Network Toxicology and Molecular Docking to Investigate the Non-AChE Mechanisms of Organophosphate-Induced Neurodevelopmental Toxicity
Abstract
:1. Introduction
2. Materials and Methods
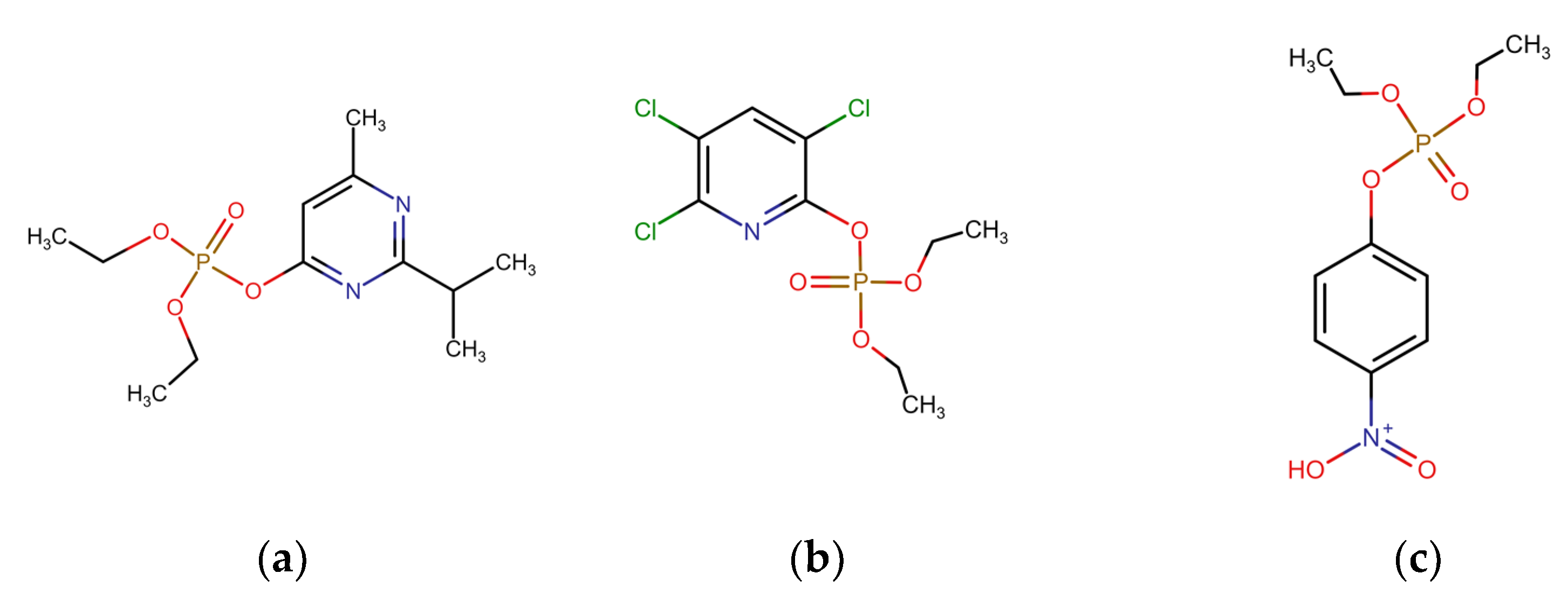
2.1. Chemical Structures of OPs
2.2. Acquisition of OP Targets
2.3. Prediction of NDT Targets
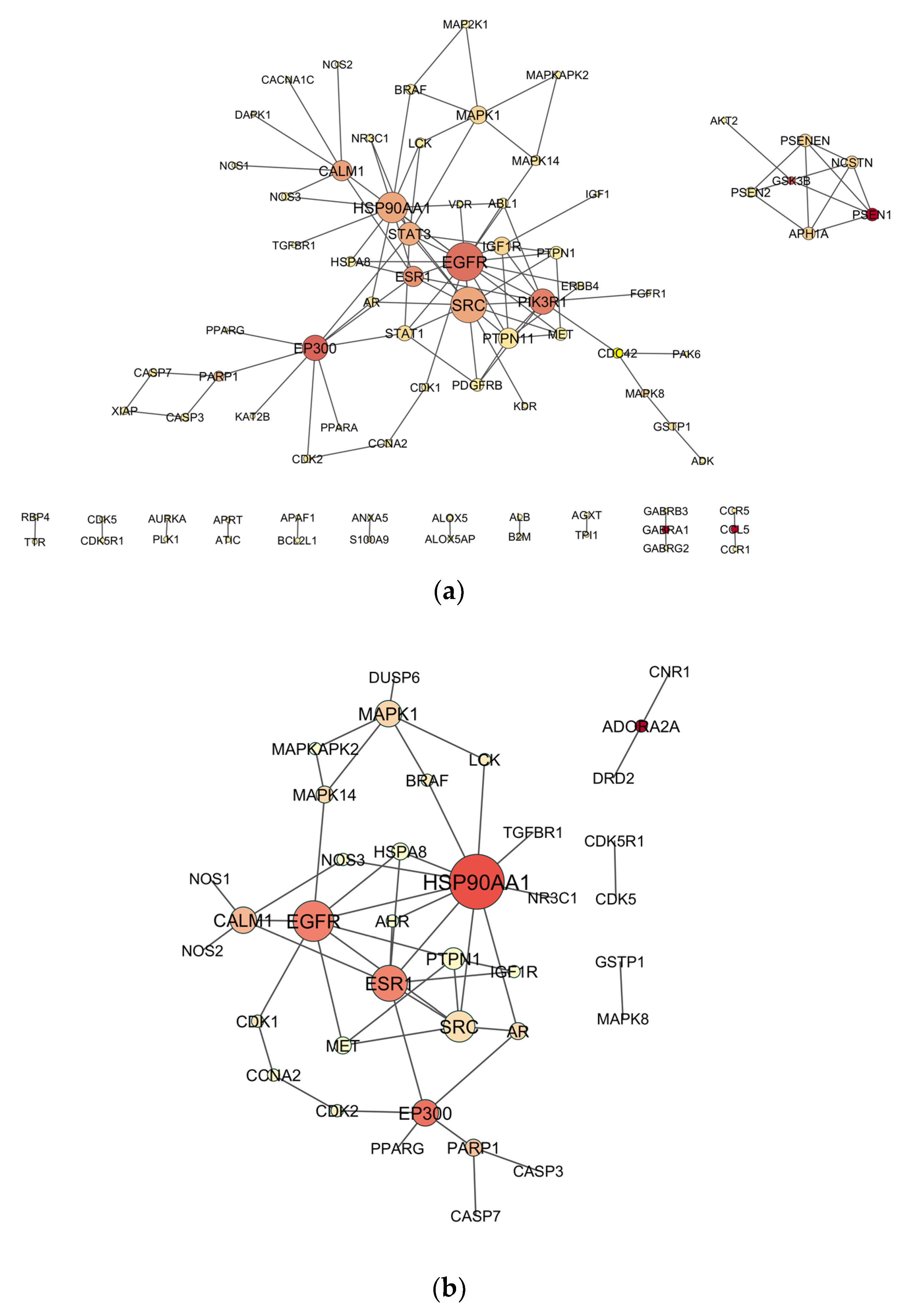
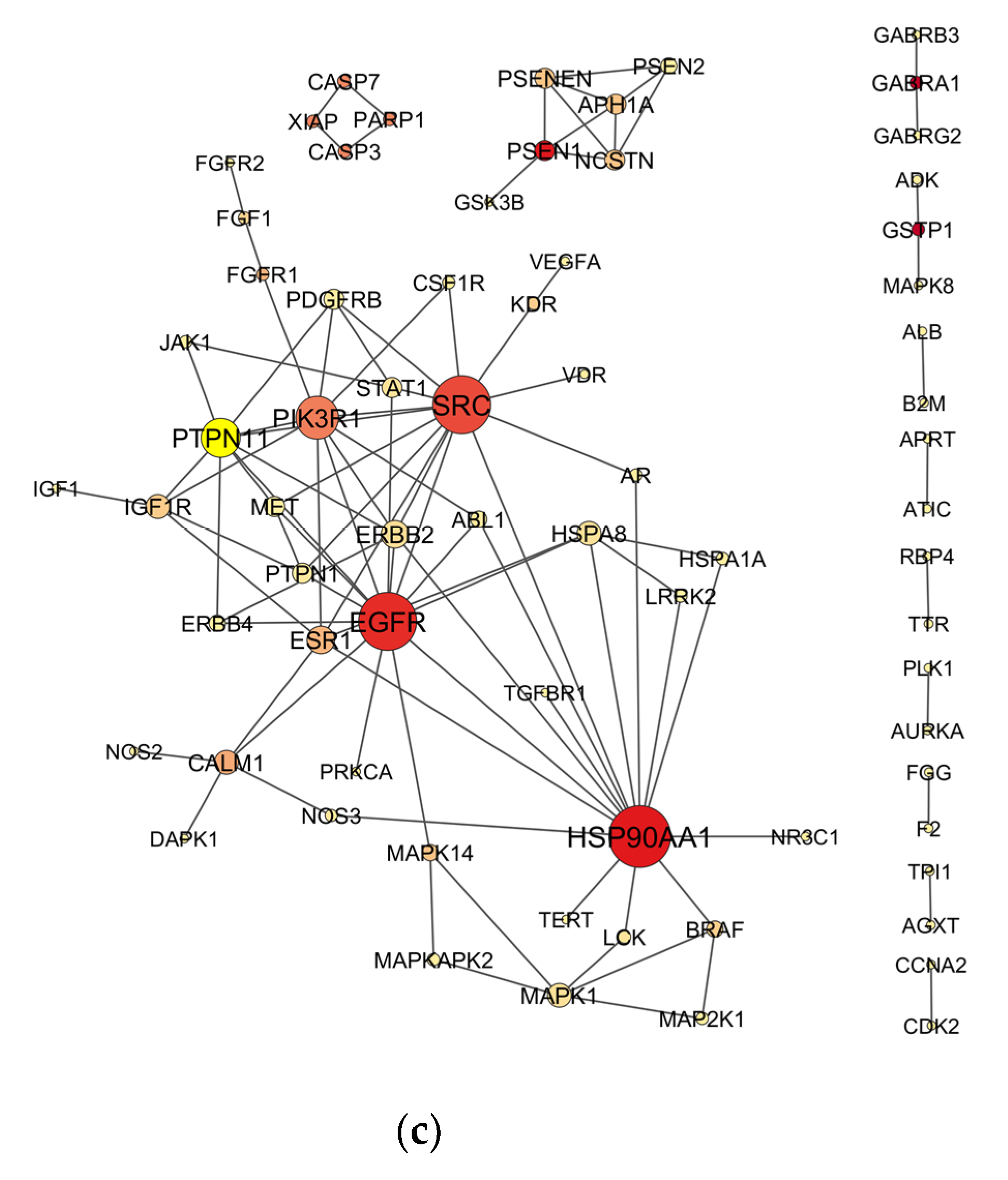
2.4. Venn Analysis and Construction of Protein–Protein Interaction (PPI) Network
2.5. Topological Analysis of PPI Networks
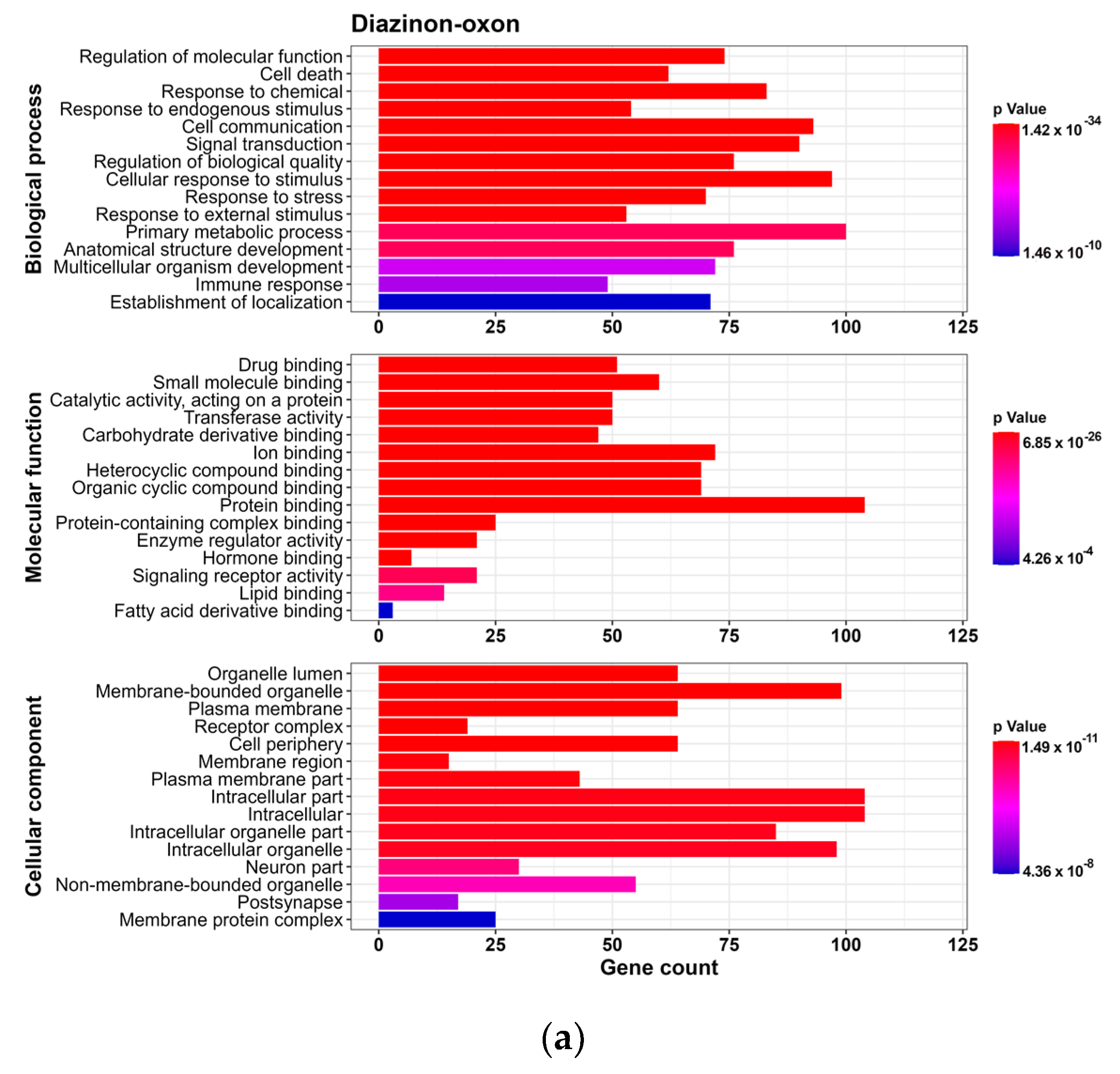
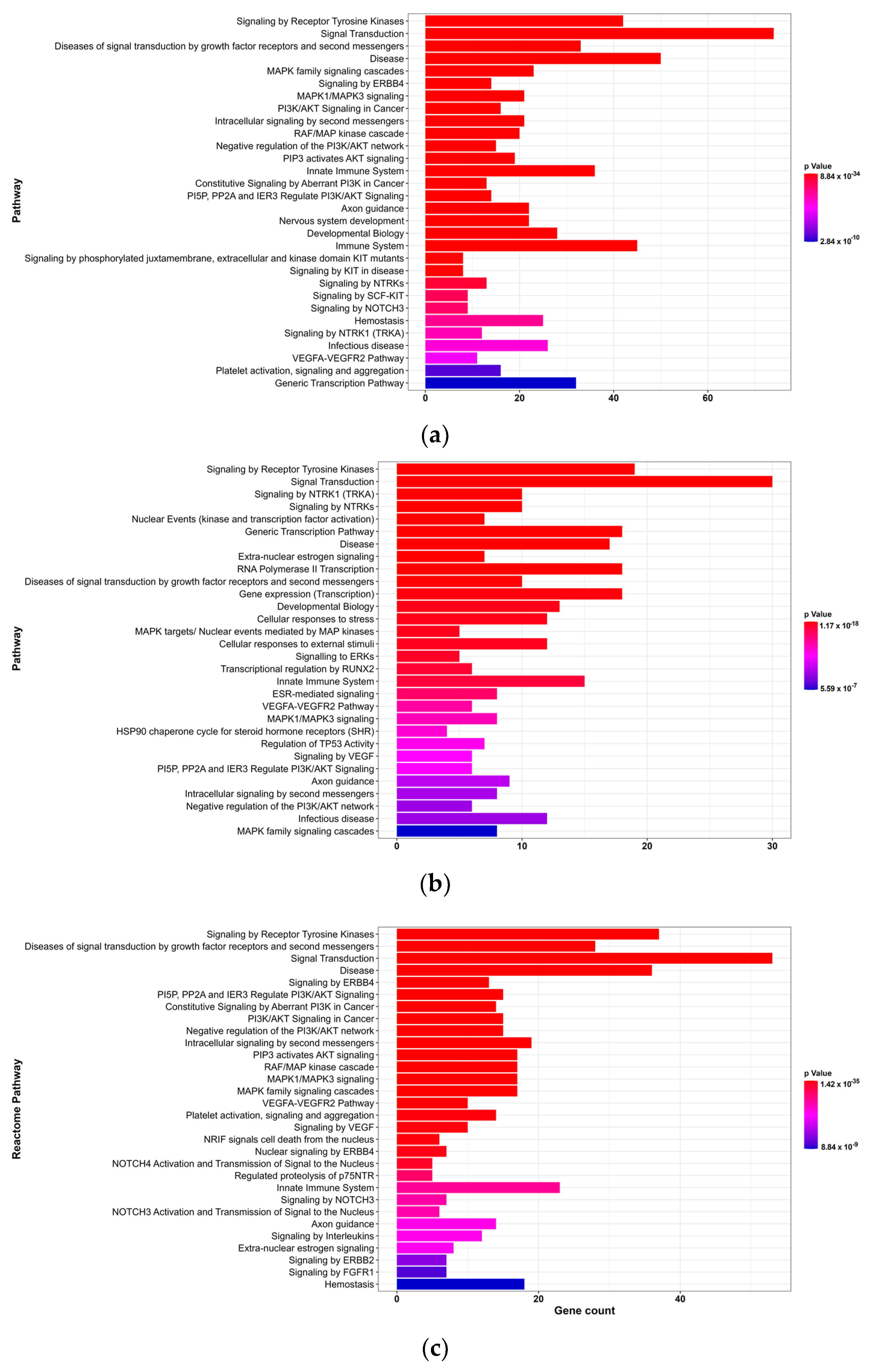
2.6. Enrichment Analysis
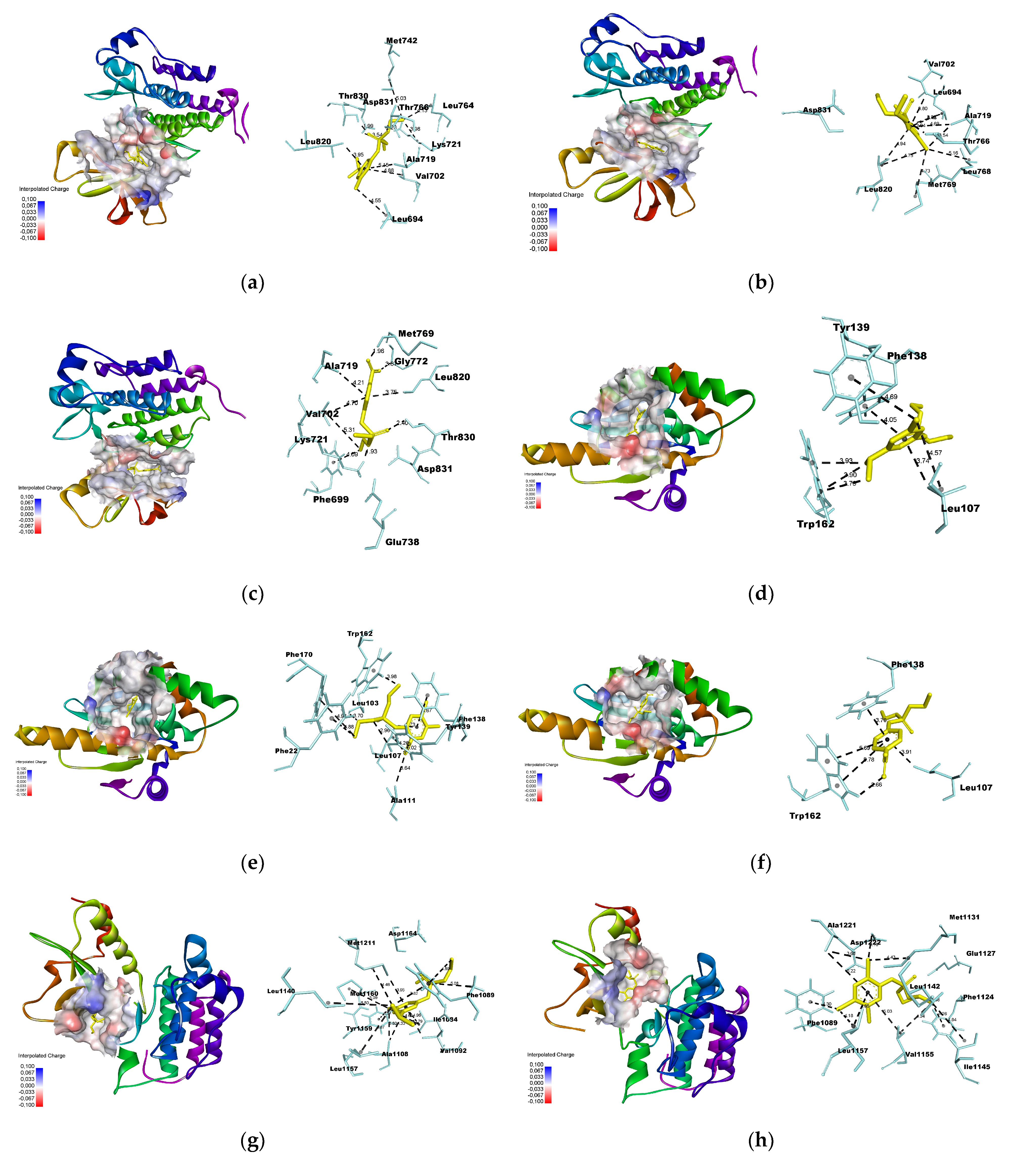
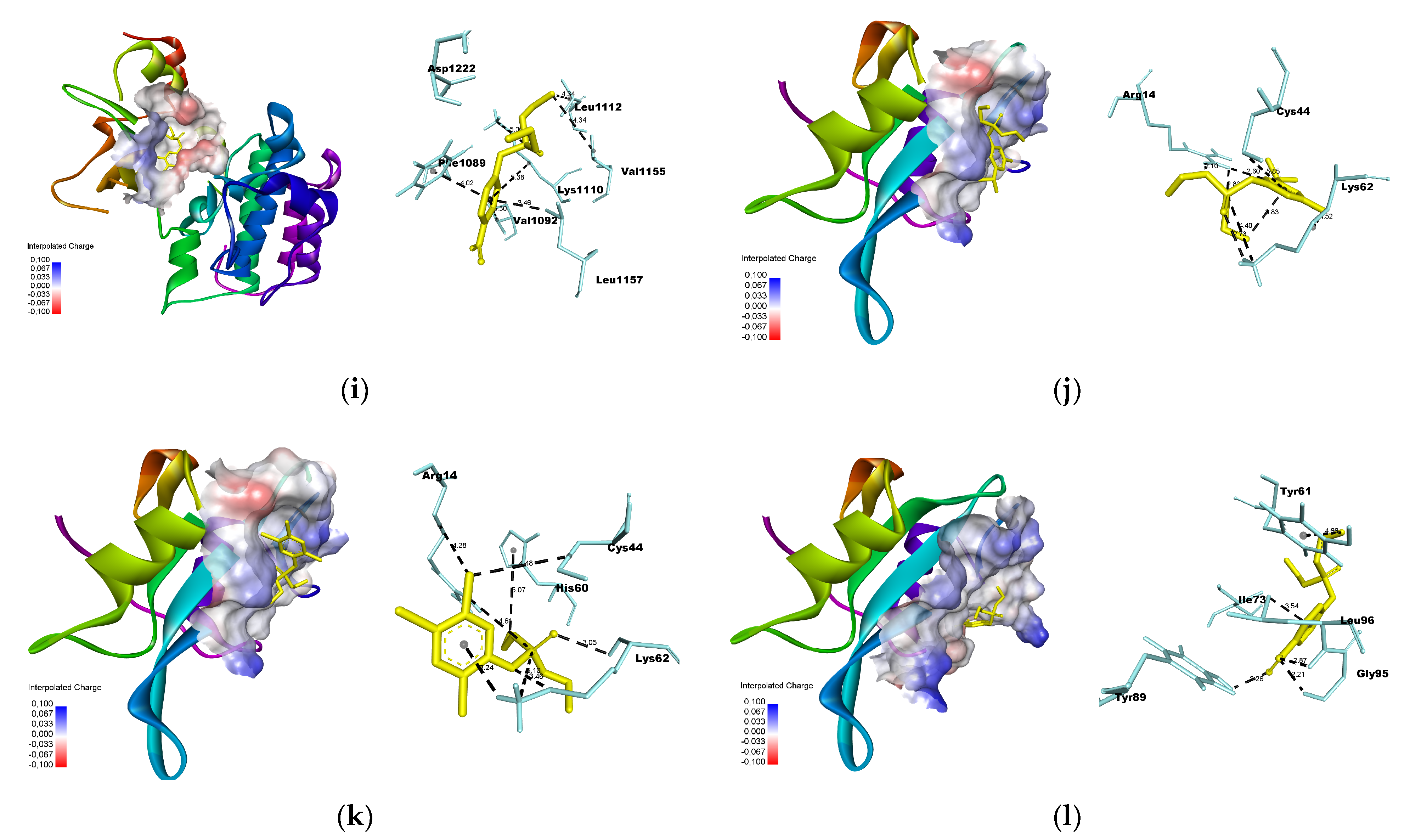
2.7. Molecular Docking
- (a)
- Macromolecule preparation: The 3D structures of the proteins were obtained from the PDB database (https://www.rcsb.org/) accessed on 28 February 2023 in PDB format. Discovery Studio 2021 was used to remove water molecules and ligands, add polar hydrogen bonds, and obtain the x, y, z coordinates for constructing the grid box. Energy minimization was performed using Swiss-PDB Viewer 4.1.0 software.
- (b)
- Ligand preparation: The 3D structures of the ligands were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) accessed on 28 February 2023 in SDF format and converted to PDB format using Open Babel.
- (c)
- Docking simulation was performed using Autodock Vina [24], using a grid box with dimensions of 25 × 25 × 25 Å. The xyz coordinates obtained from Discovery Studio were used to select the center of mass for each macromolecule (Table 1). The poses were selected based on the lowest root-mean-square deviation (RMSD) values, with a maximum threshold of 2.0 Å.
- (d)
3. Results
3.1. Candidate Targets for OP-Induced NDT
3.2. GO and Reactome Pathway Analysis
3.2.1. GO Analysis
3.2.2. Reactome Pathway Analysis
3.3. Molecular Docking
4. Discussion
4.1. Signal Transduction
4.2. Axon Guidance
4.3. Cellular Response to Stress
4.4. Activation of NMDAR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montuori, P.; De Rosa, E.; Di Duca, F.; De Simone, B.; Scippa, S.; Russo, I.; Sorrentino, M.; Sarnacchiaro, P.; Triassi, M. Occurrence, Distribution, and Risk Assessment of Organophosphorus Pesticides in the Aquatic Environment of the Sele River Estuary, Southern Italy. Toxics 2022, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Sumon, K.A.; Rashid, H.; Peeters, E.T.; Bosma, R.H.; Brink, P.J.V.D. Environmental monitoring and risk assessment of organophosphate pesticides in aquatic ecosystems of north-west Bangladesh. Chemosphere 2018, 206, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Akter, R.; Pervin, M.A.; Jahan, H.; Rakhi, S.F.; Reza, A.H.M.M.; Hossain, Z. Toxic Effects of an Organophosphate Pesticide, Envoy 50 SC on the Histopathological, Hematological, and Brain Acetylcholinesterase Activities in Stinging Catfish (Heter-opneustes fossilis). J. Basic Appl. Zool. 2020, 81, 47. [Google Scholar] [CrossRef]
- Tang, J.; Wang, W.; Jiang, Y.; Chu, W. Diazinon exposure produces histological damage, oxidative stress, immune disorders and gut microbiota dysbiosis in crucian carp (Carassius auratus gibelio). Environ. Pollut. 2020, 269, 116129. [Google Scholar] [CrossRef]
- Sandoval-Herrera, N.; Mena, F.; Espinoza, M.; Romero, A. Neurotoxicity of organophosphate pesticides could reduce the ability of fish to escape predation under low doses of exposure. Sci. Rep. 2019, 9, 10530. [Google Scholar] [CrossRef] [PubMed]
- Von Ehrenstein, O.S.; Ling, C.; Cui, X.; Cockburn, M.; Park, A.S.; Yu, F.; Wu, J.; Ritz, B. Prenatal and infant exposure to ambient pesticides and autism spectrum disorder in children: Population based case-control study. BMJ 2019, 364, 1962. [Google Scholar] [CrossRef]
- Gunier, R.B.; Bradman, A.; Harley, K.G.; Kogut, K.; Eskenazi, B. Prenatal Residential Proximity to Agricultural Pesticide Use and IQ in 7-Year-Old Children. Environ. Health Perspect. 2017, 125, 057002. [Google Scholar] [CrossRef]
- Burke, R.D.; Todd, S.W.; Lumsden, E.; Mullins, R.J.; Mamczarz, J.; Fawcett, W.P.; Gullapalli, R.P.; Randall, W.R.; Pereira, E.F.R.; Albuquerque, E.X. Developmental neurotoxicity of the organophosphorus insecticide chlorpyrifos: From clinical findings to preclinical models and potential mechanisms. J. Neurochem. 2017, 142, 162–177. [Google Scholar] [CrossRef]
- Eddleston, M.; Buckley, N.A.; Eyer, P.; Dawson, A.H. Management of Acute Organophosphorus Pesticide Poisoning. Lancet 2008, 371, 597–607. [Google Scholar] [CrossRef]
- Perez-Fernandez, C.; Morales-Navas, M.; Guardia-Escote, L.; Garrido-Cárdenas, J.A.; Colomina, M.T.; Giménez, E.; Sánchez-Santed, F. Long-term effects of low doses of Chlorpyrifos exposure at the preweaning developmental stage: A locomotor, pharmacological, brain gene expression and gut microbiome analysis. Food Chem. Toxicol. 2020, 135, 110865. [Google Scholar] [CrossRef]
- Rush, T.; Liu, X.; Hjelmhaug, J.; Lobner, D. Mechanisms of chlorpyrifos and diazinon induced neurotoxicity in cortical culture. Neuroscience 2010, 166, 899–906. [Google Scholar] [CrossRef]
- van Melis, L.V.; Heusinkveld, H.J.; Langendoen, C.; Peters, A.; Westerink, R.H. Organophosphate insecticides disturb neuronal network development and function via non-AChE mediated mechanisms. Neurotoxicology 2023, 94, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fernandez, C.; Morales-Navas, M.; Guardia-Escote, L.; Colomina, M.T.; Giménez, E.; Sánchez-Santed, F. Postnatal Ex-posure to Low Doses of Chlorpyrifos Induces Long-Term Effects on 5C-SRTT Learning and Performance, Cholinergic and GABAergic Systems and BDNF Expression. Exp. Neurol. 2020, 330, 113356. [Google Scholar] [CrossRef] [PubMed]
- Richendrfer, H.; Creton, R. Chlorpyrifos and malathion have opposite effects on behaviors and brain size that are not correlated to changes in AChE activity. Neurotoxicology 2015, 49, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Ireland, D.; Zhang, S.; Bochenek, V.; Hsieh, J.-H.; Rabeler, C.; Meyer, Z.; Collins, E.-M.S. Differences in neurotoxic outcomes of organophosphorus pesticides revealed via multi-dimensional screening in adult and regenerating planarians. Front. Toxicol. 2022, 4, 948455. [Google Scholar] [CrossRef]
- Sakle, N.S.; More, S.A.; Mokale, S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Sci. Rep. 2020, 10, 17217. [Google Scholar] [CrossRef]
- Iida, M.; Takemoto, K. A network biology-based approach to evaluating the effect of environmental contaminants on human interactome and diseases. Ecotoxicol. Environ. Saf. 2018, 160, 316–327. [Google Scholar] [CrossRef]
- Sohrabi, S.S.; Sohrabi, S.M.; Rashidipour, M.; Mohammadi, M.; Fard, J.K.; Najafgholi, H.M. Identification of common key regulators in rat hepatocyte cell lines under exposure of different pesticides. Gene 2020, 739, 144508. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, K.; Jiao, M.; Jiao, J.; Chen, D.; Yin, Y.; Zhang, J.; Li, F. Study on the Mechanism of Mesaconitine-Induced Hepatotoxicity in Rats Based on Metabonomics and Toxicology Network. Toxins 2022, 14, 486. [Google Scholar] [CrossRef]
- Flaskos, J. The developmental neurotoxicity of organophosphorus insecticides: A direct role for the oxon metabolites. Toxicol. Lett. 2012, 209, 86–93. [Google Scholar] [CrossRef]
- Androutsopoulos, V.P.; Hernandez, A.F.; Liesivuori, J.; Tsatsakis, A.M. A mechanistic overview of health associated effects of low levels of organochlorine and organophosphorous pesticides. Toxicology 2013, 307, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Cherinka, B.; Andrews, B.H.; Sánchez-Gallego, J.; Brownstein, J.; Argudo-Fernández, M.; Blanton, M.; Bundy, K.; Jones, A.; Masters, K.; Law, D.R.; et al. Marvin: A Toolkit for Streamlined Access and Visualization of the SDSS-IV MaNGA Data Set. Astron. J. 2018, 158, 74. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- McRee, D.E. Computational Techniques. In Practical Protein Crystallography; McRee, D.E., Ed.; Elsevier: San Diego, CA, USA, 1999; pp. 91–267. [Google Scholar]
- Ribas, J.; Cubero, E.; Luque, F.J.; Orozco, M. Theoretical Study of Alkyl-π and Aryl-π Interactions. Reconciling Theory and Experiment. J. Org. Chem. 2002, 67, 7057–7065. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.; Barril, X.; Dymock, B.; Sheridan, L.; Surgenor, A.; Beswick, M.; Drysdale, M.; Collier, A.; Massey, A.; Davies, N.; et al. Structure-Activity Relationships in Purine-Based Inhibitor Binding to HSP90 Isoforms. Chem. Biol. 2004, 11, 775–785. [Google Scholar] [CrossRef]
- Mphahlele, M.J.; Maluleka, M.M.; Parbhoo, N.; Malindisa, S.T. Synthesis, Evaluation for Cytotoxicity and Molecular Docking Studies of Benzo[c]Furan-Chalcones for Potential to Inhibit Tubulin Polymerization and/or EGFR-Tyrosine Kinase Phosphorylation. Int. J. Mol. Sci. 2018, 19, 2552. [Google Scholar] [CrossRef]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
- Schiering, N.; Knapp, S.; Marconi, M.; Flocco, M.M.; Cui, J.; Perego, R.; Rusconi, L.; Cristiani, C. Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a. Proc. Natl. Acad. Sci. USA 2003, 100, 12654–12659. [Google Scholar] [CrossRef]
- Damghani, T.; Sedghamiz, T.; Sharifi, S.; Pirhadi, S. Critical c-Met-inhibitor interactions resolved from molecular dynamics simulations of different c-Met complexes. J. Mol. Struct. 2020, 1203, 127456. [Google Scholar] [CrossRef]
- Lange, G. Structure-Based Drug Design—The Use of Protein Structure in Drug Discovery. Compr. Med. Chem. II 2007, 4, 597–650. [Google Scholar] [CrossRef]
- van der Geer, P. Signal Transduction. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 436–439. [Google Scholar]
- Katz, M.; Amit, I.; Yarden, Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Park, J.H.; Shin, I.C.; Koh, H.C. Reactive oxygen species regulated mitochondria-mediated apoptosis in PC12 cells exposed to chlorpyrifos. Toxicol. Appl. Pharmacol. 2012, 263, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Ki, Y.W.; Park, J.H.; Lee, J.E.; Shin, I.C.; Koh, H.C. JNK and P38 MAPK Regulate Oxidative Stress and the Inflammatory Re-sponse in Chlorpyrifos-Induced Apoptosis. Toxicol. Lett. 2013, 218, 235–245. [Google Scholar] [CrossRef]
- Jahan, S.; Kumar, D.; Singh, S.; Kumar, V.; Srivastava, A.; Pandey, A.; Rajpurohit, C.S.; Khanna, V.K.; Pant, A.B. Resveratrol Prevents the Cellular Damages Induced by Monocrotophos via PI3K Signaling Pathway in Human Cord Blood Mesenchymal Stem Cells. Mol. Neurobiol. 2018, 55, 8278–8292. [Google Scholar] [CrossRef]
- Lasagna, M.; Ventura, C.; Hielpos, M.; Mardirosian, M.; Martín, G.; Miret, N.; Randi, A.; Núñez, M.; Cocca, C. Endocrine disruptor chlorpyrifos promotes migration, invasion, and stemness phenotype in 3D cultures of breast cancer cells and induces a wide range of pathways involved in cancer progression. Environ. Res. 2022, 204, 111989. [Google Scholar] [CrossRef]
- Suriyo, T.; Tachachartvanich, P.; Visitnonthachai, D.; Watcharasit, P.; Satayavivad, J. Chlorpyrifos Promotes Colorectal Ad-enocarcinoma H508 Cell Growth through the Activation of EGFR/ERK1/2 Signaling Pathway but Not Cholinergic Pathway. Toxicology 2015, 338, 117–129. [Google Scholar] [CrossRef]
- Subramaniam, S.; Unsicker, K. ERK and Cell Death: ERK1/2 in Neuronal Death. FEBS J. 2010, 277, 22–29. [Google Scholar] [CrossRef]
- Botella, J.A.; Kretzschmar, D.; Kiermayer, C.; Feldmann, P.; Hughes, D.A.; Schneuwly, S. Deregulation of the Egfr/Ras signaling pathway induces age-related brain degeneration in the Drosophila mutant vap. Mol. Biol. Cell 2003, 14, 241–250. [Google Scholar] [CrossRef]
- Stoeckli, E.T. Understanding Axon Guidance: Are We Nearly There Yet? Development 2018, 145, dev151415. [Google Scholar] [CrossRef]
- Romano, R.; Bucci, C. Role of EGFR in the Nervous System. Cells 2020, 9, 1887. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Oh, D.; Dubey, A.K.; Yao, M.; Yang, B.; Groves, J.T.; Sheetz, M. EGFR Family and Src Family Kinase Interactions: Mechanics Matters? Curr. Opin. Cell Biol. 2018, 51, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Knöll, B.; Drescher, U. Src Family Kinases Are Involved in EphA Receptor-Mediated Retinal Axon Guidance. J. Neurosci. 2004, 24, 6248–6257. [Google Scholar] [CrossRef]
- Yam, P.T.; Langlois, S.D.; Morin, S.; Charron, F. Sonic Hedgehog Guides Axons through a Noncanonical, Src-Family-Kinase-Dependent Signaling Pathway. Neuron 2009, 62, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Poliak, S.; Morales, D.; Croteau, L.P.; Krawchuk, D.; Palmesino, E.; Morton, S.; Cloutier, J.F.; Charron, F.; Dalva, M.B.; Ackerman, S.L.; et al. Synergistic Integration of Netrin and Ephrin Axon Guidance Signals by Spinal Motor Neurons. eLife 2015, 4, e10841. [Google Scholar] [CrossRef] [PubMed]
- Desole, C.; Gallo, S.; Vitacolonna, A.; Montarolo, F.; Bertolotto, A.; Vivien, D.; Comoglio, P.; Crepaldi, T. HGF and MET: From Brain Development to Neurological Disorders. Front. Cell Dev. Biol. 2021, 9, 683609. [Google Scholar] [CrossRef]
- Howard, A.S.; Bucelli, R.; Jett, D.A.; Bruun, D.; Yang, D.; Lein, P.J. Chlorpyrifos exerts opposing effects on axonal and dendritic growth in primary neuronal cultures. Toxicol. Appl. Pharmacol. 2005, 207, 112–124. [Google Scholar] [CrossRef]
- Yang, D.; Lauridsen, H.; Buels, K.; Chi, L.-H.; La Du, J.; Bruun, D.A.; Olson, J.R.; Tanguay, R.L.; Lein, P.J. Chlorpyrifos-Oxon Disrupts Zebrafish Axonal Growth and Motor Behavior. Toxicol. Sci. 2011, 121, 146–159. [Google Scholar] [CrossRef]
- Dowell, J.; Elser, B.A.; Schroeder, R.E.; Stevens, H.E. Cellular stress mechanisms of prenatal maternal stress: Heat shock factors and oxidative stress. Neurosci. Lett. 2019, 709, 134368. [Google Scholar] [CrossRef]
- Miller, D.J.; Fort, P.E. Heat Shock Proteins Regulatory Role in Neurodevelopment. Front. Neurosci. 2018, 12, 821. [Google Scholar] [CrossRef]
- Zohn, I.E. Hsp90 and complex birth defects: A plausible mechanism for the interaction of genes and environment. Neurosci. Lett. 2020, 716, 134680. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Liu, T.; Zhang, Z.; Wang, X.; Xu, S. Acute and subchronic toxic effects of atrazine and chlorpyrifos on common carp (Cyprinus carpio L.): Immunotoxicity assessments. Fish Shellfish. Immunol. 2015, 45, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Eder, K.J.; Leutenegger, C.M.; Köhler, H.-R.; Werner, I. Effects of neurotoxic insecticides on heat-shock proteins and cytokine transcription in Chinook salmon (Oncorhynchus tshawytscha). Ecotoxicol. Environ. Saf. 2009, 72, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Slotkin, T.A.; Seidler, F.J. Developmental neurotoxicity of organophosphates targets cell cycle and apoptosis, revealed by transcriptional profiles in vivo and in vitro. Neurotoxicology Teratol. 2012, 34, 232–241. [Google Scholar] [CrossRef]
- Lin, J.-W.; Fu, S.-C.; Liu, J.-M.; Liu, S.-H.; Lee, K.-I.; Fang, K.-M.; Hsu, R.-J.; Huang, C.-F.; Liu, K.-M.; Chang, K.-C.; et al. Chlorpyrifos induces neuronal cell death via both oxidative stress and Akt activation downstream-regulated CHOP-triggered apoptotic pathways. Toxicol. Vitr. 2023, 86, 105483. [Google Scholar] [CrossRef]
- Ewald, R.C.; Cline, H.T. NMDA Receptors and Brain Development; CRC Press: Boca Raton, FL, USA; Taylor & Francis: Abingdon, UK, 2009. [Google Scholar]
- Burnashev, N.; Szepetowski, P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr. Opin. Pharmacol. 2015, 20, 73–82. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, X.; Ashraf, U.; Zheng, B.; Ye, J.; Zhou, D.; Zhang, H.; Song, Y.; Chen, H.; Zhao, S.; et al. Activation of neuronal N-methyl-d-aspartate receptor plays a pivotal role in Japanese encephalitis virus-induced neuronal cell damage. J. Neuroinflammation 2018, 15, 238. [Google Scholar] [CrossRef]
- Jiang, S.; Li, X.; Jin, W.; Duan, X.; Bo, L.; Wu, J.; Zhang, R.; Wang, Y.; Kang, R.; Huang, L. Ketamine-Induced Neurotoxicity Blocked by N-Methyl-d-Aspartate Is Mediated through Activation of PKC/ERK Pathway in Developing Hippocampal Neu-rons. Neurosci. Lett. 2018, 673, 122–131. [Google Scholar] [CrossRef]
- Eisenkraft, A.; Falk, A.; Finkelstein, A. The Role of Glutamate and the Immune System in Organophosphate-induced CNS Damage. Neurotox. Res. 2013, 24, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, T.A.; Seidler, F.J. Comparative Developmental Neurotoxicity of Organophosphates in Vivo: Transcriptional Responses of Pathways for Brain Cell Development, Cell Signaling, Cytotoxicity and Neurotransmitter Systems. Brain Res. Bull. 2007, 72, 232–274. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Wang, H.-Y.; Borgmann-Winter, K.E.; MacDonald, M.L.; Kaprielian, H.; Stucky, A.; Kvasic, J.; Egbujo, C.; Ray, R.; Talbot, K.; et al. Src kinase as a mediator of convergent molecular abnormalities leading to NMDAR hypoactivity in schizophrenia. Mol. Psychiatry 2015, 20, 1091–1100. [Google Scholar] [CrossRef]
- Tang, Y.; Ye, M.; Du, Y.; Qiu, X.; Lv, X.; Yang, W.; Luo, J. EGFR signaling upregulates surface expression of the GluN2B-containing NMDA receptor and contributes to long-term potentiation in the hippocampus. Neuroscience 2015, 304, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.; Vitacolonna, A.; Comoglio, P.; Crepaldi, T. MET Oncogene Controls Invasive Growth by Coupling with NMDA Receptor. Cancers 2022, 14, 4408. [Google Scholar] [CrossRef] [PubMed]


| Molecule | Center Grid Box (xyz Coordinates) |
|---|---|
| HSP90AA1 | 6.63, 11.34, 24.85 |
| HSPA8 | 17.27, −0.84, 2.30 |
| ESR1 | 106.73, 15.02, 96.61 |
| EP300 | 33.20, 9.43, −14.73 |
| PIK3R1 | −20.86, 10.90, 28.28 |
| MET | 61.53, 13.21, 117.38 |
| MAPK1 | −12.97, 13.19, 40.56 |
| EGFR | 22.01, 0.25, 52.79 |
| APH1A | 121.91, 88.97, 129.43 |
| PTPN11 | 31.69, 2.17, 8.03 |
| PTPN1 | 44.80, 14.03, 2.22 |
| CALM1 | 3.70, 26.66, 105.09 |
| STAT3 | 13.12, 55.60, 0.10 |
| ERBB2 | −21.17, 86.36, 138.72 |
| SRC | 19.73, 23.31, 21.53 |
| NCSTN | 113.91, 136.83, 145.71 |
| PSENEN | 184.59, 192.05, 152.23 |
| Protein | Node | k | Clustering Coefficient | BC | ASPL | CC |
|---|---|---|---|---|---|---|
| Diazinon-oxon | ||||||
| Proto-oncogene tyrosine kinase SRC | SRC | 13 | 0.218 | 0.154 | 2.286 | 0.438 |
| Epidermal growth factor receptor | EGFR | 14 | 0.165 | 0.275 | 2.143 | 0.467 |
| Phosphatidylinositol 3-kinase regulatory subunit alpha | PIK3R1 | 9 | 0.250 | 0.238 | 2.408 | 0.415 |
| Tyrosine-protein phosphatase non-receptor type 11 | PTPN11 | 7 | 0.4286 | 0.0164 | 2.7143 | 0.368 |
| Heat shock protein HSP 90-alpha | HSP90AA1 | 11 | 0.109 | 0.154 | 2.429 | 0.412 |
| Gamma-secretase subunit APH-1A | APH1A | 4 | 0.8333 | 0.0667 | 1.5000 | 0.667 |
| Nicastrin | NCSTN | 4 | 0.833 | 0.067 | 1.500 | 0.667 |
| Gamma-secretase subunit PEN-2 | PSENEN | 4 | 0.8333 | 0.0667 | 1.5000 | 0.667 |
| Signal transducer and activator of transcription 3 | STAT3 | 8 | 0.179 | 0.142 | 2.327 | 0.430 |
| Hepatocyte growth factor receptor | MET | 4 | 0.8333 | 0.0002 | 2.9184 | 0.343 |
| Chlorpyrifos oxon | ||||||
| Heat shock protein HSP 90-alpha | HSP90AA1 | 11 | 0.109 | 0.382 | 2.034 | 0.492 |
| Epidermal growth factor receptor | EGFR | 8 | 0.179 | 0.276 | 2.172 | 0.460 |
| Proto-oncogene tyrosine kinase SRC | SRC | |||||
| Estrogen receptor | ESR1 | 7 | 0.143 | 0.269 | 2.103 | 0.475 |
| Tyrosine-protein phosphatase non-receptor type 1 | PTPN1 | 4 | 0.500 | 0.017 | 2.759 | 0.363 |
| Hepatocyte growth factor receptor | MET | 3 | 1.000 | 0.000 | 2.793 | 0.358 |
| Mitogen-activated protein kinase1 | MAPK1 | 5 | 0.100 | 0.095 | 3.172 | 0.315 |
| Calmodulin 1 | CALM1 | 5 | 0.000 | 0.158 | 2.414 | 0.414 |
| Histone acetyltransferase p300 | EP300 | 5 | 0.000 | 0.308 | 2.552 | 0.392 |
| Heat shock cognate 71 kDa protein | HSPA8 | 3 | 0.667 | 0.008 | 2.379 | 0.420 |
| Paraoxon | ||||||
| Proto-oncogene tyrosine kinase SRC | SRC | 14 | 0.220 | 0.264 | 1.875 | 0.533 |
| Epidermal growth factor receptor | EGFR | 14 | 0.198 | 0.312 | 1.800 | 0.556 |
| Tyrosine-protein phosphatase non-receptor type 11 | PTPN11 | 9 | 0.361 | 0.059 | 2.225 | 0.449 |
| Phosphatidylinositol 3-kinase regulatory subunit alpha | PIK3R1 | 10 | 0.289 | 0.180 | 2.150 | 0.465 |
| Steroid hormone receptor ERR2 | ERRB2 | 6 | 0.667 | 0.026 | 2.100 | 0.476 |
| Heat shock protein HSP 90-alpha | HSP90AA1 | 15 | 0.095 | 0.378 | 1.925 | 0.519 |
| Gamma-secretase subunit APH-1A | APH1A | 4 | 0.833 | 0.067 | 1.200 | 0.833 |
| Nicastrin | NCSTN | 4 | 0.833 | 0.067 | 1.200 | 0.833 |
| Hepatocyte growth factor receptor | MET | 4 | 0.833 | 0.001 | 2.450 | 0.408 |
| Gamma-secretase subunit PEN-2 | PSENEN | 4 | 0.833 | 0.067 | 1.200 | 0.833 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, J.A.d.C.R.; Souza, T.; Quintans, I.L.A.d.C.R.; Farias, D. Network Toxicology and Molecular Docking to Investigate the Non-AChE Mechanisms of Organophosphate-Induced Neurodevelopmental Toxicity. Toxics 2023, 11, 710. https://doi.org/10.3390/toxics11080710
Souza JAdCR, Souza T, Quintans ILAdCR, Farias D. Network Toxicology and Molecular Docking to Investigate the Non-AChE Mechanisms of Organophosphate-Induced Neurodevelopmental Toxicity. Toxics. 2023; 11(8):710. https://doi.org/10.3390/toxics11080710
Chicago/Turabian StyleSouza, Juliana Alves da Costa Ribeiro, Terezinha Souza, Isadora Louise Alves da Costa Ribeiro Quintans, and Davi Farias. 2023. "Network Toxicology and Molecular Docking to Investigate the Non-AChE Mechanisms of Organophosphate-Induced Neurodevelopmental Toxicity" Toxics 11, no. 8: 710. https://doi.org/10.3390/toxics11080710
APA StyleSouza, J. A. d. C. R., Souza, T., Quintans, I. L. A. d. C. R., & Farias, D. (2023). Network Toxicology and Molecular Docking to Investigate the Non-AChE Mechanisms of Organophosphate-Induced Neurodevelopmental Toxicity. Toxics, 11(8), 710. https://doi.org/10.3390/toxics11080710







