RNA-Seq Analysis of Testes from Mice Exposed to Neodymium Oxide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
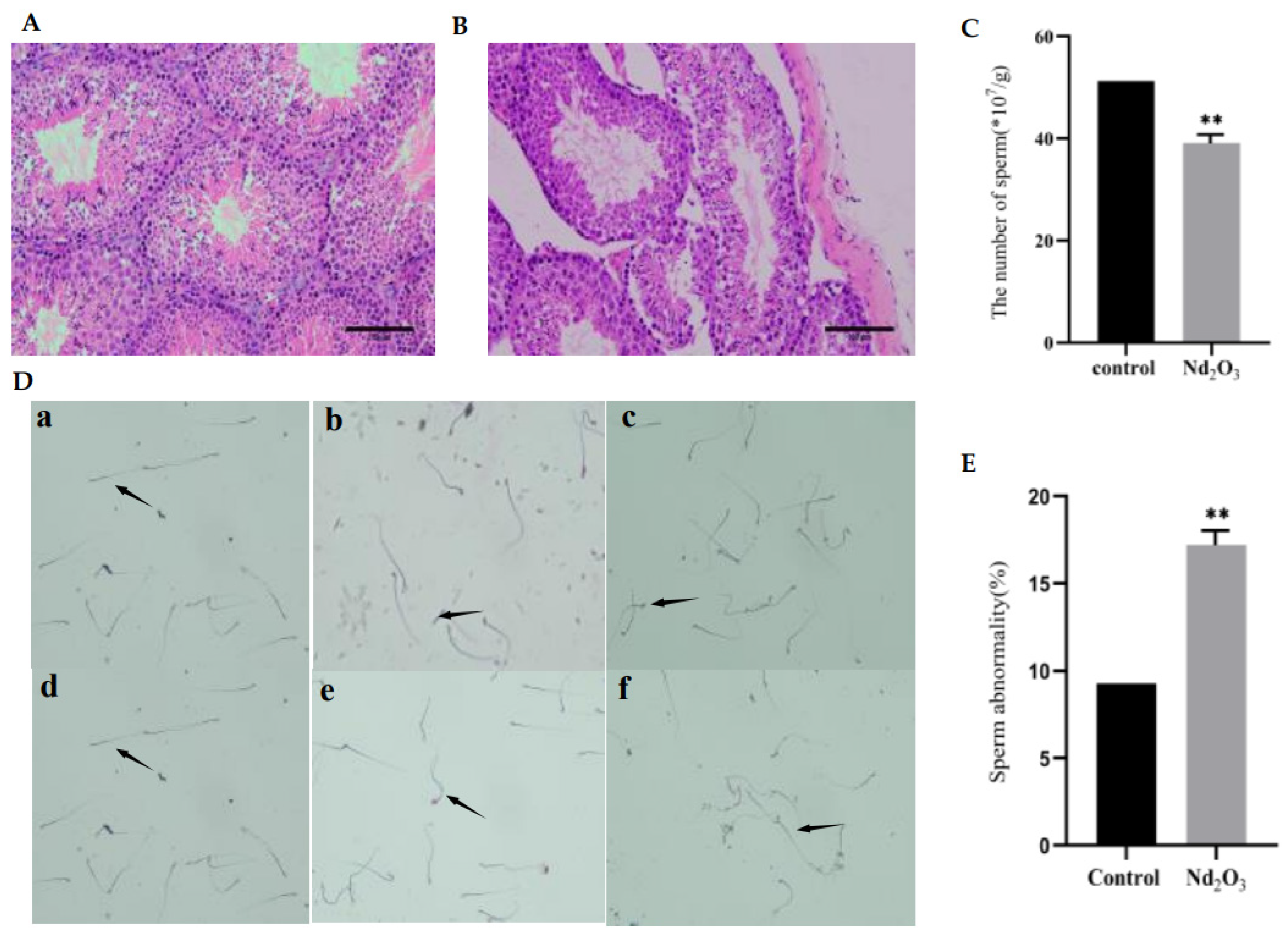
2.2. H&E Staining
2.3. Sperm Count and Morphology Assessment
2.4. RNA Library Construction and Sequencing
2.5. RNA Identification and Expression Analysis
2.6. ceRNA Networks Analysis
2.7. Protein–Protein Interaction (PPI) Network Construction
2.8. qPCR Confirmation
2.9. Statistical Analysis
3. Results
3.1. Effects of Nd2O3 Exposure on Testis and Sperm Count and Sperm Morphology
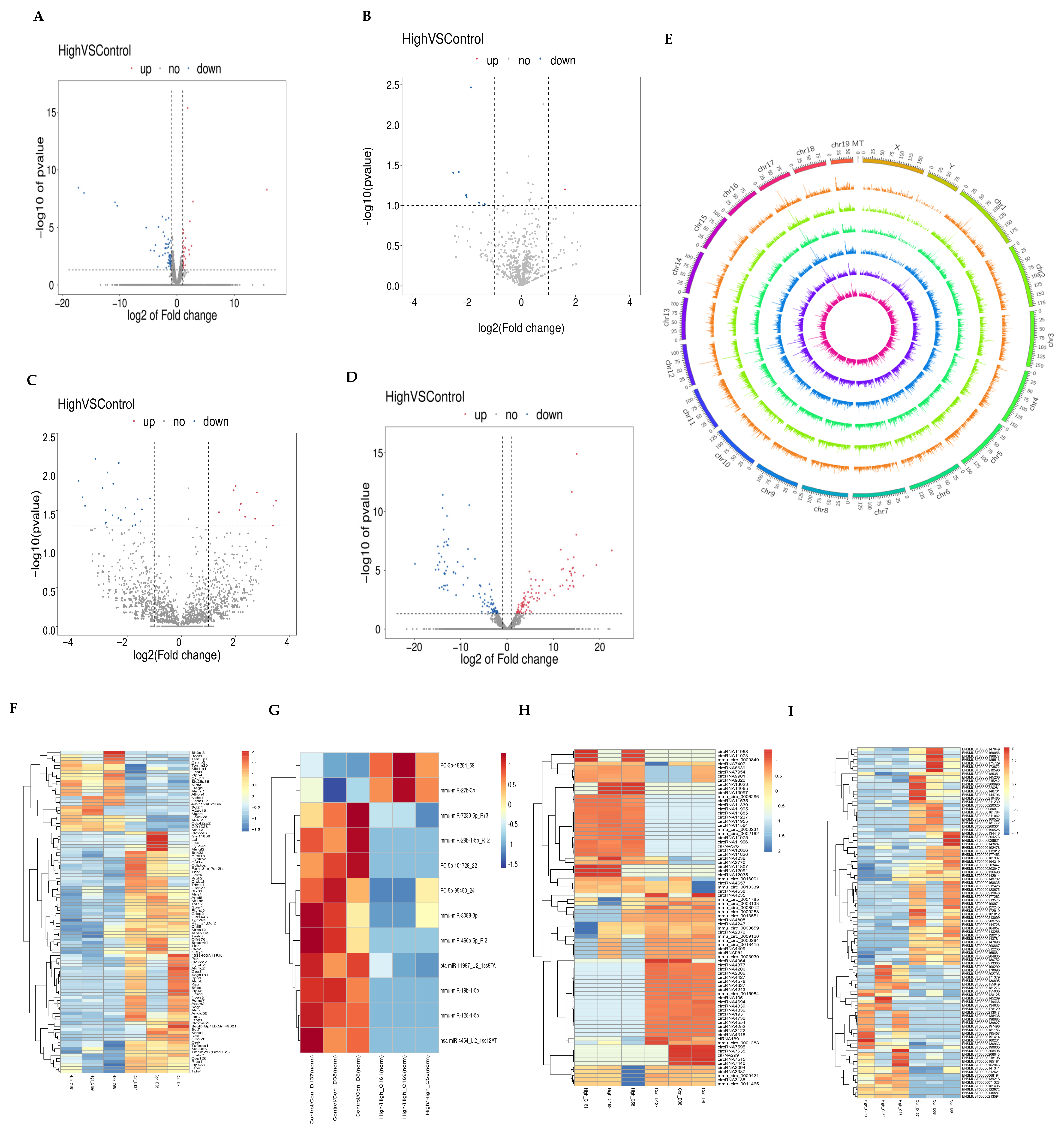
3.2. Expression Profiles of mRNA, miRNA, lncRNA, and circRNA
3.3. GO and KEGG Analysis of DEGs
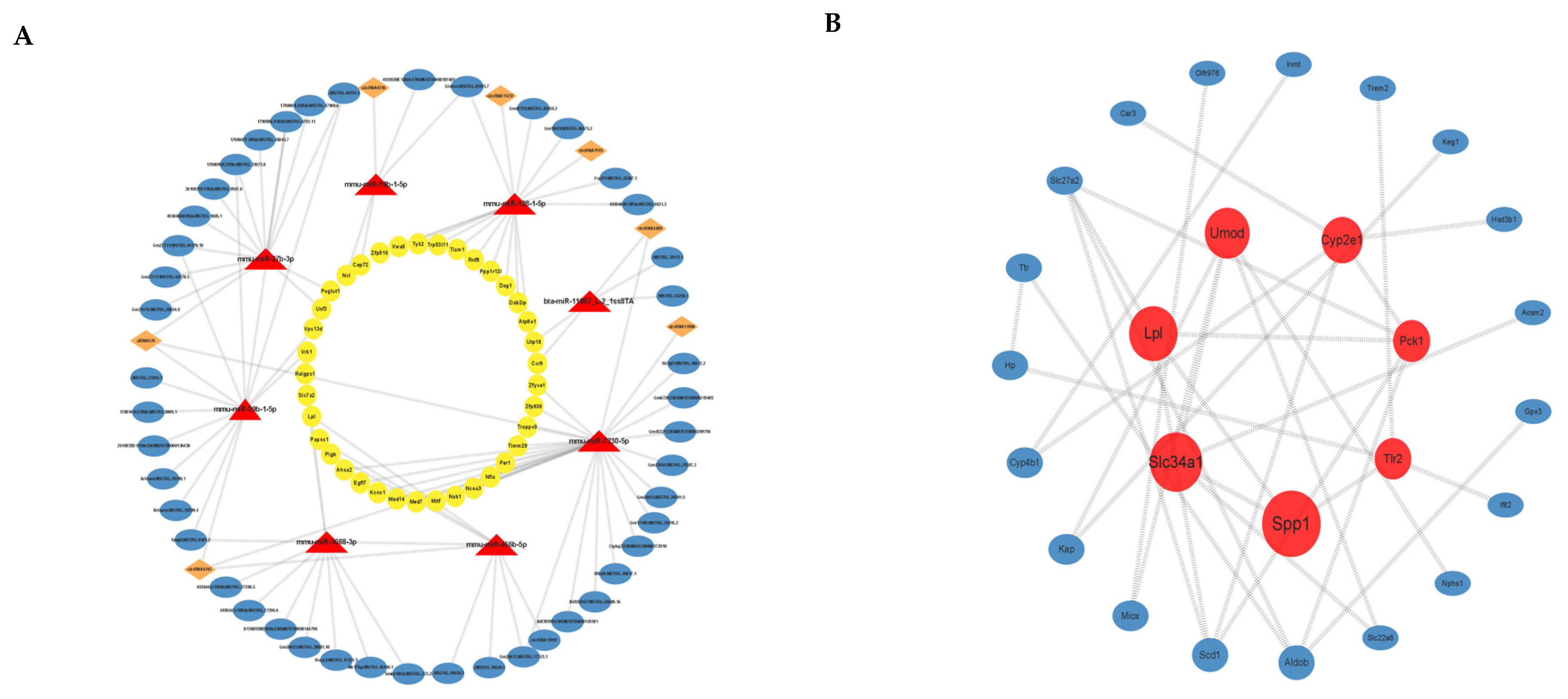
3.4. ceRNA Network Construction
3.5. PPI Analysis of Differential mRNAs
3.6. Validation of Profiles via qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alonso, E.; Sherman, A.M.; Wallington, T.J.; Everson, M.P.; Field, F.R.; Roth, R.; Kirchain, R.E. Evaluating rare earth element availability: A case with revolutionary demand from clean technologies. Environ. Sci. Technol. 2012, 46, 3406–3414. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Yang, Z.; Wang, K. Bone Mineral Density in Population Long-Term Exposed to Rare Earth Elements from a Mining Area of China. Biol. Trace Elem. Res. 2021, 199, 453–464. [Google Scholar] [CrossRef]
- Shi, X.M.; Bai, Y.C.; Gao, Y.R.; Bu, N.; Song, H.Y.; Huang, L.H.; Zhao, Y.H.; Wang, S.H. Comprehensive Analysis of Differentially Expressed lncRNAs miRNAs and mRNA and Their ceRNA Network of Patients with Rare-Earth Pneumoconiosis. Front. Genet. 2021, 12, 700398. [Google Scholar] [CrossRef]
- Huang, L.H.; Yang, H.; Su, X.; Gao, Y.R.; Xue, H.N.; Wang, S.H. Neodymium Oxide Induces Cytotoxicity and Activates NF-kappaB and Caspase-3 in NR8383 Cells. Biomed. Environ. Sci. 2017, 30, 75–78. [Google Scholar]
- Adebayo, O.A.; Akinloye, O.; Adaramoye, O.A. Cerium oxide nanoparticle elicits oxidative stress, endocrine imbalance and lowers sperm characteristics in testes of balb/c mice. Andrologia 2018, 50, e12920. [Google Scholar] [CrossRef]
- Yuan, L.; Bai, D.; Meng, L.; Wang, H.; Sun, Z.; An, T.; Chen, Z.; Deng, X.; Zhang, X. Effects of intragastric administration of La2O3 nanoparticles on mouse testes. J. Toxicol. Sci. 2020, 45, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Yang, T.; Seyler, B.C.; Wang, X.; Wang, Y.; Jiang, M.; Liu, B.; Li, F. Ambient air pollution and male fecundity: A retrospective analysis of longitudinal data from a Chinese human sperm bank (2013–2018). Environ. Res. 2020, 186, 109528. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Xia, L.; Hou, Y.; Li, Y.; Cao, D.; Liu, Y.; Chen, J.; Liu, J.; Zhang, L.; Yang, Q.; et al. Transgenerational male reproductive effect of prenatal arsenic exposure: Abnormal spermatogenesis with Igf2/H19 epigenetic alteration in CD1 mouse. Int. J. Environ. Health Res. 2022, 32, 1248–1260. [Google Scholar] [CrossRef]
- Nava-Rivera, L.E.; Betancourt-Martinez, N.D.; Lozoya-Martinez, R.; Carranza-Rosales, P.; Guzman-Delgado, N.E.; Carranza-Torres, I.E.; Delgado-Aguirre, H.; Zambrano-Ortiz, J.O.; Moran-Martinez, J. Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure. Sci. Rep. 2021, 11, 8276. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 2020, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Li, Y.; Xiang, Y.; Peng, N.; Shen, C.; Cai, Y.; Song, D.; Zhang, P.; Wang, X.; Zeng, X.; et al. Dysregulation of lncRNA and circRNA Expression in Mouse Testes after Exposure to Triptolide. Curr. Drug Metab. 2019, 20, 665–673. [Google Scholar] [CrossRef]
- Prakash, M.A.; Kumaresan, A.; Sinha, M.K.; Kamaraj, E.; Mohanty, T.K.; Datta, T.K.; Morrell, J.M. RNA-Seq analysis reveals functionally relevant coding and non-coding RNAs in crossbred bull spermatozoa. Anim. Reprod. Sci. 2020, 222, 106621. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, L.; Lv, H.; Li, Y.; Zhu, C.; Tian, W.; Zhao, L. Applying high-throughput sequencing to identify and evaluate foetal chromosomal deletion and duplication. J. Cell Mol. Med. 2020, 24, 9936–9944. [Google Scholar] [CrossRef] [PubMed]
- Gwenzi, W.; Mangori, L.; Danha, C.; Chaukura, N.; Dunjana, N.; Sanganyado, E. Sources, behaviour, and environmental and human health risks of high-technology rare earth elements as emerging contaminants. Sci. Total Environ. 2018, 636, 299–313. [Google Scholar] [CrossRef]
- Yuan, L.; Li, Q.; Bai, D.; Shang, X.; Hu, F.; Chen, Z.; An, T.; Chen, Y.; Zhang, X. La2O3 Nanoparticles Induce Reproductive Toxicity Mediated by the Nrf-2/ARE Signaling Pathway in Kunming Mice. Int. J. Nanomed. 2020, 15, 3415–3431. [Google Scholar] [CrossRef] [PubMed]
- Major, A.T.; Hogarth, C.A.; Young, J.C.; Kurihara, Y.; Jans, D.A.; Loveland, K.L. Dynamic paraspeckle component localisation during spermatogenesis. Reproduction 2019, 158, 267–280. [Google Scholar] [CrossRef]
- Zheng, H.; Huang, J.; Zhang, M.; Zhao, H.J.; Chen, P.; Zeng, Z.H. miR-27b-3p Improved High Glucose-Induced Spermatogenic Cell Damage via Regulating Gfpt1/HBP Signaling. Eur. Surg. Res. 2022, 63, 64–76. [Google Scholar] [CrossRef]
- Xue, R.; Lin, W.; Sun, J.; Watanabe, M.; Xu, A.; Araki, M.; Nasu, Y.; Tang, Z.; Huang, P. The role of Wnt signaling in male reproductive physiology and pathology. Mol. Hum. Reprod. 2021, 27, gaaa085. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Li, B. Correlative study on the JAK-STAT/PSMbeta3 signal transduction pathway in asthenozoospermia. Exp. Ther. Med. 2017, 13, 127–130. [Google Scholar] [CrossRef]
- Monrose, M.; Thirouard, L.; Garcia, M.; Holota, H.; De Haze, A.; Caira, F.; Beaudoin, C.; Volle, D.H. New perspectives on PPAR, VDR and FXRalpha as new actors in testicular pathophysiology. Mol. Aspects Med. 2021, 78, 100886. [Google Scholar] [CrossRef]
- Gan, Y.; Yang, D.; Yang, S.; Wang, J.; Wei, J.; Chen, J. Di-2-ethylhexyl phthalate (DEHP) induces apoptosis and autophagy of mouse GC-1 spg cells. Environ. Toxicol. 2020, 35, 292–299. [Google Scholar] [CrossRef]
- Shen, J.; Yang, D.; Zhou, X.; Wang, Y.; Tang, S.; Yin, H.; Wang, J.; Chen, R.; Chen, J. Role of Autophagy in Zinc Oxide Nanoparticles-Induced Apoptosis of Mouse LEYDIG Cells. Int. J. Mol. Sci. 2019, 20, 4042. [Google Scholar] [CrossRef] [PubMed]
- Lakpour, M.R.; Koruji, M.; Shahverdi, A.; Aghajanpour, S.; Rajabian Naghandar, M.; Sadighi Gilani, M.A.; Sabbaghian, M.; Aflatoonian, R. The Expression of TLR2 and TLR3 in Sertoli Cells of Azoospermic Patients. Cell J. 2017, 19, 375–385. [Google Scholar] [PubMed]
- Luo, X.; Bao, X.; Weng, X.; Bai, X.; Feng, Y.; Huang, J.; Liu, S.; Jia, H.; Yu, B. The protective effect of quercetin on macrophage pyroptosis via TLR2/Myd88/NF-kappaB and ROS/AMPK pathway. Life Sci. 2022, 291, 120064. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018, 23, 599–614.e4. [Google Scholar] [CrossRef] [PubMed]
- Salehi, N.; Karimi-Jafari, M.H.; Totonchi, M.; Amiri-Yekta, A. Integration and gene co-expression network analysis of scRNA-seq transcriptomes reveal heterogeneity and key functional genes in human spermatogenesis. Sci. Rep. 2021, 11, 19089. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Bu, N.; Yun, Y.; Shi, X.; Wang, S.; Gao, Y. RNA-Seq Analysis of Testes from Mice Exposed to Neodymium Oxide. Toxics 2023, 11, 952. https://doi.org/10.3390/toxics11120952
Wang S, Bu N, Yun Y, Shi X, Wang S, Gao Y. RNA-Seq Analysis of Testes from Mice Exposed to Neodymium Oxide. Toxics. 2023; 11(12):952. https://doi.org/10.3390/toxics11120952
Chicago/Turabian StyleWang, Shurui, Ning Bu, Yudan Yun, Xuemin Shi, Suhua Wang, and Yanrong Gao. 2023. "RNA-Seq Analysis of Testes from Mice Exposed to Neodymium Oxide" Toxics 11, no. 12: 952. https://doi.org/10.3390/toxics11120952
APA StyleWang, S., Bu, N., Yun, Y., Shi, X., Wang, S., & Gao, Y. (2023). RNA-Seq Analysis of Testes from Mice Exposed to Neodymium Oxide. Toxics, 11(12), 952. https://doi.org/10.3390/toxics11120952




