The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms
Abstract
:1. Introduction
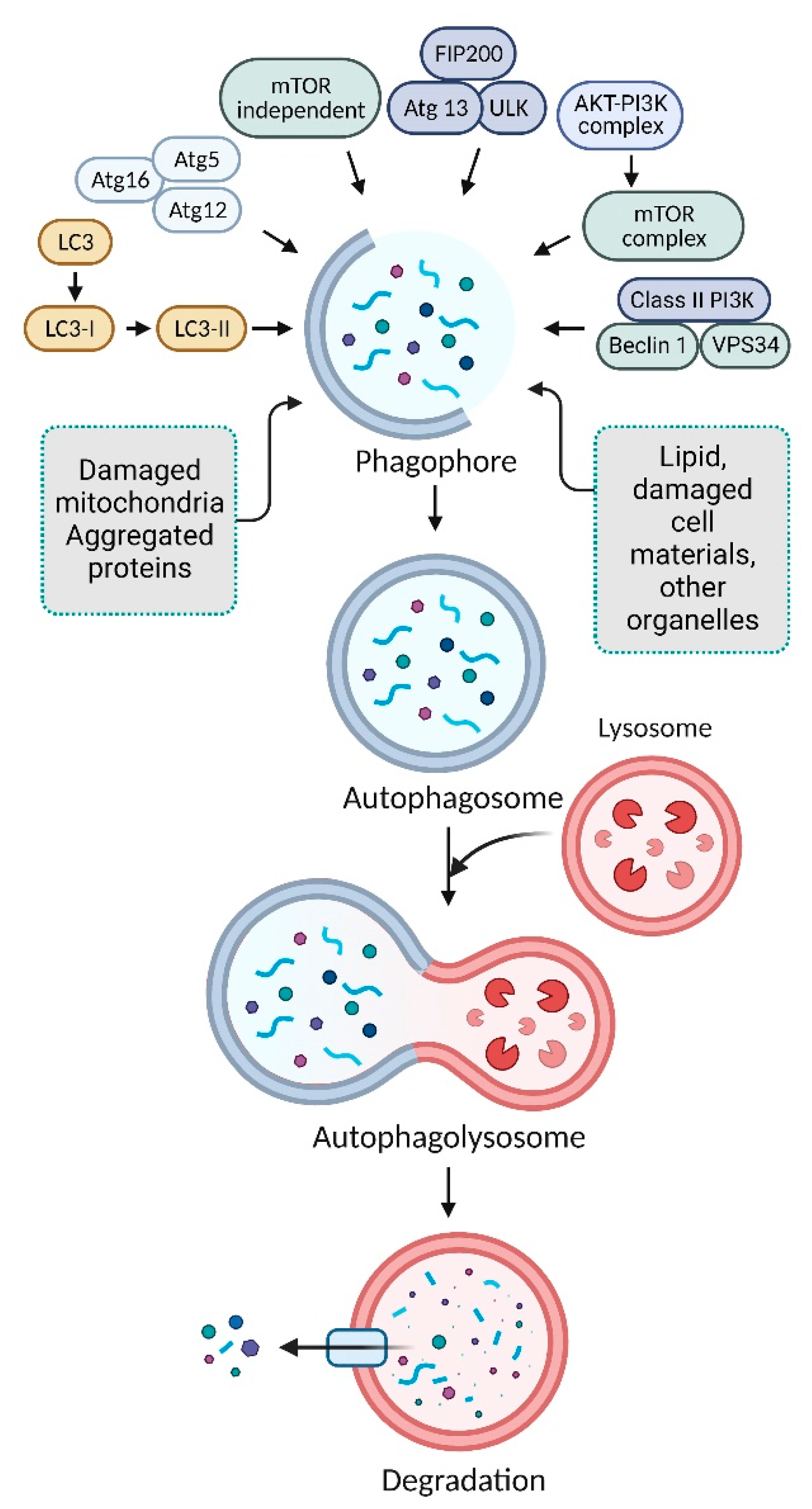
2. Mechanism of Autophagy Pathways
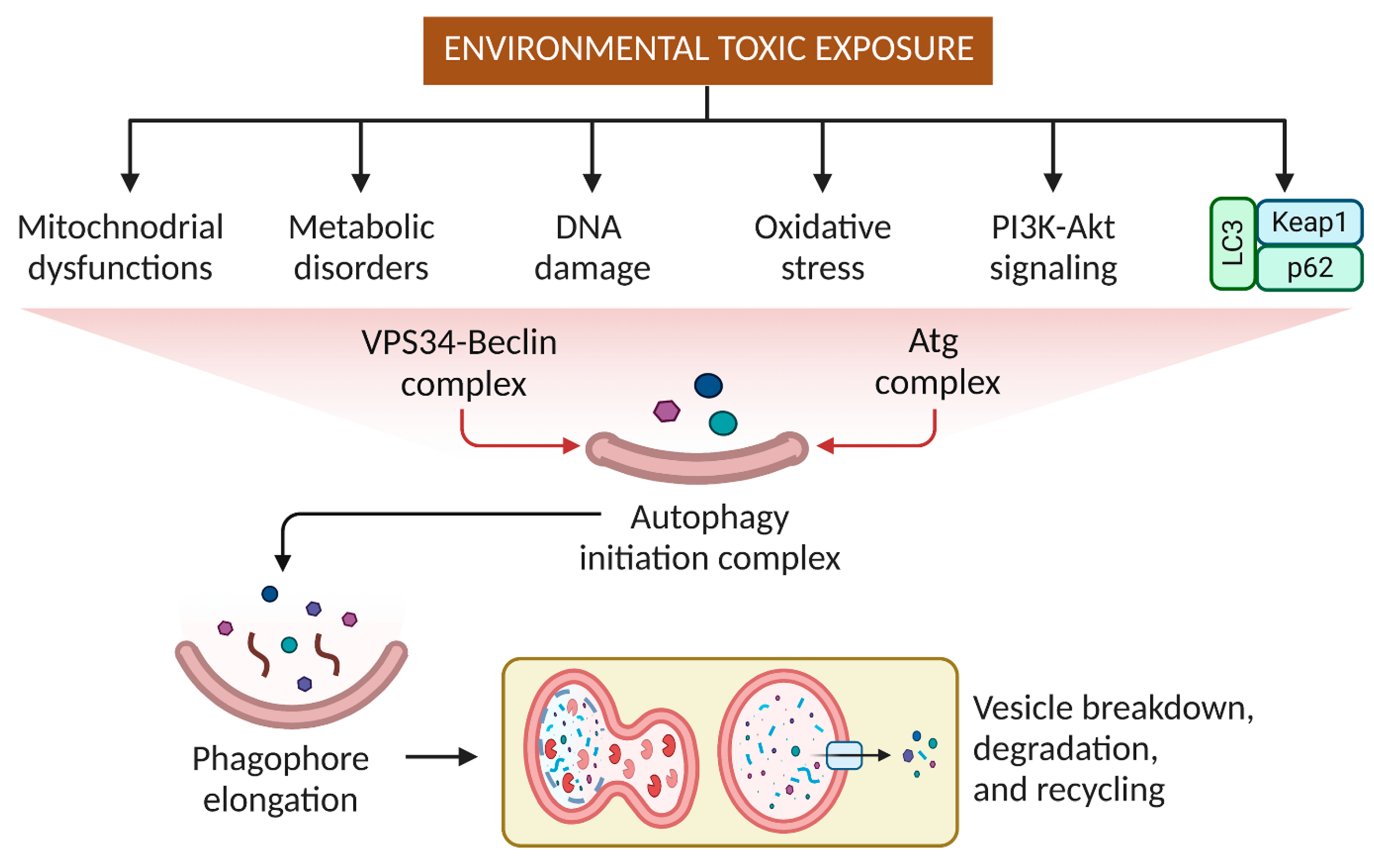
3. Effects of Pesticides and Other Small Molecular Weight Environmental Toxins on Autophagy
4. Targeting Autophagy Modulation to Eliminate Environmental Substances
4.1. Elimination of Particulate Matter by Autophagy
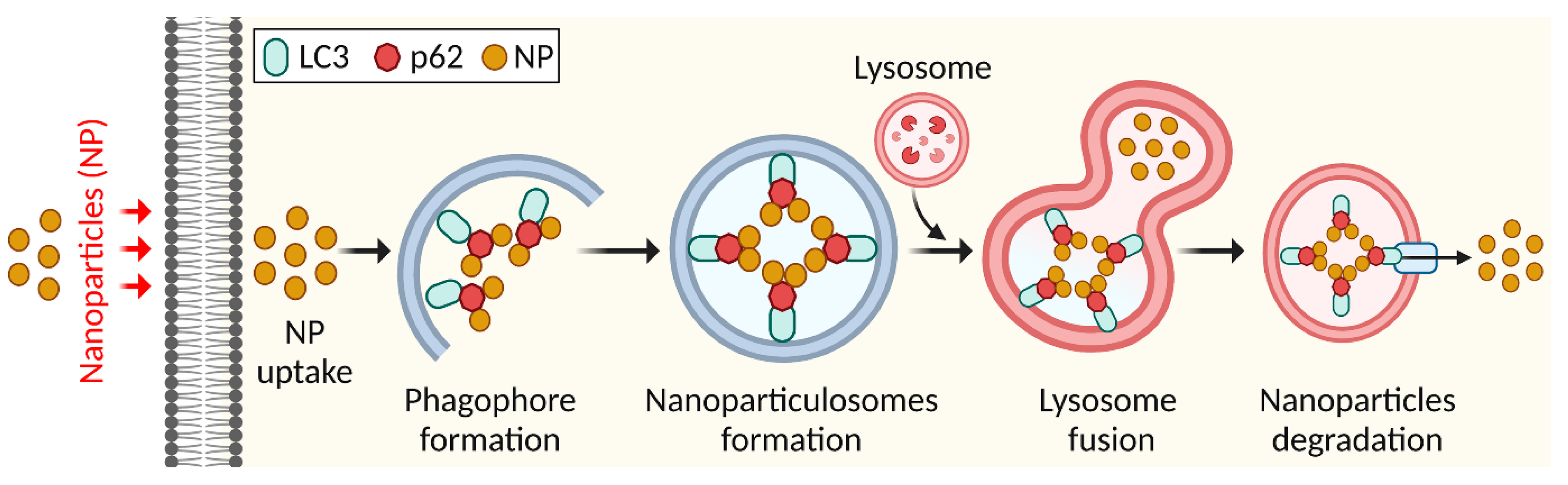
4.2. Elimination of Nanoparticles by Autophagy
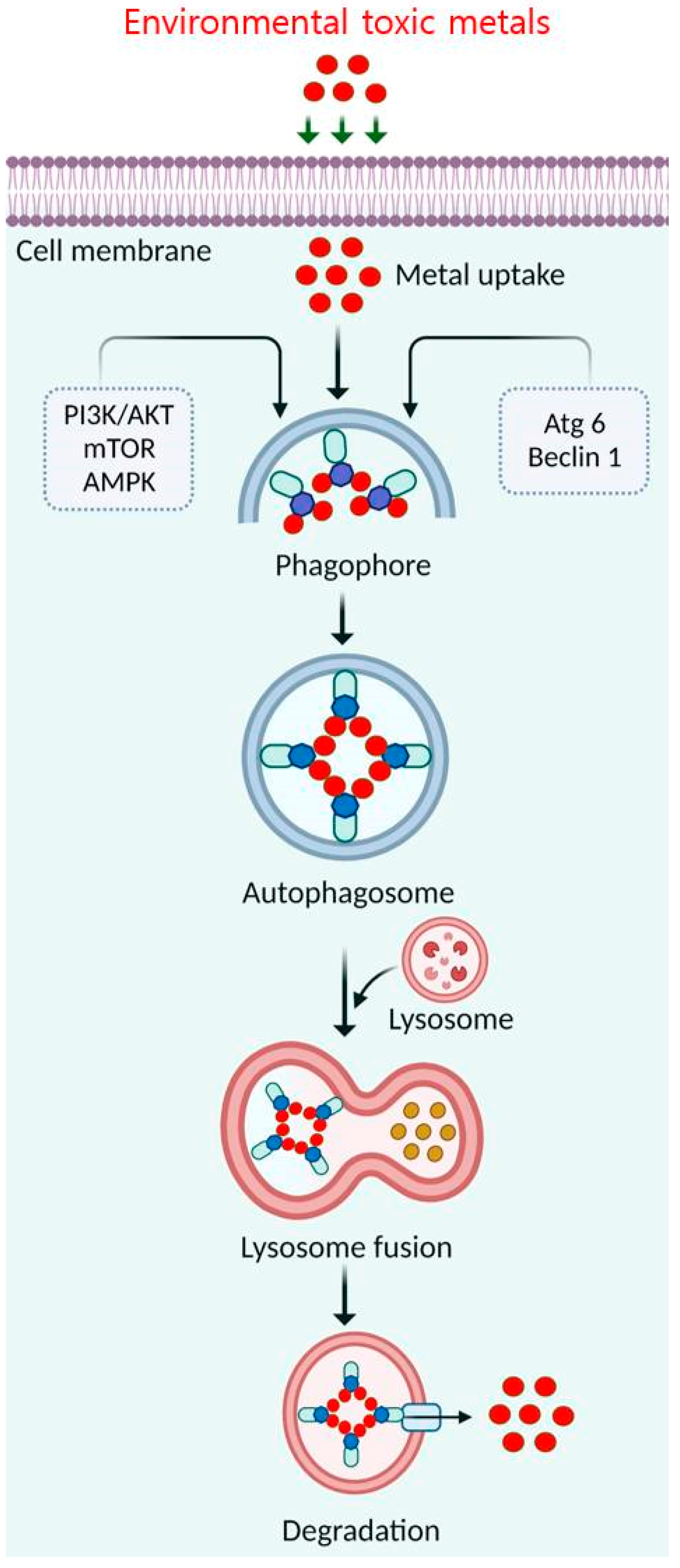
4.3. Elimination of Toxic Metals by Autophagy
4.4. Elimination of Smoke by Autophagy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, H.; Ouyang, Y.; Yin, H.; Cui, H.; Deng, H.; Liu, H.; Jian, Z.; Fang, J.; Zuo, Z.; Wang, X.; et al. Induction of autophagy via the ROS-dependent AMPK-mTOR pathway protects copper-induced spermatogenesis disorder. Redox Biol. 2022, 49, 102227. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Rahman, H.; Mamun-Or-Rashid, A.N.M.; Hwang, H.; Chung, S.; Kim, B.; Rhim, H. Autophagy Modulation in Aggresome Formation: Emerging Implications and Treatments of Alzheimer’s Disease. Biomedicines 2022, 10, 1027. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, P.; Ge, L. Targeting of biomolecular condensates to the autophagy pathway. Trends Cell Biol. 2022; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Vitto, V.A.M.; Bianchin, S.; Zolondick, A.A.; Pellielo, G.; Rimessi, A.; Chianese, D.; Yang, H.; Carbone, M.; Pinton, P.; Giorgi, C.; et al. Molecular Mechanisms of Autophagy in Cancer Development, Progression, and Therapy. Biomedicines 2022, 10, 1596. [Google Scholar] [CrossRef]
- Rahman, M.A.; Cho, Y.; Nam, G.; Rhim, H. Antioxidant compound, oxyresveratrol, inhibits APP production through the AMPK/ULK1/mTOR-mediated autophagy pathway in mouse cortical astrocytes. Antioxidants 2021, 10, 408. [Google Scholar] [CrossRef]
- Rodgers, S.J.; Jones, E.I.; Arumugam, S.; Hamila, S.A.; Danne, J.; Gurung, R.; Eramo, M.J.; Nanayakkara, R.; Ramm, G.; McGrath, M.J.; et al. Endosome maturation links PI3Kα signaling to lysosome repopulation during basal autophagy. EMBO J. 2022, 41, e110398. [Google Scholar] [CrossRef]
- Ravichandran, R.; PriyaDharshini, L.C.; Sakthivel, K.M.; Rasmi, R.R. Role and regulation of autophagy in cancer. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2022, 1868, 166400. [Google Scholar]
- Uddin, M.S.; Rahman, A.; Kabir, T.; Behl, T.; Mathew, B.; Perveen, A.; Barreto, G.E.; Bin-Jumah, M.N.; Abdel-Daim, M.M.; Ashraf, G.M. Multifarious roles of mTOR signaling in cognitive aging and cerebrovascular dysfunction of Alzheimer’s disease. Iubmb Life 2020, 72, 1843–1855. [Google Scholar] [CrossRef]
- Sarkar, S. Chemical screening platforms for autophagy drug discovery to identify therapeutic candidates for Huntington’s disease and other neurodegenerative disorders. Drug Discov. Today Technol. 2013, 10, e137–e144. [Google Scholar] [CrossRef]
- Zhou, X.; Jin, W.; Sun, H.; Li, C.; Jia, J. Perturbation of autophagy: An intrinsic toxicity mechanism of nanoparticles. Sci. Total Environ. 2022, 823, 153629. [Google Scholar] [CrossRef]
- Chatterjee, S.; Sarkar, S.; Bhattacharya, S. Toxic metals and autophagy. Chem. Res. Toxicol. 2014, 27, 1887–1900. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Wang, X.; Yang, C.; Zhu, J.; Jin, Y.; Fu, Z. The regulation of autophagy in the pesticide-induced toxicity: Angel or demon? Chemosphere 2020, 242, 125138. [Google Scholar] [CrossRef] [PubMed]
- Pesonen, M.; Vähäkangas, K. Autophagy in exposure to environmental chemicals. Toxicol. Lett. 2019, 305, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ventriglio, A.; Bellomo, A.; di Gioia, I.; Di Sabatino, D.; Favale, D.; De Berardis, D.; Cianconi, P. Environmental pollution and mental health: A narrative review of literature. CNS Spectr. 2021, 26, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Rahman, M.S.; Uddin, M.J.; Mamum-Or-Rashid, A.N.M.; Pang, M.-G.; Rhim, H. Emerging risk of environmental factors: Insight mechanisms of Alzheimer’s diseases. Environ. Sci. Pollut. Res. 2020, 27, 44659–44672. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Rhim, H. Therapeutic implication of autophagy in neurodegenerative diseases. BMB Rep. 2017, 50, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Martínez-García, G.G.; Mariño, G. Autophagy role in environmental pollutants exposure. Prog. Mol. Biol. Transl. Sci. 2020, 172, 257–291. [Google Scholar]
- Tian, L.; Ji, H.; Wang, W.; Han, X.; Zhang, X.; Li, X.; Guo, L.; Huang, L.; Gao, W. Mitochondria-targeted pentacyclic triterpenoid carbon dots for selective cancer cell destruction via inducing autophagy, apoptosis, as well as ferroptosis. Bioorganic Chem. 2023, 130, 106259. [Google Scholar] [CrossRef]
- Lin, P.-H. Highlights in Autophagy—From Basic Mechanisms to Human Disorder Treatments. Cell 2023, 12, 188. [Google Scholar] [CrossRef]
- Guan, X.; Iyaswamy, A.; Sreenivasmurthy, S.G.; Su, C.; Zhu, Z.; Liu, J.; Kan, Y.; Cheung, K.-H.; Lu, J.; Tan, J.; et al. Mechanistic Insights into Selective Autophagy Subtypes in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 3609. [Google Scholar] [CrossRef]
- Berardi, D.E.; Bock-Hughes, A.; Terry, A.R.; Drake, L.E.; Bozek, G.; Macleod, K.F. Lipid droplet turnover at the lysosome inhibits growth of hepatocellular carcinoma in a BNIP3-dependent manner. Sci. Adv. 2022, 8, eabo2510. [Google Scholar] [CrossRef] [PubMed]
- Zellner, S. Exploring Selective Autophagy Cargo and Machinery Using Proximity Proteomics. 2022. Available online: edoc.ub.uni-muenchen.de (accessed on 27 January 2023).
- Chino, H.; Mizushima, N. ER-Phagy: Quality and Quantity Control of the Endoplasmic Reticulum by Autophagy. Cold Spring Harb. Perspect. Biol. 2022, 15, a041256. [Google Scholar] [CrossRef] [PubMed]
- Trendel, J.; Aleksić, M.; Bertolini, M.; Jochem, M.; Kramer, G.; Pfeffer, S.; Bukau, B.; Krijgsveld, J. Translational Activity Controls Ribophagic Flux and Turnover of Distinct Ribosome Pools. bioRxiv 2022. [Google Scholar] [CrossRef]
- Halcrow, E.F.J.; Mazza, R.; Diversi, A.; Enright, A.; D’Avino, P.P. Midbody Proteins Display Distinct Dynamics during Cytokinesis. Cells 2022, 11, 3337. [Google Scholar] [CrossRef] [PubMed]
- Almacellas, E.; Mauvezin, C. Emerging roles of mitotic autophagy. J. Cell Sci. 2022, 135, jcs255802. [Google Scholar] [CrossRef]
- Wu, P.; Choo, C.Y.L.; Lu, H.; Wei, X.; Chen, Y.; Yago, J.I.; Chung, K. Pexophagy is critical for fungal development, stress response, and virulence in Alternaria alternata. Mol. Plant Pathol. 2022, 23, 1538–1554. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, J.; Gao, W.; Sun, Y.; Liu, X.; Lv, Z.; Li, L.; Xue, D. The lncRNA TCONS_00021785/miR-21-5p/Trim33 axis regulates VMP1-mediated zymophagy, reduces the activation of trypsinogen, and promotes acinar cell recovery. Cell Death Discov. 2022, 8, 65. [Google Scholar] [CrossRef]
- Boland, B.; Yu, W.H.; Corti, O.; Mollereau, B.; Henriques, A.; Bezard, E.; Pastores, G.M.; Rubinsztein, D.C.; Nixon, R.A.; Duchen, M.; et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018, 17, 660–688. [Google Scholar] [CrossRef]
- Tedesco, B.; Vendredy, L.; Timmerman, V.; Poletti, A. The chaperone-assisted selective autophagy complex dynamics and dysfunctions. Autophagy 2023, 1–23. [Google Scholar] [CrossRef]
- Yu, G.; Klionsky, D.J. Life and Death Decisions—The Many Faces of Autophagy in Cell Survival and Cell Death. Biomolecules 2022, 12, 866. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, M.R.; Zaman, T.; Uddin, M.S.; Islam, R.; Abdel-Daim, M.M.; Rhim, H. Emerging Potential of Naturally Occurring Autophagy Modulators Against Neurodegeneration. Curr. Pharm. Des. 2020, 26, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Wollert, T. Don’t forget to be picky–selective autophagy of protein aggregates in neurodegenerative diseases. Curr. Opin. Cell Biol. 2022, 75, 102064. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.-H.; Yang, S.; Wei, J.; Shea, C.R.; Zhong, W.; Wang, F.; Shah, P.; Kibriya, M.G.; Cui, X.; Ahsan, H.; et al. Autophagy of the m6A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat. Commun. 2021, 12, 2183. [Google Scholar] [CrossRef]
- Paskeh, M.D.A.; Entezari, M.; Clark, C.; Zabolian, A.; Ranjbar, E.; Farahani, M.V.; Saleki, H.; Sharifzadeh, S.O.; Far, F.B.; Ashrafizadeh, M.; et al. Targeted regulation of autophagy using nanoparticles: New insight into cancer therapy. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2022, 1868, 166326. [Google Scholar] [CrossRef]
- Patwa, J.; Flora, S.J.S. Heavy metal-induced cerebral small vessel disease: Insights into molecular mechanisms and possible reversal strategies. Int. J. Mol. Sci. 2020, 21, 3862. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, P.; Huang, Y.; Li, Y.-F.; Lu, J.; Li, M.; Kurihara, H.; Luo, Z.; Meng, T.; Onishi, M.; et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 2021, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Cui, Y.; Zhang, X.; Zhang, Y.; Chen, M.; Zhou, T.; Lan, X.; Dong, W.; Pan, C. Chlorpyrifos induction of testicular-cell apoptosis through generation of reactive oxygen species and phosphorylation of AMPK. J. Agric. Food Chem. 2018, 66, 12455–12470. [Google Scholar] [CrossRef]
- Janda, E.; Isidoro, C.; Carresi, C.; Mollace, V. Defective autophagy in Parkinson’s disease: Role of oxidative stress. Mol. Neurobiol. 2012, 46, 639–661. [Google Scholar] [CrossRef]
- Dagda, R.K.; Das Banerjee, T.; Janda, E. How Parkinsonian toxins dysregulate the autophagy machinery. Int. J. Mol. Sci. 2013, 14, 22163–22189. [Google Scholar] [CrossRef]
- Wills, J.; Credle, J.; Oaks, A.W.; Duka, V.; Lee, J.-H.; Jones, J.; Sidhu, A. Paraquat, but not maneb, induces synucleinopathy and tauopathy in striata of mice through inhibition of proteasomal and autophagic pathways. PLoS ONE 2012, 7, e30745. [Google Scholar] [CrossRef]
- Rahman, M.A.; Bishayee, K.; Habib, K.; Sadra, A.; Huh, S.-O. 18alpha-Glycyrrhetinic acid lethality for neuroblastoma cells via de-regulating the Beclin-1/Bcl-2 complex and inducing apoptosis. Biochem. Pharm. 2016, 117, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Bishayee, K.; Sadra, A.; Huh, S.-O. Oxyresveratrol activates parallel apoptotic and autophagic cell death pathways in neuroblastoma cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 23–36. [Google Scholar] [CrossRef]
- Rotimi, D.E.; Singh, S.K. Interaction between apoptosis and autophagy in testicular function. Andrologia 2022, 54, e14602. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, A.; Wang, X.; Zeng, X.; Xing, H. AMPK/PPAR-γ/NF-κB axis participates in ROS-mediated apoptosis and autophagy caused by cadmium in pig liver. Environ. Pollut. 2022, 294, 118659. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Akter, S.; Dorotea, D.; Mazumder, A.; Uddin, M.N.; Hannan, M.A.; Hossen, M.J.; Ahmed, M.S.; Kim, W.; Kim, B.; et al. Renoprotective potentials of small molecule natural products targeting mitochondrial dysfunction. Front. Pharmacol. 2022, 13, 925993. [Google Scholar] [CrossRef]
- Seyrek, K.; Wohlfromm, F.; Espe, J.; Lavrik, I.N. The cross-talk of autophagy and apoptosis in breast carcinoma: Implications for novel therapies? Biochem. J. 2022, 479, 1581–1608. [Google Scholar] [CrossRef]
- Riederer, E.; Cang, C.; Ren, D. Lysosomal Ion Channels: What Are They Good For and Are They Druggable Targets? Annu. Rev. Pharmacol. Toxicol. 2022, 63, 19–41. [Google Scholar] [CrossRef]
- Wu, D.-L.; Cheng, L.; Rao, Q.-X.; Wang, X.-L.; Zhang, Q.-C.; Yao, C.-X.; Chen, S.-S.; Liu, X.; Song, W.; Zhou, J.-X. Toxic Effects and Transcriptional Responses in Zebrafish Liver Cells Following Perfluorooctanoic Acid Exposure. Aquat. Toxicol. 2022, 253, 106328. [Google Scholar] [CrossRef]
- Ramalingam, M.; Jeong, H.-S.; Hwang, J.; Cho, H.-H.; Kim, B.C.; Kim, E.; Jang, S. Autophagy Signaling by Neural-Induced Human Adipose Tissue-Derived Stem Cell-Conditioned Medium during Rotenone-Induced Toxicity in SH-SY5Y Cells. Int. J. Mol. Sci. 2022, 23, 4193. [Google Scholar] [CrossRef]
- Venkatesan, R.; Park, Y.U.; Ji, E.; Yeo, E.-J.; Kim, S.Y. Malathion increases apoptotic cell death by inducing lysosomal membrane permeabilization in N2a neuroblastoma cells: A model for neurodegeneration in Alzheimer’s disease. Cell Death Discov. 2017, 3, 17007. [Google Scholar] [CrossRef]
- Elmorsy, E.; Al-Ghafari, A.; Al Doghaither, H.; Salama, M.; Carter, W.G. An Investigation of the Neurotoxic Effects of Malathion, Chlorpyrifos, and Paraquat to Different Brain Regions. Brain Sci. 2022, 12, 975. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.C.; Duarte, F.V.; Varela, A.T.I.F.; Rolo, A.P.; Palmeira, C.M.M.; Dorta, D.J. Exposure to BDE-153 induces autophagy in HepG2 cells. Toxicol. Vitr. 2017, 42, 61–68. [Google Scholar] [CrossRef]
- Zavadskiy, S.; Sologova, S.; Moldogazieva, N. Oxidative distress in aging and age-related diseases: Spatiotemporal dysregulation of protein oxidation and degradation. Biochimie 2022, 195, 114–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xia, Q.; Zhou, Y.; Li, J. Endoplasmic reticulum stress and autophagy contribute to cadmium-induced cytotoxicity in retinal pigment epithelial cells. Toxicol. Lett. 2019, 311, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Rakowski, M.; Porębski, S.; Grzelak, A. Nutraceuticals as Modulators of Autophagy: Relevance in Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 3625. [Google Scholar] [CrossRef]
- Du, Y.; Cai, Z.; Zhou, G.; Liang, W.; Man, Q.; Wang, W. Perfluorooctanoic acid exposure increases both proliferation and apoptosis of human placental trophoblast cells mediated by ER stress-induced ROS or UPR pathways. Ecotoxicol. Environ. Saf. 2022, 236, 113508. [Google Scholar] [CrossRef]
- Su, Q.; Huang, Y.; Wei, Z.; Zhu, C.; Zeng, W.; Wang, S.; Long, S.; Zhang, G.; Yang, J.; Wang, X. A novel multi-gradient PASS nanofibrous membranes with outstanding particulate matter removal efficiency and excellent antimicrobial property. Sep. Purif. Technol. 2023, 307, 122652. [Google Scholar] [CrossRef]
- Onaiwu, G.E.; Okuo, J.M. Quantification of Fine Particulate Matter (PM2. 5) and Its Correlation with Meteorological Parameters Within the Ambient Air of Automobile Workshops in Benin City. Aerosol. Sci. Eng. 2022, 1–10. [Google Scholar] [CrossRef]
- Park, S.; Ku, J.; Lee, S.-M.; Hwang, H.; Lee, N.; Kim, H.; Yoon, K.-J.; Kim, Y.; Choi, S.Q. Potential toxicity of inorganic ions in particulate matter: Ion permeation in lung and disruption of cell metabolism. Sci. Total Environ. 2022, 824, 153818. [Google Scholar] [CrossRef]
- Liu, Y.; He, X.; Liu, J.; Zhang, L.; Xiong, A.; Wang, J.; Liu, S.; Jiang, M.; Luo, L.; Li, G. Transcriptome analysis identifies IL24 as an autophagy modulator in PM2. 5 caused lung dysfunction. Ecotoxicol. Environ. Saf. 2022, 244, 114039. [Google Scholar] [CrossRef]
- Kaur, M.; Chandel, J.; Malik, J.; Naura, A.S. Particulate matter in COPD pathogenesis: An overview. Inflamm. Res. 2022, 71, 797–815. [Google Scholar] [CrossRef] [PubMed]
- Mwase, C.; Phung, T.-K.N.; O’Sullivan, M.J.; Mitchel, J.A.; De Marzio, M.; Kılıç, A.; Weiss, S.T.; Fredberg, J.J.; Park, J.-A. Mechanical Compression of Human Airway Epithelial Cells Induces Release of Extracellular Vesicles Containing Tenascin C. Cells 2022, 11, 256. [Google Scholar] [CrossRef] [PubMed]
- Costa-Beber, L.C.; Guma, F.T.C.R. The macrophage senescence hypothesis: The role of poor heat shock response in pulmonary inflammation and endothelial dysfunction following chronic exposure to air pollution. Inflamm. Res. 2022, 71, 1433–1448. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Huang, H.; Zhang, J.; Liu, Y.; Zhong, W.; Chen, W.; Lu, Y.; Qiao, Y.; Zhao, H.; Meng, X.; et al. HDM induce airway epithelial cell ferroptosis and promote inflammation by activating ferritinophagy in asthma. FASEB J. 2022, 36, e22359. [Google Scholar] [CrossRef] [PubMed]
- Das, D.N. Elucidating Mechanisms of Benzo [a] Pyrene Mediated Apoptotic and Autophagic Cell Death and Its Prevention with Phytotherapeutics. Ph.D. Thesis, National Institute of Technology, Rourkela, India, 2015. [Google Scholar]
- Colasanti, T.; Fiorito, S.; Alessandri, C.; Serafino, A.; Andreola, F.; Barbati, C.; Morello, F.; Alfè, M.; Di Blasio, G.; Gargiulo, V.; et al. Diesel exhaust particles induce autophagy and citrullination in Normal Human Bronchial Epithelial cells. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, L.K.; Rath, P.; Choudhury, M. A Comprehensive Review on the Classification, Uses, Sources of Nanoparticles (NPs) and Their Toxicity on Health. Aerosol Sci. Eng. 2022, 1–18. [Google Scholar] [CrossRef]
- Zaher, S.; Soliman, M.E.; Elsabahy, M.; Hathout, R.M. Protein nanoparticles as natural drugs carriers for cancer therapy. Adv. Tradit. Med. 2022, 1–30. [Google Scholar] [CrossRef]
- Raj, E.N.; Lin, Y.; Chen, C.; Liu, K.; Chao, J. Selective autophagy pathway of nanoparticles and nanodrugs: Drug delivery and pathophysiological effects. Adv. Ther. 2020, 3, 2000085. [Google Scholar] [CrossRef]
- Shang, M.; Niu, S.; Chang, X.; Li, J.; Zhang, W.; Guo, M.; Wu, T.; Zhang, T.; Tang, M.; Xue, Y. Silver nanoparticle-induced impaired autophagic flux and lysosomal dysfunction contribute to the microglia inflammation polarization. Food Chem. Toxicol. 2022, 170, 113469. [Google Scholar] [CrossRef]
- Negi, S.; Chaudhuri, A.; Kumar, D.N.; Dehari, D.; Singh, S.; Agrawal, A.K. Nanotherapeutics in autophagy: A paradigm shift in cancer treatment. Drug Deliv. Transl. Res. 2022, 12, 2589–2612. [Google Scholar] [CrossRef]
- Alavi, M.; Kamarasu, P.; McClements, D.J.; Moore, M.D. Metal and metal oxide-based antiviral nanoparticles: Properties, mechanisms of action, and applications. Adv. Colloid Interface Sci. 2022, 306, 102726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Tuning Autophagy-Inducing Activity and Toxicity for Lanthanide Nanocrystals; Springer Nature: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Rehman, Y.; Qutaish, H.; Kim, J.H.; Huang, X.-F.; Alvi, S.; Konstantinov, K. Microenvironmental Behaviour of Nanotheranostic Systems for Controlled Oxidative Stress and Cancer Treatment. Nanomaterials 2022, 12, 2462. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Krantz, S.; Qin, X.; Li, S.; Gunasekara, H.; Kim, Y.-M.; Zimnicka, A.; Bae, M.; Ma, K.; Toth, P.T.; et al. Caveolin-1 controls mitochondrial damage and ROS production by regulating fission-fusion dynamics and mitophagy. Redox Biol. 2022, 52, 102304. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, W.; Lu, S.-Y.; Cai, X.; Chen, C.; Zhang, Q.; Duan, Y.; Xie, D.; Zhang, Q.; Ran, H.; et al. Biodegradable doxorubicin-loaded ferric phosphate nanosheets for specific tumor elimination through autophagy inhibition-enhanced apoptosis/ferroptosis pathway. Chem. Eng. J. 2023, 454, 140455. [Google Scholar] [CrossRef]
- Xiong, P.; Huang, X.; Ye, N.; Lu, Q.; Zhang, G.; Peng, S.; Wang, H.; Liu, Y. Cytotoxicity of Metal—Based Nanoparticles: From Mechanisms and Methods of Evaluation to Pathological Manifestations. Adv. Sci. 2022, 9, 2106049. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, M.S.; Uddin, M.J.; Mamun-Or-Rashid, A.N.M. Proteostasis and Neurodegeneration: Perspectives in the Pathogenesis of Molecular and Cellular Mechanisms. In Quality Control of Cellular Protein in Neurodegenerative Disorders; IGI Global: Hershey, PA, USA, 2020; pp. 154–178. [Google Scholar]
- Uddin, M.S.; Mamun, A.A.; Rahman, M.A.; Behl, T.; Perveen, A.; Hafeez, A.; Bin-Jumah, M.N.; Abdel-Daim, M.M.; Ashraf, G.D. Emerging proof of protein misfolding and interactions in multifactorial Alzheimer’s disease. Curr. Top. Med. Chem. 2020, 20, 2380–2390. [Google Scholar] [CrossRef]
- Kumari, B.; Bharti, V.K. Environmental toxicology of arsenic: Current understanding of toxicity, detection, and remedial strategies. Res. Sq. 2022; preprint. [Google Scholar]
- Esmaeili, Y.; Yarjanli, Z.; Pakniya, F.; Bidram, E.; Łos, M.J.; Eshraghi, M.; Klionsky, D.J.; Ghavami, S.; Zarrabi, A. Targeting autophagy, oxidative stress, and ER stress for neurodegenerative diseases treatment. J. Control. Release 2022, 345, 147–175. [Google Scholar] [CrossRef]
- Zhao, X.; Shi, X.; Yao, Y.; Li, X.; Xu, S. Autophagy flux inhibition mediated by lysosomal dysfunction participates in the cadmium exposure-induced cardiotoxicity in swine. BioFactors 2022, 48, 946–958. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Oh, S.-H. Arsenite-induced cytotoxicity is regulated by p38-SQSTM1/p62 and JNK-BNIP3L/Nix signaling in lung cancer cells. Biochem. Biophys. Res. Commun. 2022, 587, 16–23. [Google Scholar] [CrossRef]
- Tang, X.; Wang, Z.; Hu, S.; Zhou, B. Assessing Drug-Induced Mitochondrial Toxicity in Cardiomyocytes: Implications for Preclinical Cardiac Safety Evaluation. Pharmaceutics 2022, 14, 1313. [Google Scholar] [CrossRef] [PubMed]
- Djavaheri-Mergny, M.; Giuriato, S.; Tschan, M.P.; Humbert, M. Therapeutic Modulation of Autophagy in Leukaemia and Lymphoma. Cells 2019, 8, 103. [Google Scholar] [CrossRef] [Green Version]
- Oršolić, N.; Jembrek, M.J. Molecular and Cellular Mechanisms of Propolis and Its Polyphenolic Compounds against Cancer. Int. J. Mol. Sci. 2022, 23, 10479. [Google Scholar] [CrossRef]
- Wang, S.H.; Shih, Y.L.; Ko, W.C.; Wei, Y.H.; Shih, C.M. Cadmium-induced autophagy and apoptosis are mediated by a calcium signaling pathway. Cell. Mol. Life Sci. 2008, 65, 3640–3652. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C. Evaluation of Protective Effect of Naringenin on Cadmium-Induced Kidney Injury in Rats. Pak. J. Zoöl. 2022. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Kolettis, N. Interplay of Apoptosis and Autophagy in Acute Lymphoblastic Leukemic Cells; California State University: Northridge, CA, USA, 2014. [Google Scholar]
- Lankford, E.; Thomas, C.S.; Marchi, S.; Brown, S.; Woski, S.A.; Vincent, J.B. Examining the Potential Formation of Ternary Chromium-Histidine-DNA Complexes and Implications for Their Carcinogenicity. Biol. Trace Elem. Res. 2022, 200, 1473–1481. [Google Scholar] [CrossRef]
- Ye, L.; Li, X.; Chen, X.; Lian, Q.; Ge, R.-S. Environmental toxicants on Leydig cell function. In Spermatogenesis; CRC Press: Boca Raton, FL, USA, 2018; pp. 245–267. [Google Scholar]
- Di Gioacchino, M.; Petrarca, C.; Perrone, A.; Farina, M.; Sabbioni, E.; Hartung, T.; Martino, S.; Esposito, D.L.; Lotti, L.V.; Mariani-Costantini, R. Autophagy as an ultrastructural marker of heavy metal toxicity in human cord blood hematopoietic stem cells. Sci. Total Environ. 2008, 392, 50–58. [Google Scholar] [CrossRef]
- Scharf, P.; Broering, M.F.; Da Rocha, G.H.O.; Farsky, S.H.P. Cellular and molecular mechanisms of environmental pollutants on hematopoiesis. Int. J. Mol. Sci. 2020, 21, 6996. [Google Scholar] [CrossRef]
- Ranjbary, A.G.; Saleh, G.K.; Azimi, M.; Karimian, F.; Mehrzad, J.; Zohdi, J. Superparamagnetic Iron Oxide Nanoparticles Induce Apoptosis in HT-29 Cells by Stimulating Oxidative Stress and Damaging DNA. Biol. Trace Elem. Res. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Tam, E.; Reno, C.; Nguyen, K.; Cho, S.; Sweeney, G. Importance of Autophagy in Mediating Cellular Responses to Iron Overload in Cardiomyocytes. Rev. Cardiovasc. Med. 2022, 23, 167. [Google Scholar] [CrossRef]
- Gubas, A.; Dikic, I. A guide to the regulation of selective autophagy receptors. FEBS J. 2022, 289, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Yeh, K.; Li, L.; Wania, F.; Abbatt, J.P. Thirdhand smoke from tobacco, e-cigarettes, cannabis, methamphetamine and cocaine: Partitioning, reactive fate, and human exposure in indoor environments. Environ. Int. 2022, 160, 107063. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, G.; Yuan, S.; Tan, C.; Lian, P.; Fu, L.; Hou, Q.; Xu, B.; Wang, H. Cigarette smoke-induced pulmonary inflammation and autophagy are attenuated in Ephx2-deficient mice. Inflammation 2017, 40, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Li, Y.; Fu, Y.; Huang, L.; Liu, B.; Zhang, L.; Shao, X.M.; Xiao, D. Inhibition of autophagy signaling via 3-methyladenine rescued nicotine-mediated cardiac pathological effects and heart dysfunctions. Int. J. Biol. Sci. 2020, 16, 1349. [Google Scholar] [CrossRef] [Green Version]
- Mercado, N.; Colley, T.; Baker, J.R.; Vuppussetty, C.; Kono, Y.; Clarke, C.; Tooze, S.; Johansen, T.; Barnes, P.J. Bicaudal D1 impairs autophagosome maturation in chronic obstructive pulmonary disease. FASEB BioAdv. 2019, 1, 688–705. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Z.; Shan, D.; Wu, Y.; Zhao, Y.; Li, C.; Shu, Y.; Linghu, X.; Wang, B. Adverse effects of exposure to fine particles and ultrafine particles in the environment on different organs of organisms. J. Environ. Sci. 2022; in press. [Google Scholar] [CrossRef]
- Koskela, A.; Manai, F.; Basagni, F.; Liukkonen, M.; Rosini, M.; Govoni, S.; Monte, M.D.; Smedowski, A.; Kaarniranta, K.; Amadio, M. Nature-Inspired Hybrids (NIH) Improve Proteostasis by Activating Nrf2-Mediated Protective Pathways in Retinal Pigment Epithelial Cells. Antioxidants 2022, 11, 1385. [Google Scholar] [CrossRef]
- Tran, I.; Ji, C.; Ni, I.; Min, T.; Tang, D.; Vij, N. Role of cigarette smoke–induced aggresome formation in chronic obstructive pulmonary disease–emphysema pathogenesis. Am. J. Respir. Cell Mol. Biol. 2015, 53, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Vij, N.; Chandramani-Shivalingappa, P.; Van Westphal, C.; Hole, R.; Bodas, M. Cigarette smoke-induced autophagy impairment accelerates lung aging, COPD-emphysema exacerbations and pathogenesis. Am. J. Physiol.-Cell Physiol. 2018, 314, C73–C87. [Google Scholar] [CrossRef] [Green Version]
- Duszka, K.; Gregor, A.; Guillou, H.; König, J.; Wahli, W. Peroxisome Proliferator-Activated Receptors and Caloric Restriction—Common Pathways Affecting Metabolism, Health, and Longevity. Cells 2020, 9, 1708. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S. The Regulation of Inflammatory Responses of Airway Epithelial Cells and Fibroblasts to Rhinoviral Infection. Ph.D. Thesis, University of Sheffield, Sheffield, UK, 2015. [Google Scholar]
- Zhang, M.; Fang, L.; Zhou, L.; Molino, A.; Valentino, M.R.; Yang, S.; Zhang, J.; Li, Y.; Roth, M. MAPK15-ULK1 signaling regulates mitophagy of airway epithelial cell in chronic obstructive pulmonary disease. Free. Radic. Biol. Med. 2021, 172, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J.; Baker, J.; Donnelly, L.E. Autophagy in asthma and chronic obstructive pulmonary disease. Clin. Sci. 2022, 136, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Dias-Teixeira, K.L.; Gh, M.S.; Romano, J.; Norouzi, F.; Laurie, G.W. Autophagy in the normal and diseased Cornea. Exp. Eye Res. 2022, 225, 109274. [Google Scholar] [CrossRef]
- Carinci, M.; Palumbo, L.; Pellielo, G.; Agyapong, E.D.; Morciano, G.; Patergnani, S.; Giorgi, C.; Pinton, P.; Rimessi, A. The Multifaceted Roles of Autophagy in Infectious, Obstructive, and Malignant Airway Diseases. Biomedicines 2022, 10, 1944. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.A.; Rahman, M.S.; Parvez, M.A.K.; Kim, B. The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms. Toxics 2023, 11, 135. https://doi.org/10.3390/toxics11020135
Rahman MA, Rahman MS, Parvez MAK, Kim B. The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms. Toxics. 2023; 11(2):135. https://doi.org/10.3390/toxics11020135
Chicago/Turabian StyleRahman, Md. Ataur, Md Saidur Rahman, Md. Anowar Khasru Parvez, and Bonglee Kim. 2023. "The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms" Toxics 11, no. 2: 135. https://doi.org/10.3390/toxics11020135
APA StyleRahman, M. A., Rahman, M. S., Parvez, M. A. K., & Kim, B. (2023). The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms. Toxics, 11(2), 135. https://doi.org/10.3390/toxics11020135








