A Novel Weissella cibaria Strain UTNGt21O Isolated from Wild Solanum quitoense Fruit: Genome Sequence and Characterization of a Peptide with Highly Inhibitory Potential toward Gram-Negative Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Selection
2.2. NGS de Novo Sequencing of UTNGt21O Isolate
2.3. Peptide Preparation, Determination of Minimum Inhibitory Concentration (MIC) and Molecular Weight
2.4. The Effect of Different Doses of Peptide UTNGt21O Alone and in Combination with EDTA on Indicator Cells Viability at Two Growth Stages
2.5. Permeation of Cytoplasmic Membrane
2.6. Cell Membrane Integrity Assay
2.7. Transmission Electron Microscope (TEM)
2.8. Statistical Analysis
3. Results and Discussion
3.1. Genome Assembly and Taxonomic Classification
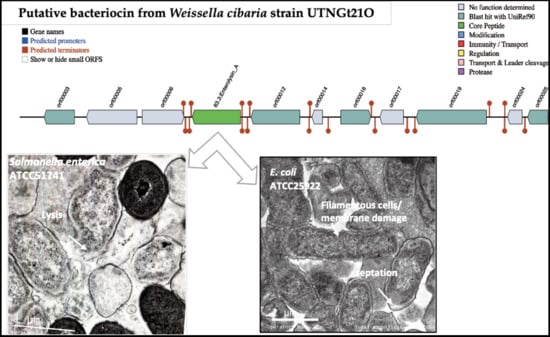
3.2. Identification of Putative Gene Cluster Encoding Bacteriocins
3.3. Peptide UTNGt21O Enhanced the Target Cell Lysis When Co-Cultivated with EDTA
3.4. Peptide UTNGt21O Permeated the Target Cell Membrane
3.5. Integrity of Target Cell Membrane Treated with the Peptide UTNGt21O
3.6. The Peptide UTNGt21O Generate Changes of the Target Cell Membrane Shape
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Linares-Morales, J.R.; Gutiérrez-Méndez, N.; Rivera-Chavira, B.E.; Pérez-Vega, S.B.; Nevárez-Moorillón, G.V. Biocontrol processes in fruits and fresh produce, the use of lactic acid bacteria as a sustainable option. Front. Sustain. Food Syst. 2018, 2, 50. [Google Scholar] [CrossRef]
- Arena, M.P.; Capozzi, V.; Russo, P.; Drider, D.; Spano, G.; Fiocco, D. Immunobiosis and probiosis: Antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 2018, 102, 9949–9958. [Google Scholar] [CrossRef]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [CrossRef]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Martínez-Hernández, J.L.; Aguilar, C.N.; Nevárez-Moorillón, G.V. Production of bioactive peptides from lactic acid bacteria: A sustainable approach for healthier foods. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1039–1051. [Google Scholar] [CrossRef]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of bioactive peptides by Lactobacillus species: From gene to application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Ricks, N.J.; Carroll, C.; Walters, A.; Newell, P.D.; Chaston, J.M. Genome sequence of Weissella cibaria DmW_103, isolated from wild Drosophila. Genome Announc. 2017, 5, e00512–e00517. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Dai, F.; Yao, F.; Tan, M.; Pan, Q. Genome sequence of Weissella cibaria M2, a potential probiotic strain isolated from the feces of a giant panda. Microbiol. Resour. Announc. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Chen, Y.S.; Lee, Y.S.; Yang, C.H.; Srionnual, S.; Wu, H.C.; Chang, C.H. Comparative genomic analysis of bacteriocin-producing Weissella cibaria 110. Appl. Microbiol. Biotechnol. 2017, 101, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Park, J.Y.; Jeong, H.R.; Heo, H.J.; Han, N.S.; Kim, J.H. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe 2012, 18, 96–102. [Google Scholar] [CrossRef]
- Benavidez, A.B.; Ulcuango, M.; Yepez, L.; Tenea, G.N. Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Rev. Arg. Microbiol. 2016, 48, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Hurtado, P.; Ortega, C. Inhibitory effect of substances produced by native Lactococcus lactis strains of tropical fruits towards food pathogens. Prev. Nutr. Food Sci. 2018, 23, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N.; Pozo Delgado, T. Antimicrobial peptides from Lactobacillus plantarum UTNGt2 prevent harmful bacteria growth on fresh tomatoes. J. Microbiol. Biotechnol. 2019, 29, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Hinestroza-Córdoba, L.I.; Duarte Serna, S.; Seguí, L.; Barrera, C.; Betoret, N. Characterization of Powdered Lulo (Solanum quitoense) Bagasse as a Functional Food Ingredient. Foods 2020, 9, 723. [Google Scholar] [CrossRef] [PubMed]
- Månberger, A.; Verbrugghe, P.; Guðmundsdóttir, E.E.; Santesson, S.; Nilsson, A.; Oli Hreggviosson, G.; Linares-Pasten, J.; Nordberg Karlsson, E. Taxogenomic assessment and genomic characterisation of Weissella cibaria strain 92 able to metabolise oligosaccharides derived from dietary fibres. Sci. Rep. 2020, 10, 5853. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. In Gene Prediction. Methods in Molecular Biology; Kollmar, M., Ed.; Humana: New York, NY, USA, 2019; Volume 1962. [Google Scholar]
- Chopra, L.; Singh, G.; Jena, K.K.; Sahoo, D.K. Sonorensin: A new bacteriocin with potential of an anti-biofilm agent and a food biopreservative. Sci. Rep. 2015, 5, 13422. [Google Scholar] [CrossRef]
- Tenea, G.N.; Lara, M.I. Antimicrobial compounds produced by Weissella confusa Cys2-2 strain inhibit Gram-negative bacteria growth. CyTA J. Food 2019, 17, 105–111. [Google Scholar] [CrossRef]
- Patra, P.; Roy, S.; Sarkar, S.; Mitra, S.; Pradhan, S.; Debnath, N.; Goswami, A. Damage of lipopolysaccharides in outer cell membrane and production of ROS-mediated stress within bacteria makes nano zinc oxide a bactericidal agent. Appl. Nanosci. 2015, 5, 857–866. [Google Scholar] [CrossRef]
- Miao, J.; Zhou, J.; Liu, G.; Chen, F.; Chen, Y.G.; Xiangyang, D.; William, S.; Mingyue, S.; Hang, X.; Yong, C. Membrane disruption and DNA binding of Staphylococcus aureus cell induced by a novel antimicrobial peptide produced by Lactobacillus paracasei subsp. tolerans FX-6. Food Control 2016, 59, 609–613. [Google Scholar] [CrossRef]
- Tenea, G.N.; Olmedo, D.; Ortega, C. Peptide-based formulation from lactic acid bacteria Impairs the pathogen growth in Ananas comosus (Pineapple). Coatings 2020, 10, 457. [Google Scholar] [CrossRef]
- Kisand, V.; Lettieri, T. Genome sequencing of bacteria: Sequencing, de novo assembly and rapid analysis using open source tools. BMC Genom. 2019, 14, 211. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Lucid, A.; Arendt, E.K.; Sleator, R.D.; Lucey, B.; Coffey, A. Genomics of Weissella cibaria with an examination of its metabolic traits. Microbiology 2015, 161, 914–930. [Google Scholar] [CrossRef] [PubMed]
- Fessard, A.; Remize, F. Genetic and technological characterization of lactic acid bacteria isolated from tropically grown fruits and vegetables. Int. J. Food Microbiol. 2019, 301, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.K.; Cho, M.S.; Park, D.S. Red pepper powder is a crucial factor that influences the ontogeny of Weissella cibaria during kimchi fermentation. Sci. Rep. 2016, 6, 28232. [Google Scholar] [CrossRef] [PubMed]
- Srionnual, S.; Yanagida, F.; Lin, L.H.; Hsiao, K.N.; Chen, Y.S. Weissellicin 110, a newly discovered bacteriocin from Weissella cibaria 110, isolated from plaa-som, a fermented fish product from Thailand. Appl. Environ. Microbiol. 2007, 73, 2247–2250. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; de Jong, A.; Montalban-Lopez, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Nielsen, T.; Nes, I.F.; Halo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Farrag, H.A.; Abdallah, N.; Shehata, M.; Awad, E.M. Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J. Biomed. Sci. 2019, 26, 69. [Google Scholar] [CrossRef]
- Alakomi, H.L.; Saarela, M.; Helander, I.M. Effect of EDTA on Salmonella enterica serovar Typhimurium involves a component not assignable to lipopolysaccharide release. Microbiology 2003, 149, 2015–2021. [Google Scholar] [CrossRef]
- Martin-Visscher, L.A.; Yoganathan, S.; Sit, S.C.; Lohans, C.T.; Vederas, J.C. The activity of bacteriocins from Carnobacterium maltaromaticum UAL307 against Gram-negative bacteria in combination with EDTA treatment. FEMS Microbiol. Lett. 2011, 317, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Branen, J.K.; Davidson, P.M. Enhancement of nisin, lysozyme, and monolaurin antimicrobial activities by ethylenediaminetetraacetic acid and lactoferrin. Int. J. Food Microbiol. 2004, 90, 63–74. [Google Scholar] [CrossRef]
- Singh, A.P.; Prabha, V.; Rishi, P. Value addition in the efficacy of conventional antibiotics by Nisin against Salmonella. PLoS ONE 2013, 8, e76844. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Teng, D.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Combined systems approaches reveal a multistage mode of action of a marine antimicrobial peptide against pathogenic Escherichia coli and its protective effect against bacterial peritonitis and endotoxemia. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.R.; Sugimoto, Y.; Aoki, T. Ovotransferrin antimicrobial peptide (OTAP-92) kills bacteria through a membrane damage mechanism. Biochim. Biophys. Acta 2000, 1523, 196–205. [Google Scholar] [CrossRef]
- Xue, R.; Liu, Y.; Zhang, Q.; Liang, C.; Qin, H.; Liu, P.; Wang, K.; Zhang, X.; Chen, L.; Wei, Y. Shape changes and interaction mechanism of Escherichia coli cells treated with sericin and use of a sericin-based hydrogel for wound healing. Appl. Environ. Microbiol. 2016, 82, 4663–4672. [Google Scholar] [CrossRef] [PubMed]
- Elliott, T.S.; Shelton, A.; Greenwood, D. The response of Escherichia coli to ciprofloxacin and norfloxacin. J. Med. Microbiol. 1987, 23, 83–88. [Google Scholar] [CrossRef]
- Goh, H.F.; Philip, K. Purification and characterization of bacteriocin produced by Weissella confusa A3 of dairy origin. PLoS ONE 2015, 10, e0140434. [Google Scholar] [CrossRef]
- Castellano, P.; Pérez Ibarreche, M.; Blanco Massani, M.; Fontana, C.; Vignolo, G.M. Strategies for pathogen biocontrol using lactic acid bacteria and their metabolites: A focus on meat ecosystems and industrial environments. Microorganisms 2017, 5, 38. [Google Scholar] [CrossRef]
- Mun, S.Y.; Chang, H.C. Characterization of Weissella koreensis SK isolated from kimchi fermented at low temperature (around 0 °C) Based on complete genome sequence and corresponding phenotype. Microorganisms 2020, 8, 1147. [Google Scholar] [CrossRef] [PubMed]
- Franco, W.; Pérez-Díaz, I.M.; Connelly, L.; Diaz, J.T. Isolation of exopolysaccharide-producing yeast and lactic acid bacteria from quinoa (Chenopodium Quinoa) sourdough fermentation. Foods 2020, 9, 337. [Google Scholar] [CrossRef] [PubMed]







| Status | # of BUSCOs | Percentage |
|---|---|---|
| Complete BUSCOs (C) | ||
| Complete and single-copy BUSCOs (S) | 141 | 95.27% |
| Complete and duplicated BUSCOs (D) | 1 | 0.68% |
| Fragmented BUSCOs (F) | 1 | 0.68% |
| Missing BUSCOs (M) | 5 | 3.38% |
| Total BUSCO groups searched | 148 | 100.00% |
| Description | Query # | Query Length | I_Pct. (%) |
|---|---|---|---|
| CP027427.1 W. cibaria strain BM2 chromosome, complete genome | 1 | 422,230 | 83 |
| CP014332.1 W. jogaejeotgali strain FOL01, complete genome | 2 | 22,017 | 87 |
| CP029684.1 Oenococcus sicerae strain UCMA 15228 chromosome, complete genome | 1 | 59,319 | 73 |
| CP027563.1 W. confusa strain VTT E-133279 chromosome, complete genome | 4 | 532,349 | 75 |
| CP041193.1 W. cibaria strain CBA3612 chromosome, complete genome | 2 | 194,652 | 81 |
| CP033608.1 W. hellenica strain 0916-4-2 chromosome, complete genome | 2 | 279,551 | 77 |
| CP026847.1 W. koreensis strain WiKim0080 chromosome, complete genome | 2 | 66,496 | 79 |
| CP023501.1 W. paramesenteroides strain FDAARGOS_414 chromosome, complete genome | 1 | 4177 | 95 |
| CP026849.1 W. koreensis strain WiKim0080 plasmid pWKW_2, complete sequence | 1 | 6178 | 84 |
| CP028160.1 Lactococcus lactis subsp. lactis strain 14B4 chromosome, complete genome | 1 | 13,239 | 80 |
| CP022606.1 W. cibaria strain CMS1, complete genome | 2 | 236,419 | 80 |
| CP035746.1 Leuconostoc mesenteroides strain SRCM103460 chromosome, complete genome | 1 | 24,997 | 89 |
| Peptide UTNGt21O Dose/(+/-) EDTA | Difference in Cell Viability Relative to Control Untreated (LOGCFU/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early Growth (OD605 = 0.2 ± 0.3) | Logarithmic Growth (OD605 = 0.7± 0.5) | |||||||||||
| Salmonella enterica subsp. enterica ATCC51741 | E. coli ATCC25922 | Salmonella enterica subsp. enterica ATCC51741 | E. coli ATCC25922 | |||||||||
| 1 h | 3 h | 6 h | 1 h | 3 h | 6 h | 1 h | 3 h | 6 h | 1 h | 3 h | 6 h | |
| 1× MIC | 0.68a | 5.14b | 6.00 | 1.68b | 8.02b | ND | 0.29a | 7.22 | ND | 0.42a | 5.67c | 5.60 |
| 1.5× MIC | 0.68a | 5.83c | 6.75 | 1.85bc | 8.24b | ND | 0.38a | 7.22 | ND | 0.34a | 5.42bc | 7.53 |
| 2× MIC | 0.68a | 7.08ef | 6.45 | 2.03c | 8.24b | ND | 0.58a | 7.22 | ND | 0.04a | 5.67c | 7.64 |
| 1× MIC + EDTA (20 mM) | 0.79a | 6.91d | ND | 1.75bc | 8.54c | ND | 0.37a | ND | ND | 0.39a | 5.37b | 6.02 |
| 1.5× MIC +EDTA (20 mM) | 0.79a | 7.97f | ND | 1.91bc | 8.54c | ND | 0.56a | ND | ND | 0.27a | 5.46bc | ND |
| 2× MIC + EDTA (20 mM) | 0.79a | 8.22h | ND | 2.21d | 8.54c | ND | 0.63a | ND | ND | 0.07a | 6.13d | ND |
| EDTA (20 mM) | 0.80a | 0.90a | 0.97 | 0.64a | 0.67a | 0.79 | 0.77a | 0.83 | 0.83 | 0.87a | 0.93a | 0.96 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenea, G.N.; Hurtado, P.; Ortega, C. A Novel Weissella cibaria Strain UTNGt21O Isolated from Wild Solanum quitoense Fruit: Genome Sequence and Characterization of a Peptide with Highly Inhibitory Potential toward Gram-Negative Bacteria. Foods 2020, 9, 1242. https://doi.org/10.3390/foods9091242
Tenea GN, Hurtado P, Ortega C. A Novel Weissella cibaria Strain UTNGt21O Isolated from Wild Solanum quitoense Fruit: Genome Sequence and Characterization of a Peptide with Highly Inhibitory Potential toward Gram-Negative Bacteria. Foods. 2020; 9(9):1242. https://doi.org/10.3390/foods9091242
Chicago/Turabian StyleTenea, Gabriela N., Pamela Hurtado, and Clara Ortega. 2020. "A Novel Weissella cibaria Strain UTNGt21O Isolated from Wild Solanum quitoense Fruit: Genome Sequence and Characterization of a Peptide with Highly Inhibitory Potential toward Gram-Negative Bacteria" Foods 9, no. 9: 1242. https://doi.org/10.3390/foods9091242
APA StyleTenea, G. N., Hurtado, P., & Ortega, C. (2020). A Novel Weissella cibaria Strain UTNGt21O Isolated from Wild Solanum quitoense Fruit: Genome Sequence and Characterization of a Peptide with Highly Inhibitory Potential toward Gram-Negative Bacteria. Foods, 9(9), 1242. https://doi.org/10.3390/foods9091242