Biodegradation Potential of C7-C10 Perfluorocarboxylic Acids and Data from the Genome of a New Strain of Pseudomonas mosselii 5(3)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Chemicals and Reagents
2.3. Genome Sequencing, Assembly and Annotation
2.4. Growing Environments and Conditions
2.5. Isolation and Identification of PFCA Biotransformation Products
2.6. Statistical Analysis
3. Results and Discussion
3.1. Identification and Functional Annotation of the Genome of Strain 5(3)
- -
- Decarboxylase gene (novR), which can carry out several successive steps of the oxidative decarboxylation of PFCA; this was previously described and shown, using a chemical method, for similar compounds [59];
- -
- Alkanesulfonate monooxygenase gene (ssuE), catalyzing the desulfonation of various organosulfonate substrates under conditions of sulfate starvation, which can also produce a protective role under conditions of oxidative stress [60];
- -
- Fluoride ion transporter gene (crcB). With an increase in fluoride levels, it enhances the transcriptional activity of genes located further downstream. It is assumes that these genes aid in reducing the detrimental impacts caused by excessive concentrations of fluoride [61], and assumes that many genes are regulated by them. According to [62], effective defluorination activity will require a high level of tolerance to elevated intracellular concentrations of F− in the microorganism.
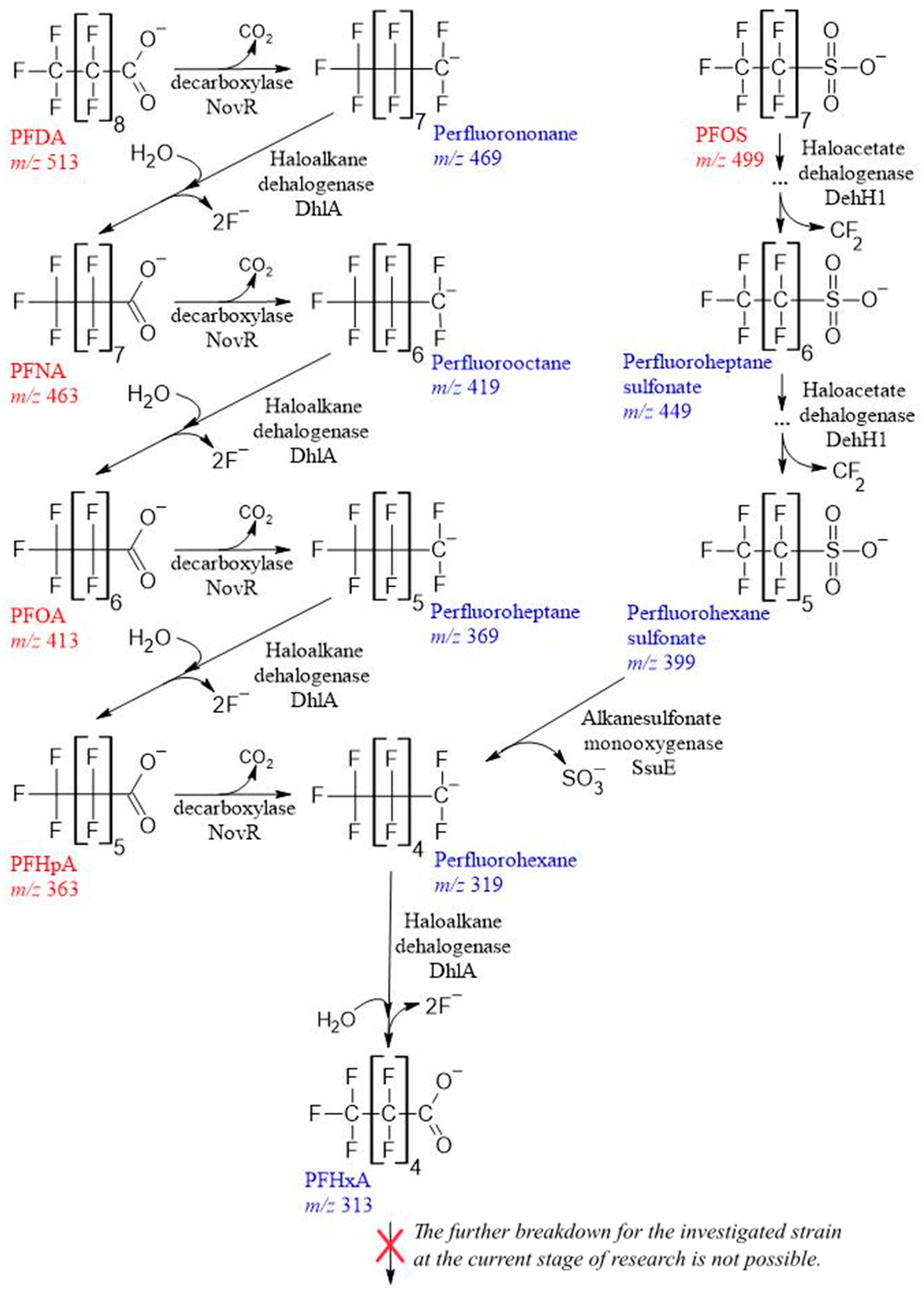
3.2. Growth on C7-C10 PFCA and Their Defluorination
3.3. Biodegradation of C7-C10 PFCAs
3.4. Fluoride Balance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, I.; McDonough, J.; Miles, J.; Storch, P. A review of emerging technologies for remediation of PFASs. Remediation 2018, 28, 101–126. [Google Scholar] [CrossRef]
- Report of the Conference of the Parties of the Stockholm Convention on Persistent Organic Pollutants on the Work of Its Fourth Meeting, 4–8 May//UNEP/POPS/COP.4/38; Stockholm Convention Secretariat: Geneva, Switzerland, 2009; pp. 66–69.
- Fujii, S.; Tanaka, S.; Hong Lien, N.P.; Qiu, Y.; Polprasert, C. New POPs in the water environment: Distribution, bioaccumulation and treatment of perfluorinated compounds: A review paper. J. Water Supply Res. Technol.-AQUA 2007, 56, 313–326. [Google Scholar] [CrossRef]
- Nascimento, R.A.; Nunoo, D.B.O.; Bizkarguenaga, E.; Schultes, L.; Zabaleta, I.; Benskin, J.P.; Spano, S.; Leonel, J. Sulfluramid use in Brazilian agriculture: A source of per- and polyfluoroalkyl substances (PFASs) to the environment. Environ. Pollut. 2018, 242, 1436–1443. [Google Scholar] [CrossRef]
- Xia, C.; Diamond, M.L.; Peaslee, G.F.; Peng, H.; Blum, A.; Wang, Z.; Shalin, A.; Whitehead, H.D.; Green, M.; Schwartz-Narbonne, H.; et al. Per- and polyfluoroalkyl substances in North American school uniforms. Environ. Sci. Technol. 2022, 56, 13845–13857. [Google Scholar] [CrossRef]
- Schaider, L.A.; Balan, S.A.; Blum, A.; Andrews, D.Q.; Strynar, M.J.; Dickinson, M.E.; Lunderberg, D.M.; Lang, J.R.; Peaslee, G.F. Fluorinated compounds in U.S. fast food packaging. Environ. Sci. Technol. Lett. 2017, 4, 105–111. [Google Scholar] [CrossRef]
- Pozo, K.; Moriera, L.B.; Karaskova, P.; Pribylova, P.; Klanova, J.; de Carvalho, M.U.; Maranho, L.A.; de Souza Abessa, D.M. Using large amounts of firefighting foams releases per- and polyfluoroalkyl substances (PFAS) into estuarine environments: A baseline study in Latin America. Mar. Pollut. Bull. 2022, 182, 113938. [Google Scholar] [CrossRef]
- Prevedouros, K.; Cousins, I.T.; Buck, R.C.; Korzeniowski, S.H. Sources, fate and transport of perfluorocarboxylates. Environ. Sci. Technol. 2006, 40, 32–44. [Google Scholar] [CrossRef]
- Wang, Y.; Munir, U.; Huang, Q. Occurrence of per- and polyfluoroalkyl substances (PFAS) in soil: Sources, fate, and remediation. Soil Environ. Health 2023, 1, 100004. [Google Scholar] [CrossRef]
- Ji, B.; Zhao, Y. World Profile of Foreseeable Strategies for the Removal of Per- and Polyfluoroalkyl Substances (PFASs) from Water. In Persistent Pollutants in Water and Advanced Treatment Technology; Springer Nature: Singapore, 2023; pp. 47–69. [Google Scholar]
- EPA 822-R-16-002; Health Effects Support Document for Perfluorooctane Sulfonate (PFOS). U.S. Environmental Protection Agency, Office of Water (4304T) Health and Ecological Criteria Division: Washington, DC, USA, 2016.
- EPA 822-R16-003; Health Effects Support Document for Perfluorooctanoic Acid (PFOA). U.S. Environmental Protection Agency, Office of Water (4304T) Health and Ecological Criteria Division: Washington, DC, USA, 2016.
- Seo, S.-H. Health risk of Human Exposure to Perfluorinated Compounds (PFASs) in Hyeongsan River, Pohang. J. Environ. Anal. Health Toxicol. 2022, 25, 77–84. [Google Scholar] [CrossRef]
- Calvert, L.; Green, M.P.; De Iuliis, G.N.; Dun, M.D.; Turner, B.D.; Clarke, B.O.; Eamens, A.L.; Roman, S.D.; Nixon, B. Assessment of the Emerging Threat Posed by Perfluoroalkyl and Polyfluoroalkyl Substances to Male Reproduction in Humans. Front. Endocrinol. 2022, 12, 799043. [Google Scholar] [CrossRef]
- Cheng, J.; Vecitis, C.D.; Park, H.; Mader, B.T.; Hoffmann, M.R. Sonochemical degradation of perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) in landfill groundwater: Environmental matrix effects. Environ. Sci. Technol. 2008, 42, 8057–8063. [Google Scholar] [CrossRef]
- Wang, F.; Shih, K.; Lu, X.; Liu, C. Mineralization behavior of fluorine in perfluorooctanesulfonate (PFOS) during thermal treatment of lime-conditioned sludge. Environ. Sci. Technol. 2013, 47, 2621–2627. [Google Scholar] [CrossRef]
- Marquínez-Marquínez, A.N.; Loor-Molina, N.S.; Quiroz-Fernández, L.S.; Maddela, N.R.; Luque, R.; Rodríguez-Díaz, J.M. Recent advances in the remediation of perfluoroalkylated and polyfluoroalkylated contaminated sites. Environ. Res. 2023, 219, 115152. [Google Scholar] [CrossRef]
- Kabiri, S.; Navarro, D.A.; Hamad, S.A.; Grimison, C.; Higgins, C.P.; Mueller, J.F.; Kookana, R.S.; McLaughlin, M.J. Physical and chemical properties of carbon-based sorbents that affect the removal of per- and polyfluoroalkyl substances from solution and soil. Sci. Total Environ. 2023, 875, 162653. [Google Scholar] [CrossRef]
- Mayer-Blackwell, K.; Sewell, H.; Fincker, M.; Spormann, A.M. Comparative physiology of organohalide-respiring bacteria. In Organohalide-Respiring Bacteria; Adrian, L., Löffler, F.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 259–280. [Google Scholar]
- Wackett, L.P. Why Is the biodegradation of polyfluorinated compounds so rare? mSphere 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Chetverikov, S.P.; Sharipov, D.A.; Korshunova, T.Y.; Loginov, O.N. Degradation of perfluorooctanyl sulfonate by strain Pseudomonas plecoglossicida 2.4-D. Appl. Biochem. Microbiol. 2017, 53, 533–538. [Google Scholar] [CrossRef]
- Shaw, D.M.J.; Munoz, G.; Bottos, E.M.; Duy, S.V.; Sauve, S.; Liu, J.; Van Hamme, J.D. Degradation and defluorination of 6:2 fluorotelomer sulfonamidoalkyl betaine and 6:2 fluorotelomer sulfonate by Gordonia sp. strain NB4-1Y under sulfur-limiting conditions. Sci. Total Environ. 2019, 647, 690–698. [Google Scholar] [CrossRef]
- Huang, S.; Jaffé, P.R. Defluorination of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) by Acidimicrobium sp. Strain A6. Environ. Sci. Technol. 2019, 53, 11410–11419. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, K.; Li, Z.; Ren, C.; Chen, J.; Lin, Y.-H.; Liu, J.; Men, Y. Microbial cleavage of c–f bonds in two C6 Per- and polyfluorinated compounds via reductive defluorination. Environ. Sci. Technol. 2020, 54, 14393–14402. [Google Scholar] [CrossRef]
- Ruiz-Urigüen, M.; Shuai, W.; Huang, S.; Jaffé, P.R. Biodegradation of PFOA in microbial electrolysis cells by Acidimicrobiaceae sp. strain A6. Chemosphere 2022, 292, 133506. [Google Scholar] [CrossRef]
- Kwon, B.G.; Lim, H.-J.; Na, S.-H.; Choi, B.-I.; Shin, D.-S.; Chung, S.-Y. Biodegradation of perfluorooctanesulfonate (PFOS) as an emerging contaminant. Chemosphere 2014, 109, 221–225. [Google Scholar] [CrossRef]
- Yi, L.B.; Chai, L.Y.; Xie, Y.; Peng, Q.J.; Peng, Q.Z. Isolation, identification, and degradation performance of a PFOA-degrading strain. Genet. Mol. Res. 2016, 15, 235–246. [Google Scholar] [CrossRef]
- Beškoski, V.P.; Yamamoto, A.; Nakano, T.; Yamamoto, K.; Matsumura, C.; Motegi, M.; Beškoski, L.S.; Inui, H. Defluorination of perfluoroalkyl acids is followed by production of monofluorinated fatty acids. Sci. Total Environ. 2018, 15, 355–359. [Google Scholar] [CrossRef]
- Chiriac, F.L.; Stoica, C.; Iftode, C.; Pirvu, F.; Petre, V.A.; Paun, I.; Pascu, L.F.; Vasile, G.G.; Nita-Lazar, M. Bacterial Biodegradation of Perfluorooctanoic Acid (PFOA) and Perfluorosulfonic Acid (PFOS) Using Pure Pseudomonas Strains. Sustainability 2023, 15, 14000. [Google Scholar] [CrossRef]
- Zhang, Z.; Sarkar, D.; Biswas, J.K.; Datta, R. Biodegradation of per- and polyfluoroalkyl substances (PFAS): A review. Bioresour. Technol. 2022, 344, 126223. [Google Scholar] [CrossRef]
- Kiel, M.; Karl-Heinrich, E. The biodegradation vs. biotransformation of fluorosubstituted aromatics. Appl. Microbiol. Biotechnol. 2015, 99, 7433–7464. [Google Scholar] [CrossRef]
- Nogales, J.; García, J.L.; Díaz, E. Degradation of Aromatic Compounds in Pseudomonas: A Systems Biology View. In Aerobic Utilization of Hydrocarbons, Oils and Lipids: Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer: Cham, Switzerland, 2017; pp. 1–49. [Google Scholar]
- Yi, L.; Tang, C.; Peng, Q.; Peng, Q.; Chai, L. Draft genome sequence of perfluorooctane acid-degrading bacterium Pseudomonas parafulva YAB-1. Genome Announc. 2015, 3, e00935-15. [Google Scholar] [CrossRef]
- Harris, J.; Gross, M.; Kemball, J.; Farajollahi, S.; Dennis, P.; Sitko, J.; Steel, J.J.; Almand, E.; Kelley-Loughnane, N.; Varaljay, V.A. Draft Genome Sequence of the Bacterium Delftia acidovorans Strain D4B, Isolated from Soil. Microbiol. Resour. Announc. 2021, 10, e0063521. [Google Scholar] [CrossRef]
- Spaan, K.M.; van Noordenburg, C.; Plassmann, M.M.; Schultes, L.; Shaw, S.; Berger, M.; Heide-Jørgensen, M.P.; Rosing-Asvid, A.; Granquist, S.M.; Dietz, R.; et al. Fluorine mass balance and suspect screening in marine mammals from the northern hemisphere. Environ. Sci. Technol. 2020, 54, 4046–4058. [Google Scholar] [CrossRef]
- Brendel, S.; Fetter, É.; Staude, C.; Vierke, L.; Biegel-Engler, A. Short-chain perfluoroalkyl acids: Environmental concerns and a regulatory strategy under REACH. Environ. Sci. Eur. 2018, 30, 9. [Google Scholar] [CrossRef]
- Li, F.; Duan, J.; Tian, S.; Ji, H.; Zhu, Y.; Wei, Z.; Zhao, D. Short-chain per- and polyfluoroalkyl substances in aquatic systems: Occurrence, impacts and treatment. Chem. Eng. J. 2020, 380, 122506. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Purification of nucleic acids by extraction with phenol: Chloroform. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4455. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.; Nikolenko, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.M.; Kwon, S.J.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Sardà Carbasse, J.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 6, 645–667. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Goker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Raymond, R.L. Microbial oxidation of n-paraffinic hydrocarbons. Dev. Ind. Microbiol. 1961, 2, 23–54. [Google Scholar]
- Bertani, G. Studies on Lysogenesis I. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Marchetto, F.; Roverso, M.; Righetti, D.; Bogialli, S.; Filippini, F.; Bergantino, E.; Sforza, E. Bioremediation of Per- and Poly-Fluoroalkyl Substances (PFAS) by Synechocystis sp. PCC 6803: A Chassis for a Synthetic Biology Approach. Life 2021, 11, 1300. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yue, Y.; Zhang, H.; Yang, Z.; Wang, H.; Tian, S.; Wang, J.; Zhang, Q.; Wang, W. Harnessing Fluoroacetate Dehalogenase for Defluorination of Fluorocarboxylic Acids: In Silico and In Vitro. Approach. Environ. Int. 2019, 131, 104999. [Google Scholar] [CrossRef]
- Trang, B.; Li, Y.; Xue, X.S.; Ateia, M.; Houk, K.N.; Dichtel, W.R. Low-temperature mineralization of perfluorocarboxylic acids. Science 2022, 377, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Shin, B.; Park, W. Protective Role of Bacterial Alkanesulfonate Monooxygenase under Oxidative Stress. Appl. Environ. Microbiol. 2020, 86, e00692-20. [Google Scholar] [CrossRef] [PubMed]
- Calero, P.; Gurdo, N.; Nikel, P.I. Role of the CrcB transporter of Pseudomonas putida in the multi-level stress response elicited by mineral fluoride. Environ. Microbiol. 2022, 24, 5082–5104. [Google Scholar] [CrossRef]
- Wackett, L.P. Pseudomonas: Versatile biocatalysts for PFAS. Environ. Microbiol. 2022, 24, 2882–2889. [Google Scholar] [CrossRef]
- Winsor, G.L.; Van Rossum, T.; Lo, R.; Khaira, B.; Whiteside, M.D.; Hancock, R.E.; Brinkman, F.S. Pseudomonas Genome Database: Facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009, 37, D483–D488. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed]
- Luz, A.L.; Anderson, J.K.; Goodrum, P.; Durda, J. Perfluorohexanoic acid toxicity, part I: Development of a chronic human health toxicity value for use in risk assessment. Regul. Toxicol. Pharmacol. 2019, 103, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Zango, Z.U.; Ethiraj, B.; Al-Mubaddel, F.S.; Alam, M.M.; Lawal, M.A.; Kadir, H.A.; Khoo, K.S.; Garba, Z.N.; Usman, F.; Zango, M.U.; et al. An overview on human exposure, toxicity, solid-phase microextraction and adsorptive removal of perfluoroalkyl carboxylic acids (PFCAs) from water matrices. Environ. Res. 2023, 231 Pt 2, 116102. [Google Scholar] [CrossRef]
- Han, J.S.; Jang, S.; Son, H.Y.; Kim, Y.B.; Kim, Y.; Noh, J.H.; Kim, M.J.; Lee, B.S. Subacute dermal toxicity of perfluoroalkyl carboxylic acids: Comparison with different carbon-chain lengths in human skin equivalents and systemic effects of perfluoroheptanoic acid in Sprague Dawley rats. Arch. Toxicol. 2020, 94, 523–539. [Google Scholar] [CrossRef]
- Ji, C.; Stockbridge, R.B.; Miller, C. Bacterial fluoride resistance, Fluc channels and the weak acid accumulation effect. J. Gen. Physiol. 2014, 144, 257–261. [Google Scholar] [CrossRef]
- McIlwain, B.C.; Michal, T.R.; Stockbridge, R.B. Membrane exporters of fluoride ion. Ann. Rev. Biochem. 2021, 90, 559–579. [Google Scholar] [CrossRef] [PubMed]
- Chetverikov, S.P.; Loginov, O.N. A new Ensifer adhaerens strain M1 is capable of transformation of perfluorocarboxylic acids. Microbiology 2019, 88, 115–117. [Google Scholar] [CrossRef]
- Stockbridge, R.B.; Lim, H.H.; Otten, R.; Williams, C.; Shane, T.; Weinberg, Z.; Miller, C. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc. Natl. Acad. Sci. USA 2012, 109, 15289–15294. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, G.; May, A.L.; Yan, J.; Brown, L.P.; Powers, J.B.; Campagna, S.R.; Löffler, F.E. Pseudomonas sp. strain 273 degrades fluorinated alkanes. Environ. Sci. Technol. 2020, 54, 14994–15003. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; May, A.L.; Chen, G.; Brown, L.P.; Powers, J.B.; Tague, E.D.; Campagna, S.R.; Löffler, F.E. Pseudomonas sp. strain 273 incorporates organofluorine into the lipid bilayer during growth with fluorinated alkanes. Environ. Sci. Technol. 2022, 56, 8155–8166. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, G.; Ramirez, D.; Yan, J.; Löffler, F.E. Complete Genome Sequence of Pseudomonas sp. Strain 273, a Haloalkane-Degrading Bacterium. Microbiol. Resour. Announc. 2023, 12, e0017623. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chetverikov, S.; Hkudaygulov, G.; Sharipov, D.; Starikov, S.; Chetverikova, D. Biodegradation Potential of C7-C10 Perfluorocarboxylic Acids and Data from the Genome of a New Strain of Pseudomonas mosselii 5(3). Toxics 2023, 11, 1001. https://doi.org/10.3390/toxics11121001
Chetverikov S, Hkudaygulov G, Sharipov D, Starikov S, Chetverikova D. Biodegradation Potential of C7-C10 Perfluorocarboxylic Acids and Data from the Genome of a New Strain of Pseudomonas mosselii 5(3). Toxics. 2023; 11(12):1001. https://doi.org/10.3390/toxics11121001
Chicago/Turabian StyleChetverikov, Sergey, Gaisar Hkudaygulov, Danil Sharipov, Sergey Starikov, and Darya Chetverikova. 2023. "Biodegradation Potential of C7-C10 Perfluorocarboxylic Acids and Data from the Genome of a New Strain of Pseudomonas mosselii 5(3)" Toxics 11, no. 12: 1001. https://doi.org/10.3390/toxics11121001
APA StyleChetverikov, S., Hkudaygulov, G., Sharipov, D., Starikov, S., & Chetverikova, D. (2023). Biodegradation Potential of C7-C10 Perfluorocarboxylic Acids and Data from the Genome of a New Strain of Pseudomonas mosselii 5(3). Toxics, 11(12), 1001. https://doi.org/10.3390/toxics11121001






