Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters
Abstract
:1. Introduction
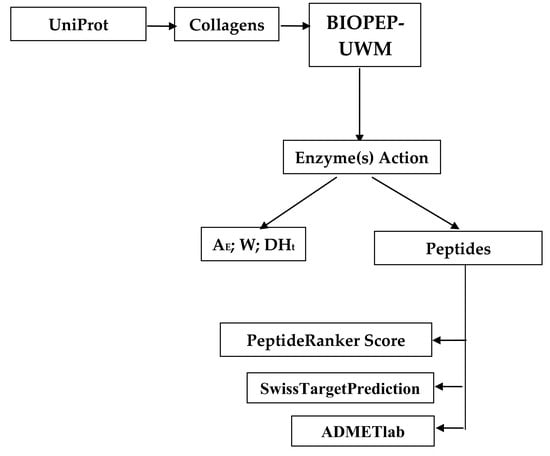
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACE | angiotensin converting enzyme (EC 3.4.15.1) |
| ACEi | angiotensin converting enzyme inhibitor |
| ADMET | absorption, distribution, metabolism, excretion, toxicity |
| BIOPEP-UWM | database of protein and bioactive peptide sequences (http://www.uwm.edu.pl/biochemia) [22] |
| A | depending on the context: alanine or the frequency of the occurrence of bioactive fragments in a protein sequence [22] described by the following equation: A = a/N where a—the number of fragments with a given activity, N—the number of amino acid residues in a protein |
| AE | The frequency of release of fragments with a given activity by selected enzymes [22,23] described by the following equation: AE = d/N where d-the number of peptides with a given activity (e.g., ACE inhibitors) released by a given enzyme (e.g., trypsin) N-the number of amino acid residues in a protein |
| B | stem bromelain (bromelain) (EC 3.4.22.32) |
| CaMPDE | calmodulin-dependent cyclic nucleotide phosphodiesterase (EC 3.1.4.17) |
| CH | collagen hydrolysate |
| Ch | chymotrypsin (EC 3.4.21.1) |
| ChEMBL | ChEMBL database of molecules with drug-like properties (https://www.ebi.ac.uk/chembl) [66] |
| CoA | coenzyme A |
| Complement factor B | alternative-complement-pathway C3/C5 convertase (EC 3.4.21.47) |
| COX-2 | cyclooxygenase-2 (prostaglandin-endoperoxide synthase; EC 1.14.99.1) |
| DHt | theoretical degree of hydrolysis (%) [22] described by the following equation: DHt = (d/D) × 100% where d—the number of hydrolyzed peptide bonds in a protein/peptide chain D—the total number of peptide bonds in a protein/peptide chain |
| DPP-III | dipeptidyl peptidase III (EC 3.4.14.4) |
| DPP-IV | dipeptidyl peptidase IV (EC 3.4.14.5) |
| DPP-IVi | dipeptidyl peptidase IV inhibitor |
| F | depending on the context: phenylalanine or ficin (EC 3.4.22.3) |
| G | glycine |
| HMG-CoA | 3-hydroxy-3-methyl-glutaryl-CoA reductase (EC 1.1.1.34) |
| HT | high throughput technology |
| Hyp | hydroxyl-proline/hydroxyl-lysine |
| IC50 | concentration of a peptide corresponding to its half-inhibitory effect (μM) |
| IUBMB | International Union of Biochemistry and Molecular Biology |
| LD50 | dose of a compound which kills 50% tested animals (mg × kg −1) |
| P | proline |
| Pap | papain (EC 3.4.22.2) |
| Pep | pepsin (EC 3.4.23.1) |
| QSAR | Quantitative Structure-Activity Relationship [19] |
| SMILES | Simplified Molecular Input Line Entry Specification [25] |
| T | depending on the context: treonine or trypsin (EC 3.4.21.4) |
| T1/2 | theoretical half-life time (h) |
| VD | volume distribution (L × kg −1) |
| W | depending on the context: tryptophan or the relative frequency of release of fragments with a given activity by selected enzymes [22] described by the following equation: W = AE/A where AE—the frequency of release of fragments with a given activity by selected enzymes (see above) A—the frequency of bioactive fragments occurrence in a protein sequence (see above). |
References
- Song, H.; Li, B. Beneficial Effects of Collagen Hydrolysate: A Review on Recent Developments. Biomed. J. Sci. Tech. Res. 2017, 1, 458–461. [Google Scholar] [CrossRef]
- Zdzieblik, D.; Oesser, S.; Baumstark, M.W.; Gollhoffer, M.; König, D. Collagen peptide supplementation in combination with resistance training improves body composition and increases muscle strength in elderly sarcopenic men: A randomised controlled trial. Br. J. Nutr. 2015, 114, 1237–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Offengenden, M.; Chakrabarti, S.; Wu, J. Chicken collagen hydrolysates differentially mediate anti-inflammatory activity and type I collagen synthesis on human dermal fibroblasts. Food Sci. Hum. Wellness 2018, 7, 138–147. [Google Scholar] [CrossRef]
- Raman, M.; Gopakumar, K. Fish Collagen and its Applications in Food and Pharmaceutical Industry: A Review. EC Nutr. 2018, 13, 752–767. [Google Scholar]
- Gómez-Guillén, M.C.; Giménez, B.; López-Caballero, M.E.; Montero, M.P. Functional and bioactive properties of collagen and gelatin from alternative sources: A review. Food Hydrocoll. 2011, 8, 1813–1827. [Google Scholar] [CrossRef] [Green Version]
- Sylvipriya, K.S.; Kumar, K.K.; Bhat, A.R.; Kumar, B.D.; John, A.; Iakshmanan, P. Collagen: Animal sources and biomedical application. J. Appl. Pharm. Sci. 2015, 5, 123–127. [Google Scholar] [CrossRef] [Green Version]
- León-López, A.; Vargas-Torres, A.; Zeugolis, D.I.; Aguirre-Álvarez, G. Hydrolyzed Collagen-Sources and Applications. Molecules 2019, 24, 4031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive peptides from muscle sources: Meat and fish. Nutrients 2011, 3, 765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein-originating peptides as tastants–Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Darewicz, M.; Minkiewicz, P. Peptides Derived from Foods as Supportive Diet Components in the Prevention of the Metabolic Syndrome. Compr. Rev. Food Sci. Food Saf. 2018, 17, 63–81. [Google Scholar] [CrossRef] [Green Version]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef] [Green Version]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-Originating ACE Inhibitors, Including Antihypertensive Peptides, as Preventive Food Components in Blood Pressure Reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Girija, A.R. Peptide nutraceuticals. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 157–181. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Miciński, J.; Darewicz, M.; Bucholska, J. Biological and chemical databases for research into the composition of animal source foods. Food Rev. Int. 2013, 29, 321–351. [Google Scholar] [CrossRef]
- Agyei, D.; Bambarandage, E.; Udenigwe, C.C. The role of bioinformatics in the discovery of bioactive peptides. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, F., Valeris, P., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 337–344. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocol Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [Green Version]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Protasiewicz, M.; Mogut, D. Chemometrics and cheminformatics in the analysis of biologically active peptides from food sources. J. Funct. Foods 2015, 16, 334–351. [Google Scholar] [CrossRef]
- He, R.; Ma, H.; Zhao, W.; Qu, W.; Zhao, J.; Luo, L.; Zhu, W. Modeling the QSAR of ACE-Inhibitory Peptides with ANN and Its Applied Illustration. Int. J. Pept. 2012, 620609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trend Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [Green Version]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Dziuba, J.; Michalska, J. Bovine meat proteins as potential precursors of biologically active peptides—A computational study based on the BIOPEP database. Food Sci. Technol. Int. 2011, 7, 39–45. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. Annotation of peptide structures using SMILES and other chemical codes–practical solutions. Molecules 2017, 22, 2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Wang, N.-N.; Dong, J.; Deng, Y.-H.; Zhu, M.-F.; Wen, M.; Yao, Z.-J.; Lu, A.-P.; Wang, J.-B.; Cao, D.-S. ADME properties evaluation in drug discovery: Prediction of Caco-2 cell permeability using a combination of NSGA-II and boosting. J. Chem. Inf. Model. 2016, 56, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.-N.; Huang, C.; Dong, J.; Yao, Z.-J.; Zhu, M.-F.; Deng, Z.-K.; Lv, B.; Lu, A.-P.; Chen, A.F.; Cao, D.-S. Predicting human intestinal absorption with modified random forest approach: A comprehensive evaluation of molecular representation, unbalanced data, and applicability domain issues. RSC Adv. 2017, 7, 19007–19018. [Google Scholar] [CrossRef] [Green Version]
- Kerns, E.H.; Di, L. Drug-like properties: Concepts, Structure Design and Methods: From ADME to Toxicity Optimization; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Lei, T.; Li, Y.; Song, Y.; Li, D.; Sun, H.; Hou, T. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J. Cheminform. 2016, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Wang, N.-N.; Yao, Z.-J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef]
- Panjaitan, F.C.A.; Gomez, H.L.R.; Chang, Y.-W. In Silico Analysis of Bioactive Peptides Released from Giant Grouper (Epinephelus lanceolatus) Roe Proteins Identified by Proteomics Approach. Molecules 2018, 23, 2910. [Google Scholar] [CrossRef] [Green Version]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980S–988S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeirssen, V.; van der Bent, A.; Van Camp, J.; van Amerongen, A.; Verstraete, W. A quantitative in silico analysis calculates angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie 2004, 86, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Nassa, M.; Anand, P.; Jain, A.; Chhabra, A.; Jaiswal, A.; Malhotra, U.; Rani, V. Analysis of human collagen sequences. Bioinformation 2012, 8, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Fu, U.; Therkildsen, M.E.; Aluko, R.E.; Lametsch, R. Exploration of collagen recovered from animal by-products as a precursor of bioactive peptides: Successes and challenges. Crit. Rev. Food Sci. Nutr. 2019, 59, 2011–2027. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 1–14. [Google Scholar] [CrossRef]
- Yu, D.; Wang, C.; Song, Y.; Zhu, J.; Zhang, X. Discovery of Novel Angiotensin-Converting Enzyme Inhibitory Peptides from Todarodes pacificus and Their Inhibitory Mechanism: In Silico and In Vitro Studies. Int. J. Mol. Sci. 2019, 20, 4159. [Google Scholar] [CrossRef] [Green Version]
- Darewicz, M.; Borawska, J.; Pliszka, M. Carp proteins as a source of bioactive peptides—An in silico approach. Czech. J. Food Sci. 2016, 34, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Borawska, J.; Darewicz, M.; Vegarud, G.E.; Iwaniak, A.; Minkiewicz, P. Ex vivo digestion of carp muscle tissue – ACE inhibitory and antioxidant activities of obtained hydrolysates. Food Funct. 2015, 6, 211–218. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [Green Version]
- Iwaniak, A.; Mogut, D. Metabolic Syndrome-Preventive Peptides Derived from Milk Proteins and Their Presence in Cheeses: A Review. Appl. Sci. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Hrynkiewicz, M.; Bucholska, J.; Darewicz, M. Hybrid Approach in the Analysis of Bovine Milk Protein Hydrolysates as a Source of Peptides Containing Di- and Tripeptide Bitterness Indicators. Pol. J. Food Nutr. Sci. 2020, 70, 139–150. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Minkiewicz, P.; Bucholska, J.; Darewicz, M. Soybean (Glycine max) Protein Hydrolysates as Sources of Peptide Bitter-Tasting Indicators: An Analysis Based on Hybrid and Fragmentomic Approaches. Appl. Sci. 2020, 10, 2514. [Google Scholar] [CrossRef] [Green Version]
- Song, J.J.; Wang, Q.; Du, M.; Ji, X.M.; Mao, X.Y. Identification of dipeptidyl peptidase-IV inhibitory peptides from mare whey protein hydrolysates. J. Dairy Sci. 2017, 100, 6885–6894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Zhang, F.; Ma, Z.; Chen, S.; Ding, G.; Tian, X.; Feng, R. Isolation and identification of the angiotensin-I converting enzyme (ACE) inhibitory peptides derived from cottonseed protein: Optimization of hydrolysis conditions. Int. J. Food Prop. 2019, 22, 1296–1309. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.-C.; Alashi, A.M.; Aluko, R.E.; Pan, B.S.; Chang, Y.-W. Antihypertensive properties of tilapia (Oreochromis spp.) frame and skin enzymatic protein hydrolysates. Food Nutr. Res. 2017, 61, 1391666. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Wu, W.; Zhu, M.; Xiao, Z. In silico assessment of the potential of the patatin as a precursor of bioactive peptides. J. Food Biochem. 2016, 40, 366–370. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Toldrá, F. The relevance of dipeptides and tripeptides in the bioactivity and taste of dry-cured ham. Food Prod. Process. Nutr. 2019, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Byun, H.-G.; Kim, S.-K. Structure and activity of angiotensin I converting enzyme inhibitory peptides derived from Alaskan Pollack skin. J. Biochem. Mol. Biol. 2001, 35, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Suetsuna, K. Isolation and characterization of angiotensin I-converting enzyme inhibitor dipeptides derived from Allium sativum L (garlic). J. Nutr. Biochem. 1998, 9, 415–419. [Google Scholar] [CrossRef]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef]
- Nogata, Y.; Nagamine, T.; Yanaka, M.; Ohta, H. Angiotensin I Converting Enzyme Inhibitory Peptides Produced by Autolysis Reactions from Wheat Bran. J. Agric. Food Chem. 2009, 57, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Hu, J.; Aita, D.Q.; Maruyama, S. Angiotensin I-converting enzyme inhibitory activity and insulin secretion stimulative activity of fermented fish sauce. J. Biosci. Bioeng. 2003, 95, 496–499. [Google Scholar] [CrossRef]
- Sentandreu, M.A.; Toldrá, F. Evaluation of ACE inhibitory activity of dipeptides generated by the action of porcine muscle dipeptidyl peptidases. Food Chem. 2007, 102, 511–515. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Meisel, H. Lactokinins: Whey protein-derived ACE inhibitory peptides. Nahrung 1999, 43, 165–167. [Google Scholar] [CrossRef]
- Välimaa, A.-L.; Mäkinen, S.; Mattila, P.; Marnila, P.; Pihlanto, A.; Mäki, M.; Hiidenhovi, J. Fish and fish side streams are valuable sources of high-value components. Food Qual. Saf. 2019, 3, 209–226. [Google Scholar] [CrossRef] [Green Version]
- Pripp, A.H.; Isaksson, T.; Stepaniak, L.; Sørhaug, T. Quantitative structure-activity relationship modeling of ACE-inhibitory peptides derived from milk proteins. Eur. Food Res. Technol. 2004, 219, 579–583. [Google Scholar] [CrossRef]
- Yi, Y.; Lv, Y.; Zhang, L.; Yang, Y.; Shi, Q. High throughput identification of antihypertensive peptides from fish proteome datasets. Mar. Drugs 2020, 16, 365. [Google Scholar] [CrossRef] [Green Version]
- Udenigwe, C.C. Bioinformatic approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Gogktug, A.N.; Chai, S.C.; Chen, T. Data analysis approaches in high throughput screening. In Drug Discovery; El-Shemy, H., Ed.; IntechOpen: Rijeka, Croatia, 2013; pp. 201–226. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Young, J.F.; Løkke, M.M.; Lametsch, R.; Aluko, R.E.; Therkildsen, M. Revalorisation of bovine collagen as a potential precursor of angiotensin I-converting enzyme (ACE) inhibitory peptides based on in silico and in vitro protein digestions. J. Funct. Foods 2016, 24, 196–206. [Google Scholar] [CrossRef]
- Rajendran, S.R.C.K.; Mason, B.; Udenigwe, C.C. Peptidomics of peptic digest of selected potato tuber proteins: Post-translational modifications and limited cleavage specificity. J. Agric. Food Chem. 2016, 64, 2432–2437. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Martinez, M.; Gonzalez de Mejia, E.; García-Lara, S.; Aguilar, O.; Lopez-Castillo, L.M.; Otero-Pappatheodorou, J.T. Antiproliferative effect of peptide fractions isolated from a quality protein maize, a white hybrid maize, and their derived peptides on hepatocarcinoma human HepG2 cells. J. Funct. Foods 2017, 34, 36–48. [Google Scholar] [CrossRef]
- Mojica, L.; Luna-Vital, D.A.; Gonzalez de Mejia, E. Black bean peptides inhibit glucose uptake in Caco-2 adenocarcinoma cells by blocking the expression and translocation pathway of glucose transporters. Toxicol. Rep. 2018, 5, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Dókus, L.E.; Yousef, M.; Bánóczi, Z. Modulators of calpain activity: Inhibitors and activators as potential drugs. Expert Opin. Drug Discov. 2020, 15, 471–486. [Google Scholar] [CrossRef]
- Wang, D.; DuBois, R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Sun, H.; Yu, F.-B.; Li, B.; Zhang, Y.; Zhu, Y.-T. The role of cyclooxygenase-2 in colorectal cancer. Int. J. Med. Sci. 2020, 17, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Liu, Y.; Zhang, Z.; Yang, G.-Y. Significance of complement system in ischemic stroke: A comprehensive review. Aging Dis. 2019, 10, 429–462. [Google Scholar] [CrossRef] [Green Version]
- Noris, M.; Donadelli, R.; Remuzzi, G. Autoimmune abnormalities of the alternative complement pathway in membranoproliferative glomerulonephritis and C3 glomerulopathy. Pediatr. Nephrol. 2019, 4, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.; Rojas-Quintero, J.; Cano, C.; Pérez, J.L.; Ramírez, P.; Carrasquero, R.; Torres, W.; Espinoza, C.; Chacín-González, M.; Bermúdez, V. Neprilysin: A potential therapeutic target of arterial hypertension? Curr. Cardiol. Rev. 2020, 16, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Książczyk, M.; Lelonek, M. Angiotensin receptor/neprilysin inhibitor—A breakthrough in chronic heart failure therapy: Summary of subanalysis on PARADIGM-HF trial findings. Heart Fail. Rev. 2020, 25, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.-L.; Schimmel, P.; Ewalt, K.L. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem. Sci. 2004, 29, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Saiyasit, N.; Sripetchwandee, J.; Chattipakorn, N.; Chattipakorn, S.C. Potential roles of neurotensin on cognition in conditions of obese-insulin resistance. Neuropeptides 2018, 72, 12–22. [Google Scholar] [CrossRef]
- Herrera-Ruiz, D.; Knipp, G.T. Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharmaceut. Sci. 2003, 92, 691–714. [Google Scholar] [CrossRef]
- Hessler, G.; Baringhaus, K.-H. Artificial intelligence in drug design. Molecules 2020, 23, 2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Mayorga, K.; Madariaga-Mazon, A.; Medina-Franco, J.L.; Maggiora, G. The impact of chemoinformatics on drug discovery in the pharmaceutical industry. Expert Opin. Drug Discov. 2020, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Naveja, J.J.; Rico-Hidalgo, M.P.; Medina-Franco, J.L. Analysis of a large food chemical database: Chemical space, diversity, and complexity. F1000 Res. 2018, 7, 993. [Google Scholar] [CrossRef]
- Santibáñez-Morán, M.G.; Rico-Hidalgo, M.P.; Manallack, D.T.; Medina-Franco, J.L. The acid/base profile of a large food chemical database. Mol. Inf. 2019, 38, 1800171. [Google Scholar] [CrossRef]
- Santibáñez-Morán, M.G.; Medina-Franco, J.L. Analysis of the acid/base profile of natural products from different sources. Mol. Inf. 2020, 39, 1900099. [Google Scholar] [CrossRef]
- Yu, Z.; Fan, Y.; Zhao, W.; Ding, L.; Li, J.; Liu, L. Novel angiotensin-converting enzyme inhibitory peptides derived from Oncorhynchus mykiss nebulin: Virtual screening and in silico molecular docking study. J. Food Sci. 2018, 83, 2375–2383. [Google Scholar] [CrossRef]
- Zhao, W.; Xue, S.; Yu, Z.; Ding, L.; Li, J.; Liu, J. Novel ACE inhibitors derived from soybean proteins using in silico and in vitro studies. J. Food Biochem. 2019, 43, e12975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, D.; Yu, Z.; Ding, L.; Liu, J. Novel membrane peptidase inhibitory peptides with activity against angiotensin converting enzyme and dipeptidyl peptidase IV identified from hen eggs. J. Funct. Foods 2020, 64, 103649. [Google Scholar] [CrossRef]
- Fan, Y.; Yu, Z.; Zhao, W.; Ding, L.; Zheng, F.; Li, J.; Liu, J. Identification and molecular mechanism of angiotensin-converting enzyme inhibitory peptides from Larimichthys crocea titin. Food Sci. Hum. Wellness 2020. [Google Scholar] [CrossRef]
- Capecchi, A.; Awale, M.; Probst, D.; Reymond, J.-L. PubChem and ChEMBL beyond Lipinski. Mol. Inf. 2019, 38, 1900016. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Matsui, T. Intestinal absorption of small peptides: A review. Int. J. Food Sci. Technol. 2019, 54, 1942–1948. [Google Scholar] [CrossRef] [Green Version]


| Source of Collagen | Major A (A ≥ 0.500) | Moderate A (A = 0.1002−0.499) | Minor A (A = 0.001−0.099) | ||
|---|---|---|---|---|---|
| cow (Bos taurus) | 0.834 ah 1 0.854 dpp | 0.214 am;re 0.238 at | 0.002 apr;35pd | 0.003 im | 0.006 ren |
| 0.009 ne | 0.014 emb | 0.037 che | |||
| 0.041 glui | 0.057 inh | 0.059 ao | |||
| 0.069 st | 0.073 dpp3 | ||||
| pig (Sus scrofa) | 0.846 dpp 0.847 ah | 0.216 am;re 0.238 at | 0.001 emb | 0.002 apr;35pd | 0.003 im |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.038 che | 0.042 glui | 0.058 inh | |||
| 0.060 ao | 0.076 dpp3 | ||||
| sheep (Ovis aries) | 0.833 ah 0.845 dpp | 0.198 am;re 0.215 at | 0.001 emb;im | 0.002 35pd | 0.003 apr |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.033 che | 0.045 glui | 0.052 inh | |||
| 0.059 ao | 0.073 dpp3 | ||||
| chicken (Gallus gallus) | 0.843 ah 0.852 dpp | 0.210 am;re 0.223 at | 0.001 lig | 0.002 apr;35pd;emb | 0.003 im |
| 0.006 ren;is;st | 0.008 ne | 0.040 glui;che | |||
| 0.057 inh | 0.061 ao | 0.075 dpp3 | |||
| duck (Anas platyrhynchos platyrhynchos) | 0.847 ah 0.870 dpp | 0.240 at 0.210 am;re | 0.002 hypl;35pd | 0.003 lig | 0.004 emb |
| 0.007 apr | 0.011 ren | 0.033 glui;che | |||
| 0.046 inh | 0.057 ao | 0.083 dpp3 | |||
| horse (Equus caballus) | 0.843 ah;dpp | 0.215 am;re 0.238 at | 0.001 emb | 0.00235pd | 0.003 apr;im |
| 0.006 ren | 0.007 st | 0.009 ne | |||
| 0.037 che | 0.041 glui | 0.058 inh;ao | |||
| 0.073 dpp3 | |||||
| salmon (Salmo salar) | 0.798 dpp 0.799 ah | 0.170 at;am;re | 0.002 emb;is | 0.00335pd;lig;apr | 0.005 st |
| 0.008 ne;ren | 0.023 che | 0.029 glui | |||
| 0.053 ao | 0.074 dpp3 | ||||
| rainbow trout (Oncorhynchus mykiss) | 0.810 dpp 0.846 ah | 0.161 am 0.162 re 0.181 at | 0.001 hypl | 0.002 lig | 0.003 emb;35pd |
| 0.006 apr | 0.008 st | 0.009 ne | |||
| 0.013 ren | 0.015 glui | 0.018 che | |||
| 0.020 inh | 0.034 ao | 0.071 dpp3 | |||
| goat (Capra hircus) | 0.842 ah 0.849 dpp | 0.213 am;re 0.273 at | 0.001 emb | 0.002 apr;35pd | 0.003 im |
| 0.006 ren | 0.007 st | 0.009 ne | |||
| 0.037 che | 0.041 glui | 0.057 inh | |||
| 0.058 ao | 0.074 dpp3 | ||||
| rabbit (Oryctolagus cuniculus) | 0.834 ah 0.849 dpp | 0.199 am;re 0.215 at | 0.001 emb;im | 0.002 35pd | 0.003 apr |
| 0.006 ren | 0.007 st | 0.008 ne | |||
| 0.034 che | 0.045 glui | 0.052 inh | |||
| 0.059 ao | 0.073 dpp3 | ||||
| turkey (Meleagris gallopavo) | 0.822 dpp 0.841 ah | 0.192 am;re 0.220 at | 0.002 hypl | 0.003 lig;35pd | 0.004 emb |
| 0.007 apr | 0.009 ne | 0.012 st | |||
| 0.014 ren | 0.027 che | 0.030 glui | |||
| 0.041 inh | 0.059 ao | 0.082 dpp3 | |||
| Source of Collagen | Enzyme | AE | W | DHt (%) |
|---|---|---|---|---|
| cow (Bos taurus) | B 1 | 0.158 dpp2 0.097 ah 0.042 am;at;re 0.014 dpp3 0.005 glui 0.001 ao | 0.25 im 0.195 am;re 0.191 dpp3 0.186 dpp 0.175 at 0.120 glui 0.115 ah 0.113 ren 0.024 ao | 55.55 |
| F | 0.122 dpp 0.071 ah 0.021 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.143 dpp 0.118 am;re 0.114 dpp3 0.105 at 0.084 ah 0.024 ao | 45.69 | |
| Pap | 0.151 dpp 0.105 ah 0.018 am;at;re 0.008 dpp3 0.001 ren | 1.000 lig 0.226 ren 0.176 dpp 0.124 ah 0.104 dpp3 0.084 am;re 0.076 at 0.012 ao | 46.11 | |
| Pep | 0.004 ah;dpp 0.002 ren 0.001 dpp3 | 0.339 ren 0.019 dpp3 0.004 ah;dpp | 4.38 | |
| T | 0.002 ah 0.001 dpp;ao;at | 0.012 ao 0.006 at 0.003 ah 0.002 dpp | 8.75 | |
| Pep + T | 0.013 ah;dpp 0.005 dpp3 0.002 ren 0.001 at | 0.338 ren 0.067 dpp3 0.016 dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.141 ah 0.107 dpp 0.055 re;am;at 0.029 dpp3 0.005 ne 0.003 ren 0.002 ao 0.001 35pd | 0.667 35pd 0.544 ne 0.452 ren 0.409 dpp3 0.256 re;at 0.230 am 0.161 ah 0.125 dpp 0.036 ao | 45.63 | |
| pig (Sus scrofa) | B | 0.158 dpp 0.098 ah 0.041 am;at;re 0.013 dpp3 0.006 glui 0.001 ao | 0.250 im 0.192 am;re 0.187 dpp 0.176 dpp3 0.174 at 0.133 glui 0.116 ah 0.111 ren 0.024 ao | 55.43 |
| F | 0.124 dpp 0.721 ah 0.025 am;at;re 0.008 dpp3 0.001 ao;ren | 0.117 re 0.106 at 0.085 ah 0.117 am 0.024 ao 0.147 dpp 0.222 ren 0.102 dpp3 | 45.91 | |
| Pap | 0.152 dpp 0.109 ah 0.018 am;at;re 0.007 dpp3 0.001 ren | 1.000 lig 0.222 ren 0.180 dpp 0.128 ah 0.093 dpp3 0.084 am;re 0.076 at 0.012 ao | 45.83 | |
| Pep | 0.003 ah;dpp 0.002 ren | 0.333 ren 0.009 dpp3 0.004 ah 0.003 dpp | 4.41 | |
| T | 0.001 at;ah | 0.012 ao 0.006 at 0.002 ah | 8.76 | |
| Pep + T | 0.013 ah 0.012 dpp 0.004 dpp3 0.002 ren 0.001 at | 0.333 ren 0.056 dpp3 0.015 ah 0.014 dpp 0.012 ao 0006 at | 13.17 | |
| Pep + T + Ch | 0.147 ah 0.109 dpp 0.055 am;at;re 0.031 dpp3 0.005 ne 0.003 ao;ren 0.001 35pd | 0.667 35pd 0.583 ne 0.444 ren 0.407 dpp3 0.256 am;re 0.232 at 0.174 ah 0.128 dpp 0.100 st 0.005 ao | 45.83 | |
| sheep (Ovis aries) | B | 0.158 dpp 0.095 ah 0.041 am;at;re 0013 dpp3 0.005 glui 0.001 ao | 0.500 im 0.205 am;re 0.189 at 0.186 dpp 0.180 dpp3 0.114 ah 0.107 glui 0.023 ao | 55.69 |
| F | 0.118 dpp 0.070 ah 0.023 am;at;re 0.007 dpp3 0.003 ao | 0.139 dpp 0.127 ren 0.115 am;re 0.106 at 0.095 dpp3 0.084 ah 0.047 ao | 45.69 | |
| Pap | 0.145 dpp 0.103 ah 0.019 am;at;re 0.006 dpp3 | 1.000 lig 0.175 dpp 0.127 ren 0.124 ah 0.094 am;re 0.087 at 0.075 dpp3 0.016 glui 0.012 ao | 45.62 | |
| Pep | 0.003 ah;dpp 0.001 dpp3;ren | 0.255 ren 0.019 dpp3 0.004 ah 0.003 dpp | 4.55 | |
| T | 0.001 ah | 0.012 ao 0.003 at 0.002 ah | 9.10 | |
| Pep + T | 0.012 ah;dpp 0.005 dpp3 0.001 ren | 0.255 ren 0.066 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.003 at | 13.65 | |
| Pep + T + Ch | 0.137 ah 0.107 dpp 0.050 am;at;re 0.030 dpp3 0.004 ne 0.003 ren 0.002 ao 0.001 35pd | 0.667 35pd 0.509 ren 0.494 ne 0.410 dpp3 0.250 am;re 0.231 at 0.165 ah 0.127 dpp 0.101 st 0.036 ao | 46.04 | |
| chicken (Gallus gallus) | B | 0.154 dpp 0.096 ah 0.043 am;at,re 0.012 dpp3 0.006 glui 0.001 ao;im | 0.500 im 0.203 am;re 0.185 at 0.180 dpp 0.159 dpp3 0.141 glui 0.114 ah 0.111 ren 0.023 ao | 55.59 |
| F | 0.120 dpp 0.068 ah 0.025 am;at;re 0.008 dpp3 0.003 ao 0.001 ren | 0.222 ren 0.141 dpp 0.116 am;re 0.107 at 0.103 dpp3 0.080 ah 0.047 ao | 44.97 | |
| Pap | 0.158 dpp 0.108 ah 0.019 am;at;re 0.007 dpp3 0.001 ren | 0.500 lig 0.250 im 0.222 ren 0.185 dpp 0.128 ah 0.094 dpp3 0.090 am;re 0.082 at 0.012 ao | 46.43 | |
| Pep | 0.003 ah;dpp 0.002 ren | 0.333 ren 0.009 dpp3 0.003 ah;dpp | 4.55 | |
| T | 0.002 ah 0.001 dpp | 0.012 ao 0.003 ah;at 0.001 dpp | 8.60 | |
| Pep + T | 0.013 ah;dpp 0.004 dpp3 0.002 ren | 0.333 ren 0.056 dpp3 0.016 ah 0.012 ao 0.003 at | 13.15 | |
| Pep + T + Ch | 0.143 ah 0.112 dpp 0.055 am;at;re 0.033 dpp3 0.005 ao 0.004 ne 0.003 ren 0.001 35pd | 0.667 35pd 0.500 ne 0.444 ren 0.439 dpp3 0.259 am;re 0.237 at 0.170 ah 0.131 dpp 0.125 st 0.082 ao | 45.38 | |
| duck (Anas platyrhynchos platyrhynchos) | B | 0.146 dpp 0.091 ah 0.034 am;at;re 0.015 dpp3 0.005 ren 0.003 glui 0.002 ao; st 0.001 35pd | 0.652 35pd 0.533 hyp 0.397 ren 0.247 st 0.178 dpp3 0.172 dpp 0.163 am;re 0.144 at 0.104 ah 0.096 glui 0.041 ao | 57.79 |
| F | 0.123 dpp 0.083 ah 0.025 am;at;re 0.012 dpp3 0.005 ren 0.002 st 0.001 ao; hyp,35pd | 0.533 hyp 0.397 ren 0.348 35pd 0.248 st 0.146 dpp 0.130 dpp3 0.126 re 0.119 am 0.105 at 0.095 ah 0.014 ao | 48.57 | |
| Pap | 0.154 dpp 0.113 ah 0.019 am;at;re 0.010 dpp3 0.004 ren 0.002 ao; st 0.001 hyp;lig;35pd | 0.533 hyp 0.348 35pd 0.336 ren 0.258 lig 0.182 dpp 0.161 st 0.130 ah 0.122 dpp3 0.092 am;re 0.082 at 0.027 ao | 47.87 | |
| Pep | 0.005 ah 0.004 dpp 0.001 dpp3;ren | 0.070 ren 0.010 dpp3 0.006 ah 0.005 dpp | 5.58 | |
| T | 0.001 ah;at;dpp | 0.034 at 0.001 ah;dpp | 8.83 | |
| Pep + T | 0.013 ah;dpp 0.002 at;dpp3 0.001 ren | 0.070 ren 0.018 dpp3 0.016 dpp 0.015 ah 0.006 at | 14.41 | |
| Pep + T + Ch | 0.024 dpp 0.023 ah 0.003 at 0.001 ao;dpp3;glui;reg;ren | 0.070 ren 0.028 dpp 0.027 ah;ao 0.025 glui 0.013 at 0.010 dpp3 0.004 re | 20.99 | |
| horse (Equus caballus) | B | 0.155 dpp 0.096 ah 0.042 am;at;re 0.013 dpp3 0.006 glui 0.001 ao;im;ren | 0.25 im 0.193 am;re 0.184 dpp 0.181 dpp3 0.175 at 0.115 glui 0.114 ah 0.113 ren 0.024 ao | 55.55 |
| F | 0.124 dpp 0.074 ah 0.026 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.147 dpp 0.123 am;re 0.112 at 0.104 dpp3 0.088 ah 0.012 ao | 45.83 | |
| Pap | 0.151 dpp 0.108 ah 0.018 am;at;re 0.007 dpp3 0.001 ao;lig;ren | 1.000 lig 0.226 ren 0.179 dpp 0.128 ah 0.095 dpp3 0.084 am;re 0.077 at 0.012 ao | 45.90 | |
| Pep | 0.004 ah 0.003 dpp 0.021 ren 0.001 dpp3 | 0.339 ren 0.010 dpp3 0.004 ah 0.003 dpp | 4.31 | |
| T | 0.001 dpp;ah;ao,at | 0.012 ao 0.006 at 0.002 ah 0.001 dpp | 8.82 | |
| Pep + T | 0.013 ah 0.012 dpp 0.004 dpp3 0.002 ren 0.001 ao;at | 0.339 ren 0.058 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.022 dpp 0.019 ah 0.005 dpp3 0.003 at;ren 0.002 ao 0.001 glui; st;35pd | 0.452 ren 0.333 35pd 0.101 st 0.067 dpp3 0.036 ao 0.026 dpp 0.023 ah 0.014 glui 0.012 at | 17.85 | |
| salmon (Salmo salar) | B | 0.143 dpp 0.093 ah 0.036 am;at;re 0.008 glui 0.006 dpp3 0.003 ren 0.002 ao 0.001 st;35pd | 0.333 ren 0.250 35pd 0.223 glui 0.219 at 0.218 re 0.198 at 0.179 dpp 0.143 st 0.116 ah 0.086 dpp3 0.040 ao | 57.85 |
| F | 0.115 dpp 0.071 ah 0.023 am;at;re 0.006 dpp3 0.004 ao;ren 0.001 HMGi;35pd | 1.000 HMGi 0.417 ren 0.250 35pd 0.144 dpp 0.139 am 0.139 re 0.126 at 0.089 ah 0.086 dpp3 0.067 ao | 47.48 | |
| Pap | 0.144 dpp 0.110 ah 0.0203 am;at;re 0.006 dpp3 0.003 ren 0.001 ao;HMGi;35pd | 1.000 HMGi 0.333 ren 0.250 35pd 0.181 dpp 0.138 ah 0.122 am;re 0.112 at 0.076 dpp3 0.027 ao | 46.91 | |
| Pep | 0.003 ah 0.002 dpp 0.001 ao;dpp3;ren | 0.167 ren 0.013 ao 0.010 dpp3 0.003 ah;dpp | 4.07 | |
| T | 0.002 ah 0.001 dpp | 0.003 ah 0.002 dpp | 8.77 | |
| Pep + T | 0.010 dpp 0.009 ah 0.006 dpp3 0.001 ren | 0.167 ren 0.076 dpp3 0.012 dpp 0.001 ah | 12.83 | |
| Pep + T + Ch | 0.019 ah 0.018 dpp 0.008 dpp3 0.003 ao 0.002 at;ren 0.001 glui; is;35pd | 0.333 is 0.250 ren;35pd 0.105 dpp3 0.053 ao 0.024 ah 0.023 dpp 0.021 glui 0.011 at | 18.93 | |
| rainbow trout (Oncorhynchus mykiss) | B | 0.142 dpp 0.096 ah 0.034 am;at;re 0.013 dpp3 0.003 ao;glui;ren 0.002 35pd 0.001 hyp; st | 0.500 35pd 0.467 hyp 0.236 ren 0.209 am 0.208 re 0.187 at 0.178 dpp3 0.143 glui 0.113 ah 0.093 st 0.082 ao | 61.89 |
| F | 0.118 dpp 0.098 ah 0.027 am;at;re 0.008 dpp3 0.003 ao 0.002 ren 0.001 HMGi; st;35pd | 1.000 HMGi 0.333 35pd 0.173 ren 0.171 re 0.168 am 0.149 at 0.147 dpp 0.115 ah;dpp3 0.093 st 0.082 ao | 52.44 | |
| Pap | 0.153 dpp 0.137 ah 0.025 am;at;re 0.008 dpp3 0.004 ren 0.002 ao;glui;35pd 0.001 HMGi; hyp; st;lig | 1.000 HMGi 0.500 35pd 0.467 hyp 0.318 lig 0.291 ren 0.190 dpp 0.162 ah 0.153 am;re 0.138 at 0.115 dpp3 0.105 glui 0.093 st 0.060 ao | 51.46 | |
| Pep | 0.003 ah;dpp 0.001 dpp3;ren | 0.055 ren 0.010 dpp3 0.004 ah;dpp | 5.48 | |
| T | 0.002 at 0.001 ah | 0.008 at 0.001 ah | 8.93 | |
| Pep + T | 0.011 ah;dpp 0.002 at;dpp3 0.001 ren | 0.055 ren 0.031 dpp3 0.014 dpp 0.013 ah 0.008 at | 14.40 | |
| Pep + T + Ch | 0.022 ah 0.020 dpp 0.002 ao;at;dpp3 0.001 glui; st;re;ren | 0.093 st 0.060 ao;ren 0.033 glui 0.026 ah 0.021 dpp3 0.008 at 0.004 re | 21.61 | |
| go at (Capra hircus) | B | 0.156 dpp 0.096 ah 0.042 am;at;re 0.014 dpp3 0.005 glui 0.001 ao;im;ren | 0.250 im 0.195 am;re 0.189 dpp3 0.184 dpp 0.175 at 0.114 ah 0.113 ren 0.104 glui 0.024 ao | 55.55 |
| F | 0.120 dpp 0.069 ah 0.024 am;at;re 0.008 dpp3 0.001 ao;ren | 0.226 ren 0.114 dpp 0.113 dpp3 0.111 am;re 0.010 at 0.082 ah 0.024 ao | 45.55 | |
| Pap | 0.152 dpp 0.105 ah 0.019 am;at;re 0.008 dpp3 0.001 ao;lig;ren | 1.000 lig 0.226 ren 0.179 dpp 0.125 ah 0.103 dpp3 0.088 am;re 0.079 at 0.012 ao | 46.18 | |
| Pep | 0.040 ah;dpp 0.002 ren 0.001 dpp3 | 0.339 ren 0.019 dpp3 0.004 ah;dpp | 4.38 | |
| T | 0.01 ah;ao;at;dpp | 0.117 ao 0.006 at 0.02 ah 0.001 dpp | 8.75 | |
| Pep + T | 0.013 ah;dpp 0.005 dpp3 0.002 ren 0.001 ao;at | 0.339 ren 0.067 dpp3 0.015 ah;dpp 0.012 ao 0.006 at | 13.13 | |
| Pep + T + Ch | 0.023 dpp 0.021 ah 0.006 dpp3 0.003 am;at;re 0.001 glui; st;35pd | 0.452 ren 0.333 35pd 0.101 st 0.008 dpp3 0.045 ao 0.027 dpp 0.025 ah 0.015 glui 0.012 at | 18.06 | |
| rabbit (Oryctolagus cuniculus) | B | 0.158 dpp 0.095 ah 0.041 am;at;re 0.013 dpp3 0.005 glui 0.001 ao;im | 0.500 im 0.205 am;re 0.189 at 0.186 dpp 0.179 dpp3 0.114 ah 0.097 glui 0.024 ao | 55.72 |
| F | 0.118 dpp 0.070 ah 0.023 am;at;re 0.007 dpp3 0.003 ao 0.001 ren | 0.134 dpp 0.127 ren 0.114 am;re 0.106 at 0.094 dpp3 0.084 ah 0.047 ao | 45.72 | |
| Pap | 0.149 dpp 0.103 ah 0.019 am;at;re 0.006 dpp3 0.001 ao;glui;lig;ren | 1.000 lig 0.175 dpp 0.127 ren 0.124 ah 0.094 am;re 0.087 at 0.075 dpp3 0.014 glui 0.012 ao | 45.66 | |
| Pep | 0.003 ah;dpp 0.001 ren;dpp3 | 0.255 ren 0.019 dpp3 0.004 ah 0.003 dpp | 4.55 | |
| T | 0.001 ah;ao;at;dpp | 0.012 ao 0.003 at 0.002 at 0.001 dpp | 9.10 | |
| Pep + T | 0.012 ah;dpp 0.005 dpp3 0.001 ao;at;ren | 0.255 ren 0.066 dpp3 0.015 ah 0.014 dpp 0.012 ao 0.003 at | 13.66 | |
| Pep + T + Ch | 0.021 dpp 0.020 ah 0.006 dpp3 0.003 ao 0.002 at;ren 0.001 glui; st;35pd | 0.382 ren 0.333 35pd 0.101 st 0.075 dpp3 0.057 ao 0.025 dpp 0.024 ah 0.014 glui 0.001 at | 18.83 | |
| turkey (Meleagris gallopavo) | B | 0.141 dpp 0.092 ah 0.032 am;at;re 0.016 dpp3 0.005 ren 0.004 ao 0.003 glui 0.002 st;35pd 0.001 hyp | 0.767 35pd 0.533 hyp 0.387 ren 0.196 dpp3 0.166 am;re 0.146 at 0.110 ah 0.080 glui 0.065 ao | 57.74 |
| F | 0.118 dpp 0.082 ah 0.022 re 0.021 am;at 0.011 dpp3 0.005 ren 0.003 st 0.002 ao 0.001 hyp;35pd | 0.533 hyp 0.500 35pd 0.336 ren 0.246 st 0.144 dpp 0.131 dpp3 0.115 reg 0.120 am 0.098 ah 0.097 at 0.039 ao | 48.82 | |
| Pap | 0.147 dpp 0.110 ah 0.019 am;at;re 0.011 dpp3 0.005 ren 0.003 ao 0.002 glui; st;35pd 0.001 hyp;lig | 0.533 hyp 0.500 35pd 0.336 ren 0.267 lig 0.179 dpp 0.131 ah;dpp3 0.123 st 0.100 re 0.087 at 0.087 glui 0051 ao | 47.52 | |
| Pep | 0.006 ah 0.005 dpp 0.001 ao;ren;dpp3 | 0.058 ren 0.018 dpp3 0.014 ao 0.007 ah 0.006 dpp | 6.18 | |
| T | 0.002 ah 0.001 dpp | 0.003 ah 0.002 dpp | 9.15 | |
| Pep + T | 0.017 dpp 0.016 ah 0.003 dpp3 0.001 ao;at;ren | 0.058 ren 0.037 dpp3 0.204 dpp 0.019 ah 0.014 ao 0.004 at | 15.33 | |
| Pep + T + Ch | 0.136 ah 0.106 dpp 0.047 re 0.046 am;at 0.030 dpp3 0.003 ne 0.002 ao 0.001 ren; st | 0.364 dpp3 0.330 ne 0.241 re 0.238 am 0.208 at 0.161 ah 0.129 dpp 0.066 st 0.058 ren 0039 ao | 45.69 |
| Peptide Sequence | Peptideranker Score | Collagen Source | Enzyme Applied |
|---|---|---|---|
| GF ACEi;DPP-IVi 1 | 0.994 | cow (Bos Taurus)/sheep (Ovis aries) | Pep 2 |
| SFACEi;DPP-IVi | 0.948 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus) | Pep |
| QF DPP-IVi | 0.946 | cow (Bos taurus)/chicken (Gallus gallus)/ | Pep |
| DF ACEi | 0.942 | horse (Equus caballus) | Pep |
| PGL ACEi | 0.855 | cow (Bos taurus)/pig (Sus scrofa)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) 3 | Pep |
| TF ACEi;DPP-IVi | 0.826 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) | Pep |
| GR ACEi | 0.766 | rainbow trout (Oncorhynchus mykiss) | T 4 |
| RL ACEi;DPP-IVi 4 | 0.626 | cow (Bos taurus)/pig (Sus scrofa)/sheep (Ovis aries)/chicken (Gallus gallus)/horse (Equus caballus)/salmon (Salmo salar) | Pep |
| DR DPP-IVi | 0.289 | horse (Equus caballus) | T |
| Peptide Sequence | Protein 1 | Protein 2 | Protein 3 |
|---|---|---|---|
| PGL | Dipeptidyl peptidase IV (UniProt—P27487 1; ChEMBL—CHEMBL284 2) Probability: 0.526 3 | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probability: 0.445 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.420 |
| RL | Neurotensin receptor 2 (UNiProt—O95665; ChEMBL—CHEMBL2514) Probability: 0.166 | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL573) Probability: 0.166 | Subtilisin/kexin type 6 (UniProt—P29122; ChEMBL—CHEMBL2951) Probability: 0.133 |
| GF | Oligopeptide transporter small intestine isoform (UniProt—P46059; ChEMBL—CHEMBL4605) Probability: 0.130 | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL389) Probability: 0.112 | Neprilysin (UniProt—P08473; ChEMBL—CHEMBL1944) Probability: 0.104 |
| SF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.081 | Oligopeptide transporter small intestine isoform (UniProt—P46059; ChEMBL—CHEMBL4605) Probability: 0.072 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.063 |
| TF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.238 | Tyrosyl-tRNA synthetase (UniProt—P54577; ChEMBL—CHEMBL3179) Probability: 0.143 | Cyclooxygenase-2 (UniProt—P35354; ChEMBL—CHEMBL230) Probability: 0.143 |
| QF | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probablity: 0.238 | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.230 | Tyrosyl-tRNA synthetase (UniProt—P54577; ChEMBL—CHEMBL3179) Probability: 0.140 |
| DF | Calpain 1 (UniProt—P07384; ChEMBL—CHEMBL3891) Probability: 0.150 | Angiotensin converting enzyme (UniProt—P12821; ChEMBL—CHEMBL1808) Probablity: 0.117 | Neprilysin (UniProt—P08473; ChEMBL—CHEMBL1944) Probability: 0.109 |
| DR | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL5731) Probability: 0.109 | Furin (UniProt—P09958; ChEMBL—CHEMBL2611) Probability: 0.109 | Integrin alpha-IIb/beta-3 (UniProt—P08514; P05106; ChEMBL—CHEMBL2093869) Probability: 0.109 |
| GR | Complement factor B (UniProt—P00751; ChEMBL—CHEMBL5731) Probability: 0.112 | Furin (UniProt—P09958; ChEMBL—CHEMBL2611) Probability: 0.104 | Neurotensin receptor 2 (UNiProt—O95665; ChEMBL—CHEMBL2514) Probability: 0.086 |
| Sequence | Rule of 5 | Log Caco-2 Permeability (Permeability Expressed in cm × s −1) | Human Intestinal Absorption Probability | VD 1 (L × kg −1) | T 1/2 2 (h) | LD50 3 of Acute Toxicity (mg × kg −1) |
|---|---|---|---|---|---|---|
| PGL | + | −5.643 | 0.309 | 0.149 | 0.701 | 1589 |
| RL | + | −6.203 | 0.398 | 0.160 | 1.184 | 45,963 |
| GF | + | −5.354 | 0.482 | 0.209 | 0.691 | 1344 |
| SF | + | −5.818 | 0.281 | 0.130 | 0.663 | 1513 |
| TF | + | −5.781 | 0.310 | 0.103 | 0.660 | 1385 |
| QF | + | −5.929 | 0.368 | 0.090 | 0.578 | 1592 |
| DF | + | −5.625 | 0.385 | 0.072 | 0.580 | 1672 |
| DR | + | −6.407 | 0.275 | 0.054 | 0.811 | 1494 |
| GR | + | −6.292 | 0.335 | 0.150 | 0.962 | 1140 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaniak, A.; Minkiewicz, P.; Pliszka, M.; Mogut, D.; Darewicz, M. Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods 2020, 9, 965. https://doi.org/10.3390/foods9070965
Iwaniak A, Minkiewicz P, Pliszka M, Mogut D, Darewicz M. Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods. 2020; 9(7):965. https://doi.org/10.3390/foods9070965
Chicago/Turabian StyleIwaniak, Anna, Piotr Minkiewicz, Monika Pliszka, Damir Mogut, and Małgorzata Darewicz. 2020. "Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters" Foods 9, no. 7: 965. https://doi.org/10.3390/foods9070965
APA StyleIwaniak, A., Minkiewicz, P., Pliszka, M., Mogut, D., & Darewicz, M. (2020). Characteristics of Biopeptides Released In Silico from Collagens Using Quantitative Parameters. Foods, 9(7), 965. https://doi.org/10.3390/foods9070965