Effects of Nonstarch Genetic Modifications on Starch Structure and Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Maize Grain Starch Purification
2.3. Starch Extraction, Debranching, and Characterization by Size-Exclusion Chromatography (SEC)
2.4. Differential Scanning Calorimetry (DSC)
2.5. In Vitro Starch Digestion
2.6. Fitting Amylose and Number Amylopectin CLDs
2.7. Statistical Analysis
3. Results
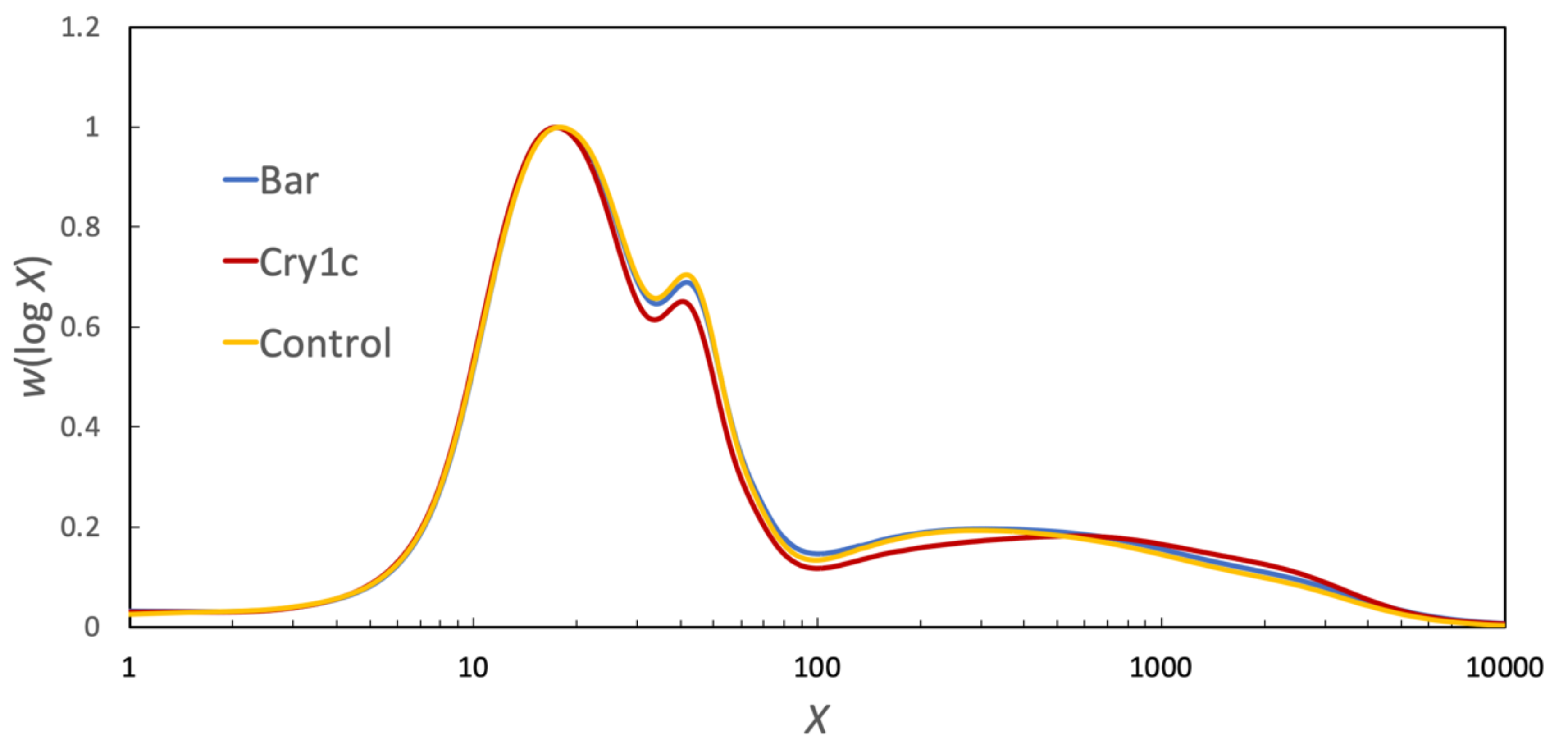
3.1. Chain-Length Distributions
3.2. Gelatinization Properties
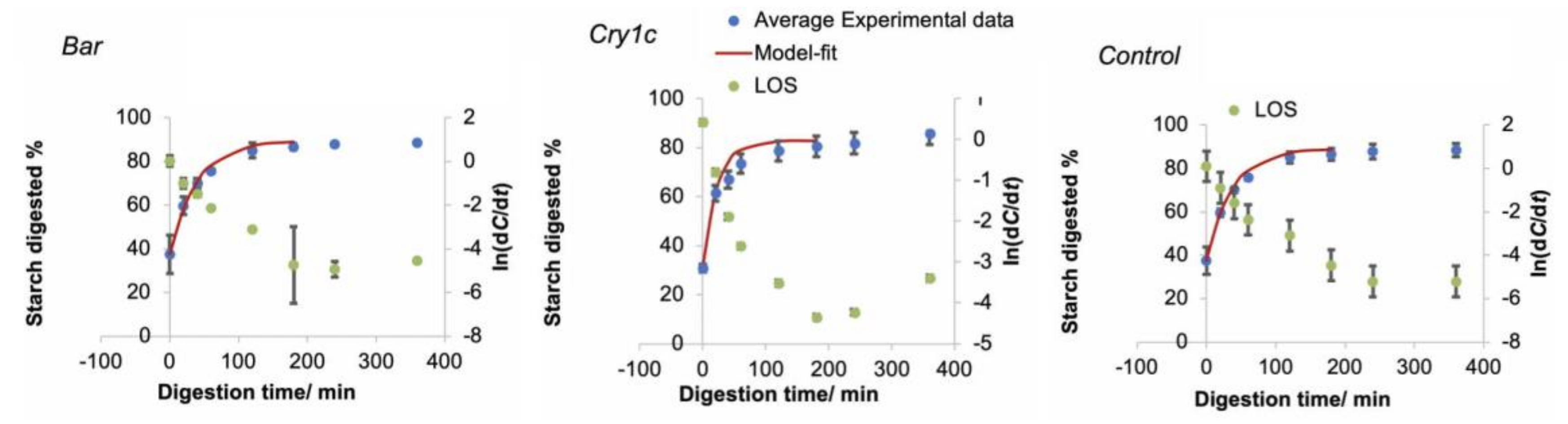
3.3. In Vitro Digestibility
3.4. Parameterizing Amylopectin and Amylose CLD with Biosynthesis Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Miguel, C.; Marum, L. An epigenetic view of plant cells cultured In Vitro: Somaclonal variation and beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar] [CrossRef] [PubMed]
- Villegas-Aguilar, M.D.C.; Fernández-Ochoa, Á.; Cádiz-Gurrea, M.D.L.L.; Pimentel-Moral, S.; Lozano-Sánchez, J.; Arráez-Román, D.; Segura-Carretero, A. Pleiotropic biological effects of dietary phenolic compounds and their metabolites on energy metabolism, inflammation and aging. Molecules 2020, 25, 596. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.M.; Wagler, T.N.; Quijada, P.; Doebley, J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006, 38, 594–597. [Google Scholar] [CrossRef]
- Kermicle, J. Pleiotropic effects on seed development of the indeterminate gametophyte gene in maize. Am. J. Bot. 1971, 58, 1–7. [Google Scholar] [CrossRef]
- Yu, W.; Li, H.; Zou, W.; Tao, K.; Zhu, J.; Gilbert, R.G. Using starch molecular fine structure to understand biosynthesis-structure-property relations. Trends Food Sci. Technol. 2019, 86, 530–536. [Google Scholar] [CrossRef]
- Noda, T.; Takahata, Y.; Sato, T.; Suda, I.; Morishita, T.; Ishiguro, K.; Yamakawa, O. Relationships between chain length distribution of amylopectin and gelatinization properties within the same botanical origin for sweet potato and buckwheat. Carbohydr. Polym. 1998, 37, 153–158. [Google Scholar] [CrossRef]
- Tao, K.; Li, C.; Yu, W.; Gilbert, R.G.; Li, E. How amylose molecular fine structure of rice starch affects pasting and gelatinization properties. Carbohydr. Polym. 2019, 204, 24–31. [Google Scholar] [CrossRef]
- Mallik, A.U. Allelopathy: Advances, challenges and opportunities. In Allelopathy in Sustainable Agriculture and Forestry; Zeng, R.S., Mallik, A.U., Luo, S.M., Eds.; Springer: New York, NY, USA, 2008; pp. 25–38. [Google Scholar] [CrossRef]
- Wu, D.X.; Shu, Q.Y.; Wang, Z.H.; Cui, H.R.; Xia, Y.W. Quality variations in transgenic rice with a synthetic cry1Ab gene from Bacillus thuringiensis. Plant. Breed. 2002, 121, 198–202. [Google Scholar] [CrossRef]
- Du, D.; Geng, C.; Zhang, X.; Zhang, Z.; Zheng, Y.; Zhang, F.; Lin, Y.; Qiu, F. Transgenic maize lines expressing a cry1C* gene are resistant to insect pests. Plant. Mol. Biol. Report 2014, 32, 549–557. [Google Scholar] [CrossRef]
- Wang, K.; Hasjim, J.; Wu, A.C.; Henry, R.J.; Gilbert, R.G. Variation in amylose fine structure of starches from different botanical sources. J. Agric. Food Chem. 2014, 62, 4443–4453. [Google Scholar] [CrossRef]
- Li, C.; Godwin, I.D.; Gilbert, R.G. Diurnal changes in sorghum leaf starch molecular structure. Plant. Sci. 2015, 239, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wambugu, P.W.; Zhang, B.; Wu, A.C.; Henry, R.J.; Gilbert, R.G. The biosynthesis, structure and gelatinization properties of starches from wild and cultivated African rice species (Oryza barthii and Oryza glaberrima). Carbohydr. Polym. 2015, 129, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Vilaplana, F.; Gilbert, R.G. Characterization of branched polysaccharides using multiple-detection size separation techniques. J. Sep. Sci. 2010, 33, 3537–3554. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, B.; Ruiter, G.; Visser, J.; Lonsmann Iversen, J. Determination of first order rate constants by natural logarithm of the slope plot exemplified by analysis of Aspergillus niger in batch culture. Biotechnol. Lett. 2003, 25, 565–571. [Google Scholar] [CrossRef]
- Butterworth, P.J.; Warren, F.J.; Grassby, T.; Patel, H.; Ellis, P.R. Analysis of starch amylolysis using plots for first-order kinetics. Carbohydr. Polym. 2012, 87, 2189–2197. [Google Scholar] [CrossRef]
- Yu, W.; Tao, K.; Gilbert, R.G. Improved methodology for analyzing relations between starch digestion kinetics and molecular structure. Food Chem. 2018, 264, 284–292. [Google Scholar] [CrossRef]
- Nada, S.S.; Zou, W.; Li, C.; Gilbert, R.G. Parameterizing amylose chain-length distributions for biosynthesis-structure-property relations. Anal. Bioanal. Chem. 2017, 409, 6813–6819. [Google Scholar] [CrossRef]
- Wu, A.C.; Morell, M.K.; Gilbert, R.G. A parameterized model of amylopectin synthesis provides key insights into the synthesis of granular starch. PLoS ONE 2013, 8, e65768. [Google Scholar] [CrossRef]
- Noda, T.; Nishiba, Y.; Sato, T.; Suda, I. Properties of starches from several low-amylose rice cultivars. Cereal Chem. 2003, 80, 193–197. [Google Scholar] [CrossRef]
- Cui, R.; Oates, C.G. The effect of retrogradation on enzyme susceptibility of sago starch. Carbohydr. Polym. 1997, 32, 65–72. [Google Scholar] [CrossRef]
- Shi, Y.-C.; Seib, P.A. The structure of four waxy starches related to gelatinization and retrogradation. Carbohydr. Res. 1992, 227, 131–145. [Google Scholar] [CrossRef]
- Gidley, M.J.; Cooke, D.; Darke, A.H.; Hoffmann, R.A.; Russell, A.L.; Greenwell, P. Molecular order and structure in enzyme-resistant retrograded starch. Carbohydr. Polym. 1995, 28, 23–31. [Google Scholar] [CrossRef]
- Syahariza, Z.A.; Sar, S.; Tizzotti, M.; Hasjim, J.; Gilbert, R.G. The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem. 2013, 136, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Hellmich, R.L.; Hellmich, K.A. Use and impact of Bt maize. Nat. Educ. Knowl. 2012, 3, 4. [Google Scholar]
- Poste, G.; Rosenberg, M.; Westpheling, J. Streptomyces: A host for heterologous gene expression. Curr. Opin. Biotechnol. 1991, 2, 674–681. [Google Scholar] [CrossRef]
- Payne, G.F.; Delacruz, N.; Coppella, S.J. Improved production of heterologous protein from Streptomyces lividans. Appl. Microbiol. Biotechnol. 1990, 33, 395–400. [Google Scholar] [CrossRef]
- Amrhein, V.; Greenland, S.; McShane, B. Retire statistical significance. Nature 2019, 567, 305–307. [Google Scholar] [CrossRef]
- Zou, W.; Sissons, M.; Warren, F.J.; Gidley, M.J.; Gilbert, R.G. Compact structure and proteins of pasta retard In Vitro digestive evolution of branched starch molecular structure. Carbohydr. Polym. 2016, 152, 441–449. [Google Scholar] [CrossRef]
- Li, C.; Powell, P.O.; Gilbert, R.G. Recent progress toward understanding the role of starch biosynthetic enzymes in the cereal endosperm. Amylase 2017, 1, 59–74. [Google Scholar] [CrossRef]
- Gao, M.; Fisher, D.K.; Kim, K.N.; Shannon, J.C.; Guiltinan, M.J. Independent genetic control of maize starch-branching enzymes IIa and IIb. Isolation and characterization of a Sbe2a cDNA. Plant. Physiol. 1997, 114, 69–78. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Kaeppler, H.F.; Rhee, Y. Epigenetic aspects of somaclonal variation in plants. Plant. Mol. Biology 2000, 43, 179–188. [Google Scholar] [CrossRef] [PubMed]
| Properties | Bar | Cry1c | Control |
|---|---|---|---|
| To (°C) | 43 ± 1 a | 43.6 ± 0.4 a | 44 ± 1 a |
| Tp (°C) | 54 ± 1 a | 51.1 ± 0.3 b | 53 ± 1 a |
| Tc (°C) | 65 ± 2a | 64.3 ± 0.2 a | 64.6 ± 0.2 a |
| Tc-To (°C) | 26 ± 2 a | 21 ± 1 a | 27 ± 1 a |
| ∆H (J g−1) | 3.6 ± 0.8 a | 3.0 ± 0.1 a | 3.7 ± 0.5 a |
| Digestion Parameter | Bar | Cry1c | Control |
|---|---|---|---|
| k/min–1 | 0.025 ± 0.001 a | 0.043 ± 0.000 b | 0.027 ± 0.005 a |
| C∞/% | 10 ± 1 a | 17 ± 4 b | 11 ± 3 a |
| - | Amylose Content (%) | βAm,i/10−3 | βAm,ii/10−3 | βAm,iii/10−3 | hAm,ii | hAm,iii | hAp,iii | hAp,ii | βAp,ii/10−2 | βAp,iii/10−2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Bar | 26 ± 1 a | 8.8 ± 0.2 a | 2.8 ± 0.1 a | 1.2 ± 0.1 ab | 10.6 ± 0.1 a | 13.6 ± 0.1 a | 1.01 ± 0.01 a | 0.35 ± 0.01 a | 3.89 ± 0.04 a | 4.6 ± 0.2 a |
| Cry1c | 25 ± 1 a | 6.6 ± 0.1 b | 2.5 ± 0.1 b | 1.2 ± 0.03 b | 11.3 ± 0.1 b | 14.2 ± 0.1 b | 1.02 ± 0.00 b | 0.32 ± 0.01 b | 4.1 ± 0.1 b | 4.6 ± 0.2 a |
| Control | 25.3 ± 0.3 a | 9.1 ± 0.1 a | 3.1 ± 0.2 c | 1.5 ± 0.1a | 10.4 ± 0.1 a | 13.4 ± 0.2 a | 1.02 ± 0.01 b | 0.34 ± 0.01 ab | 3.8 ± 0.1 a | 4.4 ± 0.03 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Du, D.; Wu, A.C.; Bai, Y.; Wu, P.; Li, C.; Gilbert, R.G. Effects of Nonstarch Genetic Modifications on Starch Structure and Properties. Foods 2020, 9, 222. https://doi.org/10.3390/foods9020222
Yu S, Du D, Wu AC, Bai Y, Wu P, Li C, Gilbert RG. Effects of Nonstarch Genetic Modifications on Starch Structure and Properties. Foods. 2020; 9(2):222. https://doi.org/10.3390/foods9020222
Chicago/Turabian StyleYu, Shiyao, Dengxiang Du, Alex C. Wu, Yeming Bai, Peng Wu, Cheng Li, and Robert G. Gilbert. 2020. "Effects of Nonstarch Genetic Modifications on Starch Structure and Properties" Foods 9, no. 2: 222. https://doi.org/10.3390/foods9020222
APA StyleYu, S., Du, D., Wu, A. C., Bai, Y., Wu, P., Li, C., & Gilbert, R. G. (2020). Effects of Nonstarch Genetic Modifications on Starch Structure and Properties. Foods, 9(2), 222. https://doi.org/10.3390/foods9020222






