mildew by Array Fiber Spectrometer. Spectrosc. Spectr. Anal. 2018, 38, 3729–3735. [Google Scholar]
- Ge, Y.; Atefi, A.; Zhang, H.; Miao, C.; Ramamurthy, R.K.; Sigmon, B.; Yang, J.; Schnable, J.C. High-throughput analysis of leaf physiological and chemical traits with VIS–NIR–SWIR spectroscopy: A case study with a maize diversity panel. Plant Methods 2019, 15, 66. [Google Scholar] [CrossRef] [Green Version]
- ElMasry, G.; Sun, D.W.; Allen, P. Chemical-free assessment and mapping of major constituents in beef using hyperspectral imaging. J. Food Eng. 2013, 117, 235–246. [Google Scholar] [CrossRef]
- Barbin, D.F.; Sun, D.W.; Su, C. NIR hyperspectral imaging as non-destructive evaluation tool for the recognition of fresh and frozen–thawed porcine longissimus dorsi muscles. Innov. Food Sci. Emerg. Technol. 2013, 18, 226–236. [Google Scholar] [CrossRef]
- Feng, C.H.; Makino, Y.; Oshita, S.; Martín, J.F.G. Hyperspectral imaging and multispectral imaging as the novel techniques for detecting defects in raw and processed meat products: Current state-of-the-art research advances. Food Control 2018, 84, 165–176. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Wang, W. Feasibility of the Detection of Carrageenan Adulteration in Chicken Meat Using Visible/Near-Infrared (Vis/NIR) Hyperspectral Imaging. Appl. Sci. 2019, 9, 3926. [Google Scholar] [CrossRef] [Green Version]
- Ropodi, A.I.; Pavlidis, D.E.; Mohareb, F.; Panagou, E.Z.; Nychas, G.J. Multispectral image analysis approach to detect adulteration of beef and pork in raw meats. Food Res. Int. 2015, 67, 12–18. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Hyperspectral imaging in tandem with multivariate analysis and image processing for non-invasive detection and visualization of pork adulteration in minced beef. Anal. Methods 2015, 7, 7496–7502. [Google Scholar] [CrossRef]
- Wu, D.; Shi, H.; He, Y.; Yu, X.; Bao, Y. Potential of hyperspectral imaging and multivariate analysis for rapid and non-invasive detection of gelatin adulteration in prawn. J. Food Eng. 2013, 119, 680–686. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Sun, D.W.; ElMasry, G.; Allen, P. Fast detection and visualization of minced lamb meat adulteration using NIR hyperspectral imaging and multivariate image analysis. Talanta 2013, 103, 130–136. [Google Scholar] [CrossRef]
- Ropodi, A.I.; Panagou, E.Z.; Nychas, G.J.E. Multispectral imaging (MSI): A promising method for the detection of minced beef adulteration with horsemeat. Food Control 2017, 73, 57–63. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S.; Liu, S. Assessment of visible near-infrared hyperspectral imaging as a tool for detection of horsemeat adulteration in minced beef. Food Bioprocess Technol. 2015, 8, 1054–1062. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Y.; Wei, W.; Peng, Y. Detection of adulteration with duck meat in minced lamb meat by using visible near-infrared hyperspectral imaging. Meat Sci. 2019, 149, 55–62. [Google Scholar] [CrossRef]
- Menesatti, P.; Zanella, A.; D’Andrea, S.; Costa, C.; Paglia, G.; Pallottino, F. Supervised multivariate analysis of hyper-spectral NIR images to evaluate the starch index of apples. Food Bioprocess Technol. 2009, 2, 308–314. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Dou, X.; Yuan, B.; Zhao, H.; Yin, G.; Ozaki, Y. Generalized two-dimensional correlation spectroscopy. Sci. China Ser. B 2004, 47, 257–266. [Google Scholar] [CrossRef]
- Noda, I. Generalized two-dimensional correlation method applicable to infrared, Raman, and other types of spectroscopy. Appl. Spectrosc. 1993, 47, 1329–1336. [Google Scholar] [CrossRef]
- He, H.J.; Wu, D.; Sun, D.W. Potential of hyperspectral imaging combined with chemometric analysis for assessing and visualising tenderness distribution in raw farmed salmon fillets. J. Food Eng. 2014, 126, 156–164. [Google Scholar] [CrossRef]
- Kapper, C.; Klont, R.E.; Verdonk, J.M.A.J.; Urlings, H.A.P. Prediction of pork quality with near infrared spectroscopy (NIRS): 1. Feasibility and robustness of NIRS measurements at laboratory scale. Meat Sci. 2012, 91, 294–299. [Google Scholar] [CrossRef]
- Mamani-Linares, L.W.; Gallo, C.; Alomar, D. Identification of cattle, llama and horse meat by near infrared reflectance or transflectance spectroscopy. Meat Sci. 2012, 90, 378–385. [Google Scholar] [CrossRef]
- Liu, Y.L.; Chen, Y.R. Two-dimensional correlation spectroscopy study of visible and near-infrared spectral variations of chicken meats in cold storage. Appl. Spectrosc. 2000, 54, 1458–1470. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.W. Application of visible and near infrared hyperspectral imaging for non-invasively measuring distribution of water-holding capacity in salmon flesh. Talanta 2013, 116, 266–276. [Google Scholar] [CrossRef]
- Bowker, B.; Hawkins, S.; Zhuang, H. Measurement of water-holding capacity in raw and freeze-dried broiler breast meat with visible and near-infrared spectroscopy. Poult. Sci. 2014, 93, 1834–1841. [Google Scholar] [CrossRef]
- Zhao, M.; O’Donnell, C.; Downey, G. Detection of offal adulteration in beefburgers using near infrared reflectance spectroscopy and multivariate modelling. J. Near Infrared Spectrosc. 2013, 21, 237. [Google Scholar] [CrossRef]
- Barbin, D.F.; ElMasry, G.; Sun, D.W.; Allen, P. Non-destructive determination of chemical composition in intact and minced pork using near-infrared hyperspectral imaging. Food Chem. 2013, 138, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Gondal, M.A.; Seddigi, Z.S.; Nasr, M.M.; Gondal, B. Spectroscopic detection of health hazardous contaminants in lipstick using laser induced breakdown spectroscopy. J. Hazard. Mater. 2010, 175, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Bilge, G.; Velioglu, H.M.; Sezer, B.; Eseller, K.E.; Boyaci, I.H. Identification of meat species by using laser-induced breakdown spectroscopy. Meat Sci. 2016, 119, 118–122. [Google Scholar] [CrossRef] [PubMed]

| Pretreatments | LVs | Calibration | Cross-Validation | Prediction | ||||
|---|---|---|---|---|---|---|---|---|
| Rc2 | RMSEC | Rcv2 | RMSECV | Rp2 | RMSEP | RPD | ||
| None | 12 | 0.9866 | 3.64% | 0.9779 | 4.71% | 0.9458 | 7.50% | 4.27 |
| Normalization | 14 | 0.9898 | 3.18% | 0.9821 | 4.24% | 0.9493 | 7.25% | 4.41 |
| SNV | 12 | 0.9864 | 3.68% | 0.9787 | 4.60% | 0.9549 | 7.04% | 4.54 |
| MSC | 13 | 0.9878 | 3.50% | 0.9801 | 4.45% | 0.9536 | 7.06% | 4.53 |
| SNV + Detrend | 12 | 0.9870 | 3.59% | 0.9797 | 4.51% | 0.9512 | 7.47% | 4.28 |
| 1st derivative | 13 | 0.9886 | 3.36% | 0.9815 | 4.30% | 0.9528 | 7.19% | 4.45 |
| 2nd derivative | 14 | 0.9896 | 3.23% | 0.9797 | 4.50% | 0.9425 | 10.18% | 3.14 |
| Method | Number | LVs | Calibration | Cross-Validation | Prediction | ||||
|---|---|---|---|---|---|---|---|---|---|
| Rc2 | RMSEC | Rcv2 | RMSECV | Rp2 | RMSEP | RPD | |||
| 2D-COS | 3 | 3 | 0.2283 | 27.78% | 0.1920 | 28.45% | 0.2720 | 27.45% | 1.17 |
| PC loadings | 9 | 6 | 0.8981 | 10.09% | 0.8344 | 10.80% | 0.7475 | 17.31% | 1.85 |
| RC | 10 | 9 | 0.9610 | 6.24% | 0.9520 | 6.93% | 0.9063 | 13.93% | 2.30 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, H.; Cheng, F.; Shi, M. Rapid Identification and Visualization of Jowl Meat Adulteration in Pork Using Hyperspectral Imaging. Foods 2020, 9, 154. https://doi.org/10.3390/foods9020154
Jiang H, Cheng F, Shi M. Rapid Identification and Visualization of Jowl Meat Adulteration in Pork Using Hyperspectral Imaging. Foods. 2020; 9(2):154. https://doi.org/10.3390/foods9020154
Chicago/Turabian StyleJiang, Hongzhe, Fengna Cheng, and Minghong Shi. 2020. "Rapid Identification and Visualization of Jowl Meat Adulteration in Pork Using Hyperspectral Imaging" Foods 9, no. 2: 154. https://doi.org/10.3390/foods9020154
APA StyleJiang, H., Cheng, F., & Shi, M. (2020). Rapid Identification and Visualization of Jowl Meat Adulteration in Pork Using Hyperspectral Imaging. Foods, 9(2), 154. https://doi.org/10.3390/foods9020154





