A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects Recruitment
2.2. Metabolomic Analysis
2.3. Statistical Analysis and Bioinformatics Analysis
3. Results
3.1. Population Characteristics
3.2. The Altered Metabolites
3.3. The Co-Owned Metabolic Changes
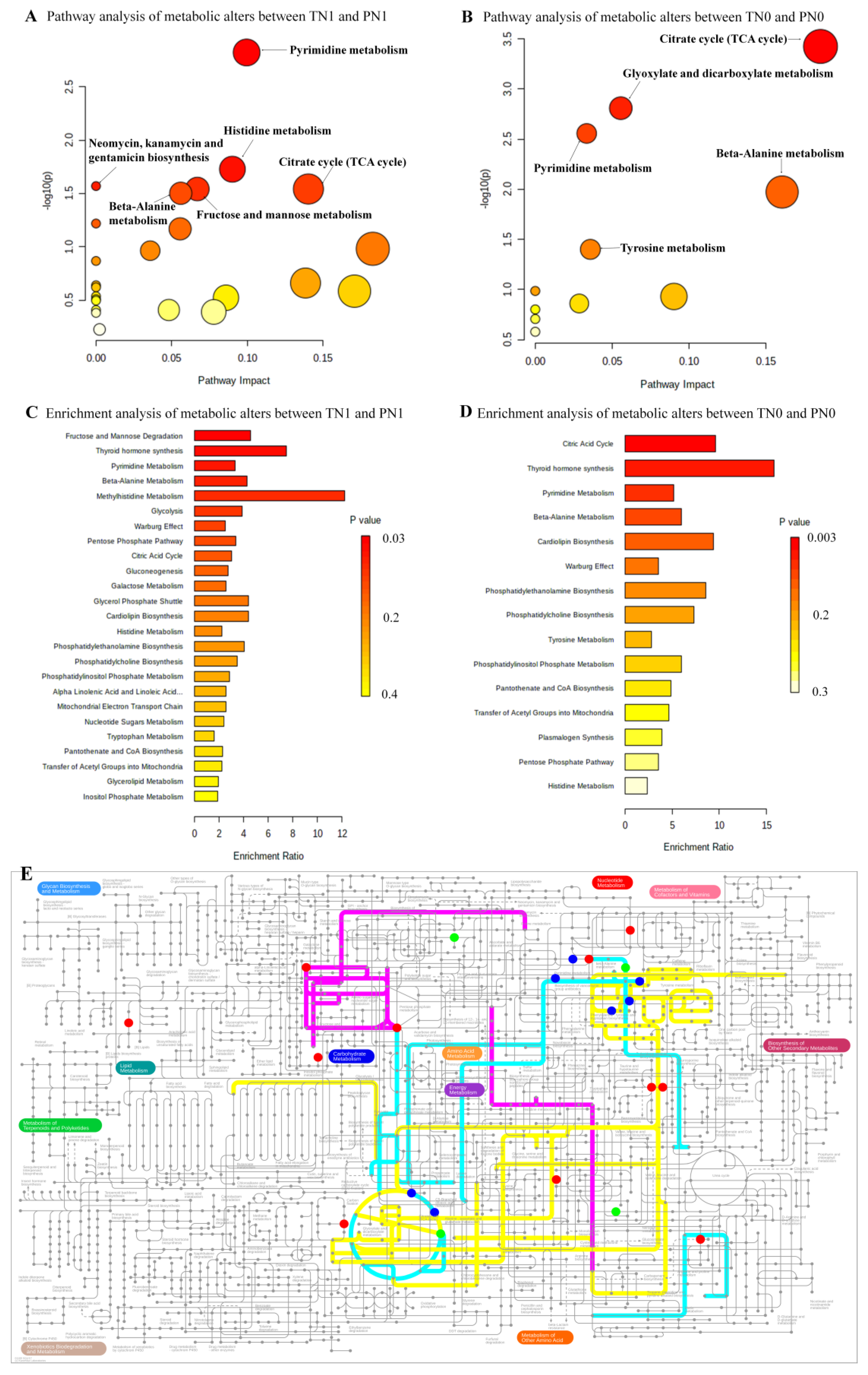
3.4. Differential Metabolites between Two Comparisons (TN1 and PN1, TN0 and PN0)
3.5. The Altered Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabanillas, M.E.; McFadden, D.G.; Durante, C. Thyroid cancer. Lancet 2016, 388, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Horiuchi, K.; Tokumitsu, H.; Sakamoto, A.; Noguchi, E.; Ueda, Y.; Okamoto, T. Time-Varying Pattern of Mortality and Recurrence from Papillary Thyroid Cancer: Lessons from a Long-Term Follow-Up. Thyroid 2019, 29, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Caron, N.R.; Clark, O.H. Papillary thyroid cancer: Surgical management of lymph node metastases. Curr. Treat. Options Oncol. 2005, 6, 311–322. [Google Scholar] [CrossRef]
- Sivanandan, R.; Soo, K.C. Pattern of cervical lymph node metastases from papillary carcinoma of the thyroid. Br. J. Surg. 2001, 88, 1241–1244. [Google Scholar] [CrossRef]
- Ito, Y.; Kudo, T.; Kobayashi, K.; Miya, A.; Ichihara, K.; Miyauchi, A. Prognostic factors for recurrence of papillary thyroid carcinoma in the lymph. World J. Surg. 2012, 36, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Lew, J.I.; Solorzano, C.C. Use of ultrasound in the management of thyroid cancer. Oncologist. 2010, 15, 253–258. [Google Scholar] [CrossRef][Green Version]
- Sipos, J.A. Advances in ultrasound for the diagnosis and management of thyroid cancer. Thyroid 2009, 19, 1363–1372. [Google Scholar] [CrossRef]
- Wong, K.T.; Ahuja, A.T. Ultrasound of thyroid cancer. Cancer Imaging 2005, 5, 157–166. [Google Scholar] [CrossRef]
- Lew, J.I.; Rodgers, S.E.; Solorzano, C.C. Developments in the use of ultrasound for thyroid cancer. Curr. Opin. Oncol. 2010, 22, 11–16. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.L.; Zhao, C.K.; Wang, D.; Wang, Q.; Li, M.X.; Wei, Q.; Guo, J.; Xu, H.X. Conventional Ultrasound, Immunohistochemical Factors and BRAF(V600E) Mutation in Predicting Central Cervical Lymph Node Metastasis of Papillary Thyroid Carcinoma. Ultrasound Med. Biol. 2018, 44, 2296–2306. [Google Scholar] [CrossRef]
- Chen, Y.J.; Zhao, R.M.; Zhao, Q.; Li, B.Y.; Ma, Q.Y.; Li, X.; Chen, X. Diagnostic significance of elevated expression of HBME-1 in papillary thyroid carcinoma. Tumour Biol. 2016, 37, 8715–8720. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Sang, W.; Zheng, S.; Ma, Y.; Liu, X.; Zhang, W. Usefulness of cytokeratin-19, galectin-3, and Hector Battifora mesothelial-1 in the diagnosis of benign and malignant thyroid nodules. Clin. Lab. 2012, 58, 673–680. [Google Scholar] [PubMed]
- O’Neill, C.J.; Bullock, M.; Chou, A.; Sidhu, S.B.; Delbridge, L.W.; Robinson, B.G.; Gill, A.J.; Learoyd, D.L.; Clifton-Bligh, R.; Sywak, M.S. BRAF(V600E) mutation is associated with an increased risk of nodal recurrence requiring reoperative surgery in patients with papillary thyroid cancer. Surgery 2010, 148, 1139–1145, discussion 1145-6. [Google Scholar] [CrossRef] [PubMed]
- Dutenhefner, S.E.; Marui, S.; Santos, A.B.; de Lima, E.U.; Inoue, M.; Neto, J.S.B.; Shiang, C.; Fukushima, J.T.; Cernea, C.R.; Friguglietti, C.U. BRAF: A tool in the decision to perform elective neck dissection? Thyroid 2013, 23, 1541–1546. [Google Scholar] [CrossRef]
- Han, P.A.; Kim, H.S.; Cho, S.; Fazeli, R.; Najafian, A.; Khawaja, H.; McAlexander, M.; Dy, B.; Sorensen, M.; Aronova, A.; et al. Association of BRAF V600E Mutation and MicroRNA Expression with Central Lymph Node Metastases in Papillary Thyroid Cancer: A Prospective Study from Four Endocrine Surgery Centers. Thyroid 2016, 26, 532–542. [Google Scholar]
- Ren, H.; Shen, Y.; Hu, D.; He, W.; Zhou, J.; Cao, Y.; Mao, Y.; Dou, Y.; Xiong, W.; Xiao, Q.; et al. Co-existence of BRAF(V600E) and TERT promoter mutations in papillary thyroid. Cancer Manag. Res. 2018, 10, 1005–1013. [Google Scholar] [CrossRef]
- Liu, R.; Li, Y.; Chen, W.; Cong, J.; Zhang, Z.; Ma, L.; Chu, L.; Xiao, H.; Zhang, Y.; Liu, Y.; et al. Mutations of the TERT promoter are associated with aggressiveness and recurrence/distant metastasis of papillary thyroid carcinoma. Oncol. Lett. 2020, 20, 50. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Gomase, V.S.; Changbhale, S.S.; Patil, S.A.; Kale, K.V. Metabolomics. Curr. Drug Metab. 2008, 9, 89–98. [Google Scholar] [CrossRef]
- Puchades-Carrasco, L.; Pineda-Lucena, A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shen, M.; Li, Y.; Liu, C.; Zhou, K.; Hu, W.; Xu, B.; Xia, Y.; Tang, W. GC-MS-based metabolomic analysis of human papillary thyroid carcinoma tissue. Int. J. Mol. Med. 2015, 36, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, M.; Liu, C.; Xia, Y.; Xu, B.; Hu, Y.; Chen, T.; Shen, M.; Tang, W. Metabolic changes associated with papillary thyroid carcinoma: A nuclear magnetic resonance-based metabolomics study. Int. J. Mol. Med. 2018, 41, 3006–3014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Böttcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef]
- DeLellis, R.A.; Heitz, L.R.; Eng, C. Pathology and Genetics of Tumours of Endocrine Organs, Vol. 8; World Health Organization; IARC Press: Lyon, France, 2004. [Google Scholar]
- Zhou, K.; Ding, X.; Yang, J.; Hu, Y.; Song, Y.; Chen, M.; Sun, R.; Dong, T.; Xu, B.; Han, X.; et al. Metabolomics Reveals Metabolic Changes Caused by Low-Dose 4-Tert-Octylphenol in mice liver. Int. J. Environ. Res. Public Health 2018, 15, 2686. [Google Scholar] [CrossRef]
- Song, M.; Huang, Z.; Wang, S.; Huang, J.; Shi, H.; Liu, Y.; Huang, Y.; Yin, Y.; Wu, Z. Predictive factors of lateral lymph node metastasis in conventional papillary thyroid carcinoma. Gland Surg. 2020, 9, 1000–1007. [Google Scholar] [CrossRef]
- Marzook, F.; Marzook, E.; El-Sonbaty, S. Allantoin may modulate aging impairments, symptoms and cancers. Pak. J. of Pharm. Sci. 2021, 34, 1377–1384. [Google Scholar]
- Buszewska-Forajta, M.; Monedeiro, F.; Gołębiowski, A.; Adamczyk, P.; Buszewski, B. Citric Acid as a Potential Prostate Cancer Biomarker Determined in Various Biological Samples. Metabolites. 2022, 12, 268. [Google Scholar] [CrossRef]
- Fiore, E.; Rago, T.; Provenzale, M.A.; Scutari, M.; Ugolini, C.; Basolo, F.; Coscio, G.D.; Miccoli, P.; Grasso, L.; Pinchera, A.; et al. L-thyroxine-treated patients with nodular goiter have lower serum TSH and lower frequency of papillary thyroid cancer: Results of a cross-sectional study on 27 914 patients. Endocr. Relat. Cancer 2010, 17, 231–239. [Google Scholar] [CrossRef]
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef]
- Budzeń, S.; Rymaszewska, J. The biological role of carnosine and its possible applications in medicine. Adv. Clin. Exp. Med. 2013, 22, 739–744. [Google Scholar] [PubMed]
- Prokopieva, V.D.; Yarygina, E.G.; Bokhan, N.A.; Ivanova, S.A. Use of Carnosine for Oxidative Stress Reduction in Different Pathologies. Oxid. Med. Cell. Longev. 2016, 2016, 2939087. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mao, C.; Wang, M.; Liu, N.; Ouyang, L.; Liu, S.; Tang, H.; Cao, Y.; Liu, S.; Wang, X.; et al. Cancer progression is mediated by proline catabolism in non-small cell lung. Oncogene 2020, 39, 2358–2376. [Google Scholar] [CrossRef]
- Lemos, H.; Huang, L.; Prendergast, G.C.; Mellor, A.L. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat. Rev. Cancer 2019, 19, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Shikawa, S.; Sugimoto, M.; Konta, T.; Kitabatake, K.; Ueda, S.; Edamatsu, K.; Okuyama, N.; Yusa, K.; Iino, M. Salivary Metabolomics for Prognosis of Oral Squamous Cell Carcinoma. Front. Oncol. 2022, 11, 789248. [Google Scholar] [CrossRef]
- Lin, R.; Zhan, M.; Yang, L.; Wang, H.; Shen, H.; Huang, S.; Huang, X.; Xu, S.; Zhang, Z.; Li, W.; et al. Deoxycholic acid modulates the progression of gallbladder cancer through N6-methyladenosine-dependent microRNA maturation. Oncogene 2020, 39, 4983–5000. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Elmaci, İ.; Hacimuftuoglu, A.; Ozpinar, A.; Hacker, E.; Ozpinar, A. PPARδ and its ligand erucic acid may act anti-tumoral, neuroprotective, and myelin protective in neuroblastoma, glioblastoma, and Parkinson’s disease. Mol. Asp. Med. 2021, 78, 100871. [Google Scholar] [CrossRef] [PubMed]
- Abaza, M.S.; Al-Attiyah, R.A.; Bhardwaj, R.; Abbadi, G.; Koyippally, M.; Afzal, M. Syringic acid from Tamarix aucheriana possesses antimitogenic and chemo-sensitizing activities in human colorectal cancer cells. Pharm. Biol. 2013, 51, 1110–1124. [Google Scholar] [CrossRef] [PubMed]
- Iorio, E.; Ricci, A.; Bagnoli, M.; Pisanu, M.E.; Castellano, G.; Di Vito, M. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005, 65, 9369–9376. [Google Scholar] [CrossRef]
- Wang, R.; Ma, L.; Weng, D.; Yao, J.; Liu, X.; Jin, F. Gallic acid induces apoptosis and enhances the anticancer effects of cisplatin in human small cell lung cancer H446 cell line via the ROS-dependent mitochondrial apoptotic pathway. Oncol. Rep. 2016, 35, 3075–3083. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.L.; Chiu, Y.M.; Ho, T.Y.; Hsieh, C.T.; Shieh, D.C.; Lee, Y.J.; Tsay, G.J.; Wu, Y.Y. Gallic Acid Induces Apoptosis in Human Gastric Adenocarcinoma Cells. Anticancer Res. 2018, 38, 2057–2067. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, C.; Wang, Y.K.; Jiang, K.; Gai, X.D. Sorbitol induces apoptosis of human colorectal cancer cells via p38 MAPK signal transduction. Oncol. Lett. 2014, 7, 1992–1996. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ding, B.; Ali, M.R.; Zhao, L.; Zang, X.; Lv, Z. Dual Effect of Tryptamine on Prostate Cancer Cell Growth Regulation: A Pilot Study. Int. J. Mol. Sci. 2022, 23, 11087. [Google Scholar] [CrossRef]
- Jiang, W.G.; Hiscox, S.; Hallett, M.B.; Scott, C.; Horrobin, D.F.; Puntis, M.C.A. Inhibition of hepatocyte growth factor-induced motility and in vitro invasion of human colon cancer cells by gamma-linolenic acid. Br. J. Cancer 1995, 71, 744–752. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, X.; Qiu, Y.; Jia, W.; Wang, J.; Yin, S. Distinct Metabolomic Profiles of Papillary Thyroid Carcinoma and Benign Thyroid adenoma. J. Proteome Res. 2015, 14, 3315–3321. [Google Scholar] [CrossRef]
- Xie, Z.; Li, X.; He, Y.; Wu, S.; Wang, S.; Sun, J.; He, Y.; Lun, Y.; Zhang, J. Immune Cell Confrontation in the Papillary Thyroid Carcinoma Microenvironment. Front. Endocrinol. 2020, 11, 570604. [Google Scholar] [CrossRef]
- Budczies, J.; Brockmöller, S.F.; Müller, B.M.; Barupal, D.K.; Richter-Ehrenstein, C.; Kleine-Tebbe, A.; Griffin, J.L.; Orešič, M.; Dietel, M.; Denkert, C.; et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: Alterations in glutamine and beta-alanine metabolism. J. Proteom. 2013, 94, 279–288. [Google Scholar] [CrossRef]
- Brix, K.; Führer, D.; Biebermann, H. Molecules important for thyroid hormone synthesis and action-known facts and future perspectives. Thyroid Res. 2011, 4 (Suppl. 1). [Google Scholar] [CrossRef]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.S.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef]
- Taylor, N.J.; Gaynanova, I.; Eschrich, S.A.; Welsh, E.A.; Garrett, T.J.; Beecher, C.; Sharma, R.; Koomen, J.M.; Smalley, K.S.M.; Messina, J.L.; et al. Metabolomics of primary cutaneous melanoma and matched adjacent extratumoral microenvironment. PLoS ONE 2020, 15, e0240849. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Kelekar, N.; Bhalla, P.; Kim, J. Fructose and Mannose in Inborn Errors of Metabolism and Cancer. Metabolites 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
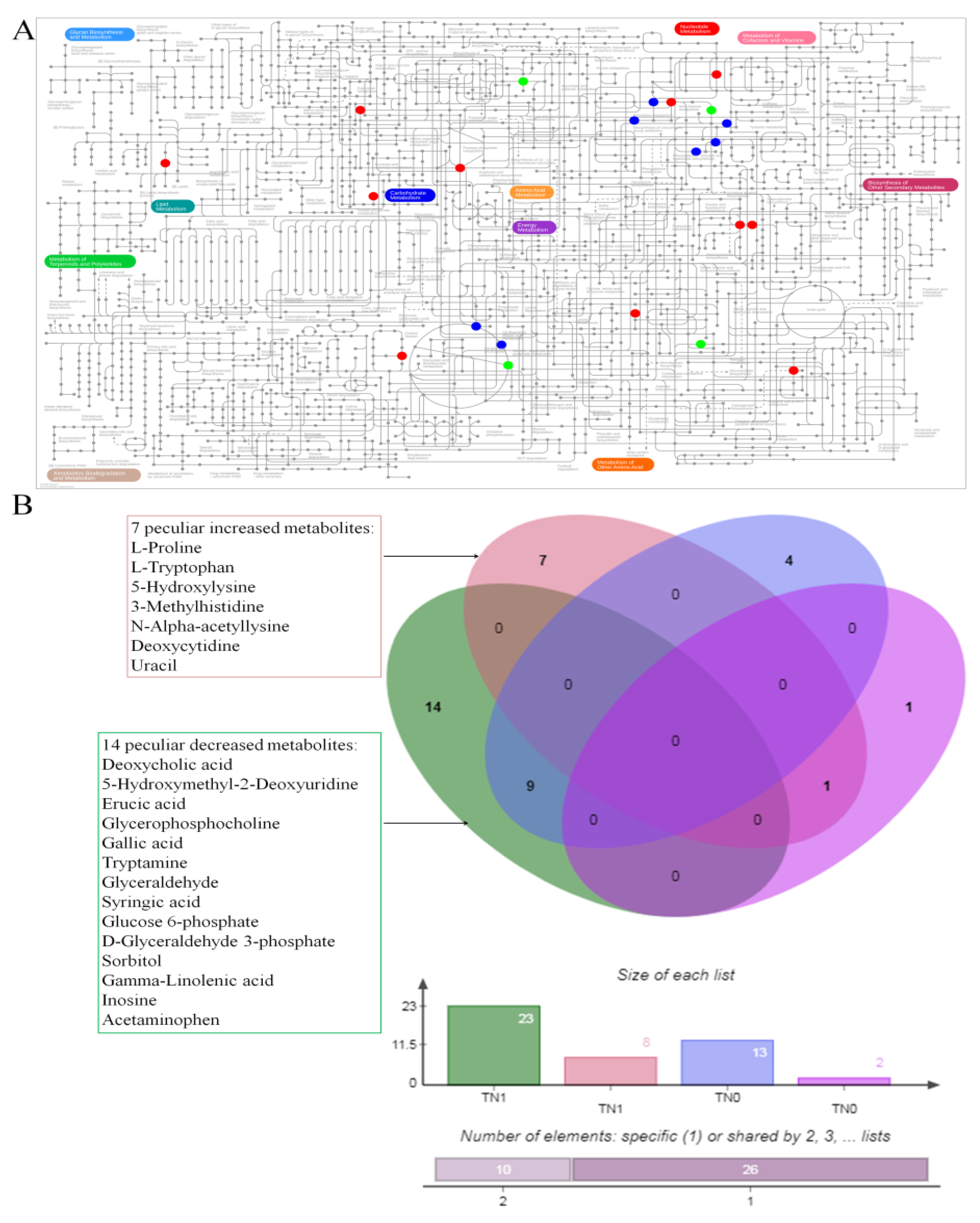
| Metabolites | Fold Change | p | FDR |
|---|---|---|---|
| Thyroxine | 0.020 | 4.815 × 10−3 | 3.485 × 10−2 |
| Deoxycholic acid | 0.023 | 2.784 × 10−3 | 2.645 × 10−2 |
| Allantoin | 0.024 | 6.246 × 10−3 | 3.921 × 10−2 |
| Iodotyrosine | 0.027 | 4.068 × 10−4 | 1.030 × 10−2 |
| 5-Hydroxymethyl-2-Deoxyuridine | 0.033 | 1.370 × 10−5 | 1.041 × 10−3 |
| Erucic acid | 0.041 | 1.027 × 10−3 | 1.562 × 10−2 |
| Cytidine monophosphate | 0.062 | 3.140 × 10−3 | 2.808 × 10−2 |
| Cis-Aconitic acid | 0.071 | 1.573 × 10−3 | 2.146 × 10−2 |
| Citric acid | 0.085 | 2.250 × 10−5 | 1.140 × 10−3 |
| Glycerophosphocholine | 0.150 | 6.354 × 10−3 | 3.921 × 10−2 |
| Uridine | 0.159 | 4.870 × 10−5 | 1.480 × 10−3 |
| Glycolic acid | 0.183 | 8.537 × 10−4 | 1.442 × 10−2 |
| Gallic acid | 0.242 | 2.610 × 10−3 | 2.645 × 10−2 |
| Tryptamine | 0.261 | 9.285 × 10−3 | 4.599 × 10−2 |
| Glyceraldehyde | 0.262 | 3.640 × 10−5 | 1.383 × 10−3 |
| Syringic acid | 0.267 | 4.436 × 10−3 | 3.393 × 10−2 |
| Glucose 6-phosphate | 0.269 | 8.055 × 10−3 | 4.535 × 10−2 |
| D-Glyceraldehyde 3-phosphate | 0.284 | 6.449 × 10−3 | 3.921 × 10−2 |
| Sorbitol | 0.320 | 4.972 × 10−4 | 1.080 × 10−2 |
| Gamma-Linolenic acid | 0.324 | 9.380 × 10−3 | 4.599 × 10−2 |
| Rhamnose | 0.332 | 6.122 × 10−4 | 1.163 × 10−2 |
| Inosine | 0.375 | 8.844 × 10−3 | 4.599 × 10−2 |
| Acetaminophen | 0.585 | 1.912 × 10−3 | 2.146 × 10−2 |
| L-Proline | 1.539 | 7.248 × 10−3 | 4.237 × 10−2 |
| L-Tryptophan | 1.747 | 4.464 × 10−3 | 3.393 × 10−2 |
| 5-Hydroxylysine | 2.817 | 6.362 × 10−3 | 3.921 × 10−2 |
| 3-Methylhistidine | 2.839 | 4.259 × 10−3 | 3.393 × 10−2 |
| N-Alpha-acetyllysine | 3.222 | 1.877 × 10−3 | 2.146 × 10−2 |
| Deoxycytidine | 3.236 | 8.773 × 10−3 | 4.599 × 10−2 |
| Uracil | 8.363 | 1.270 × 10−5 | 1.041 × 10−3 |
| Carnosine | 32.324 | 1.977 × 10−3 | 2.146 × 10−2 |
| Metablolites | Fold Change | p | FDR |
|---|---|---|---|
| cis-Aconitic acid | 0.051 | 3.330 × 10−5 | 2.386 × 10−3 |
| Allantoin | 0.096 | 3.694 × 10−4 | 7.019 × 10−3 |
| Iodotyrosine | 0.128 | 1.420 × 10−5 | 2.158 × 10−3 |
| Isocitric acid | 0.144 | 2.484 × 10−3 | 2.696 × 10−2 |
| Thyroxine | 0.192 | 1.255 × 10−3 | 1.734 × 10−2 |
| Cytidine monophosphate | 0.216 | 7.922 × 10−4 | 1.204 × 10−2 |
| Trizma Acetate | 0.226 | 1.865 × 10−4 | 4.724 × 10−3 |
| Uridine | 0.259 | 4.710 × 10−5 | 2.386 × 10−3 |
| Glycolic acid | 0.274 | 2.966 × 10−3 | 3.005 × 10−2 |
| Citric acid | 0.299 | 3.094 × 10−4 | 6.718 × 10−3 |
| Gluconolactone | 0.329 | 1.368 × 10−4 | 4.724 × 10−3 |
| N-Acetylglutamic acid | 0.360 | 1.447 × 10−3 | 1.833 × 10−2 |
| Rhamnose | 0.378 | 7.166 × 10−4 | 1.204 × 10−2 |
| Ureidopropionic acid | 2.808 | 2.299 × 10−3 | 2.688 × 10−2 |
| Carnosine | 4.734 | 1.807 × 10−4 | 4.724 × 10−3 |
| Metabolite Name | Fold Change | p | FDR |
|---|---|---|---|
| L-Malic acid | 0.187 | 2.310 × 10−2 | 0.923 |
| Thyroxine | 0.649 | 2.102 × 10−2 | 0.923 |
| Carnosine | 1.605 | 4.933 × 10−2 | 0.923 |
| Docosahexaenoic acid | 1.632 | 4.614 × 10−2 | 0.923 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, B.; Gao, W.; Xu, T.; Liu, C.; Wu, D.; Tang, W. A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer. Toxics 2023, 11, 44. https://doi.org/10.3390/toxics11010044
Xu B, Gao W, Xu T, Liu C, Wu D, Tang W. A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer. Toxics. 2023; 11(1):44. https://doi.org/10.3390/toxics11010044
Chicago/Turabian StyleXu, Bo, Wei Gao, Ting Xu, Cuiping Liu, Dan Wu, and Wei Tang. 2023. "A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer" Toxics 11, no. 1: 44. https://doi.org/10.3390/toxics11010044
APA StyleXu, B., Gao, W., Xu, T., Liu, C., Wu, D., & Tang, W. (2023). A UPLC Q-Exactive Orbitrap Mass Spectrometry-Based Metabolomic Study of Serum and Tumor Tissue in Patients with Papillary Thyroid Cancer. Toxics, 11(1), 44. https://doi.org/10.3390/toxics11010044





