Soil Actinobacteria Exhibit Metabolic Capabilities for Degrading the Toxic and Persistent Herbicide Metribuzin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Actinobacteria Isolation
2.2. Screening for Metribuzin-Degrading Actinobacteria
2.3. Actinobacterial Identification
2.3.1. Morphological, Biochemical, and Physiological Characterization
2.3.2. Molecular Identification
2.4. Metribuzin Biodegradation Kinetics and Optimization
2.5. Chemical Analysis
2.5.1. Metribuzin Residue Detection
2.5.2. Total Organic Carbon Analysis
2.5.3. ATR-FTIR Analysis of Metribuzin
2.6. Kinetic Analysis
2.7. Statistical Analysis
3. Results
3.1. Isolation and Preliminary Screening of Metribuzin-Degrading Actinobacteria
3.2. Phenotypic Characterization of the Active Isolates
3.3. Molecular Characterization of Metribuzin-Degrading Actinobacterial Isolates
3.4. Growth of Isolates in the Presence of Metribuzin
3.5. Biodegradation Kinetics and Optimization Insights
3.6. Analysis of Metribuzin Biodegradation by GC-MS
3.7. Evaluation of Total Organic Carbon (TOC)
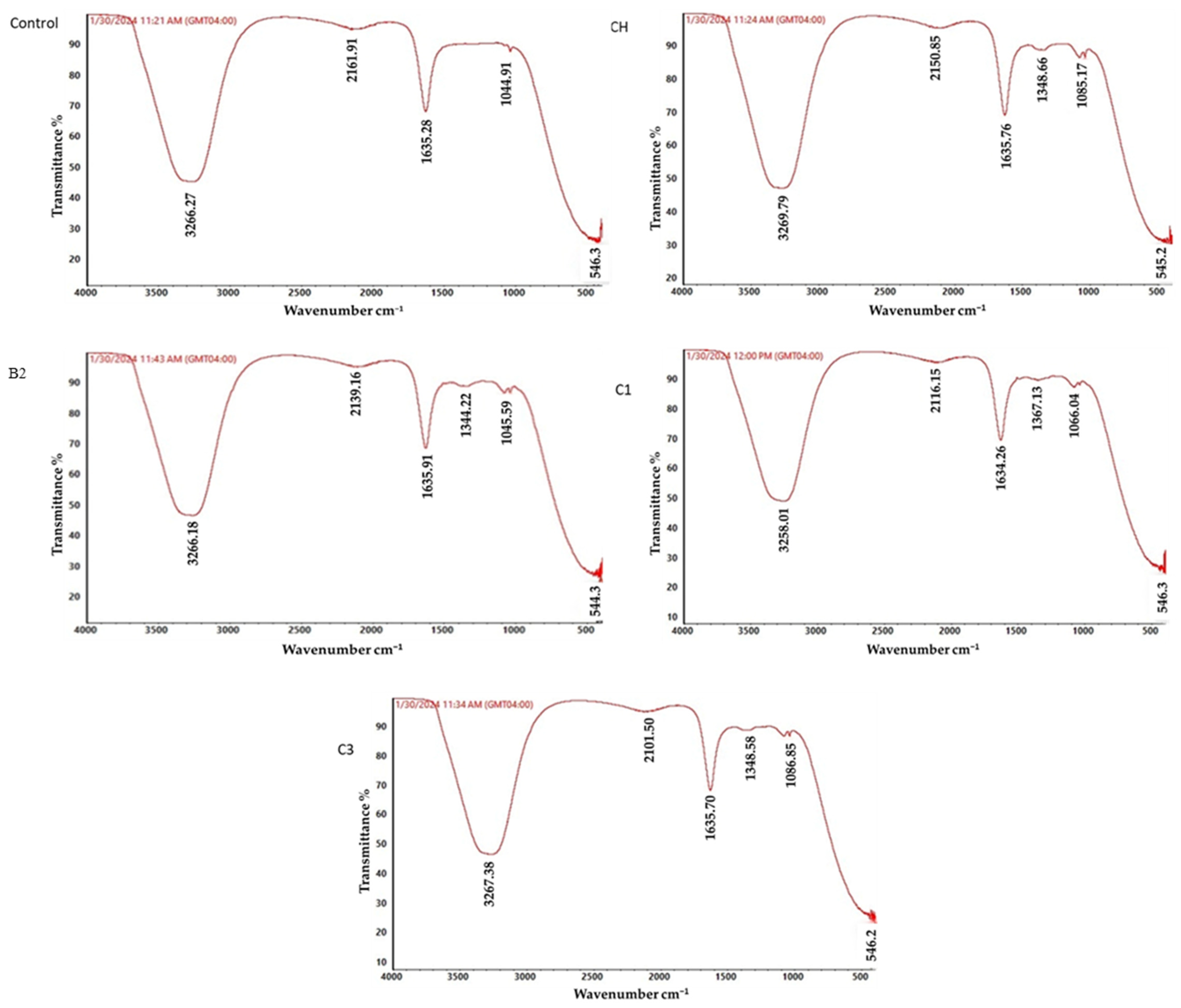
3.8. ATR-FTIR Analysis of Metribuzin Biodegradation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arsenault, W.J.; Ivany, J.A. Response of several potato cultivars to metribuzin and diquat. Crop Prot. 2001, 20, 547–552. [Google Scholar] [CrossRef]
- Haq, A.u.; Saeed, M.; Muneer, M.; Jamal, M.A.; Maqbool, T.; Tahir, T. Biosorption of metribuzin pesticide by Cucumber (Cucumis sativus) peels-zinc oxide nanoparticles composite. Sci. Rep. 2022, 12, 5840. [Google Scholar] [CrossRef]
- Kadam, S.R.; Jadhav, N.L.; Pandit, A.B.; Pejaver, M.K. Degradation kinetics and mechanism of hazardous metribuzin herbicide using advanced oxidation processes (HC & HC+ H2O2). Chem. Eng. Process.-Process Intensif. 2021, 166, 108486. [Google Scholar]
- Senseman, S. Herbicide Handbook Weed Science Society of America; Weed Science Society of America: Champaign, IL, USA, 2007. [Google Scholar]
- Xiang, D.; Zhu, L.; Yang, S.; Hou, X. Scrutinizing the interaction between metribuzin with glutathione reductase 2 from Arabidopsis thaliana: Insight into the molecular toxicity in agriculture. Environ. Sci. Pollut. Res. 2023, 30, 11936–11945. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Hou, Z.; Wu, X.; Gao, H.; Sun, F.; Pan, H. Biodegradation of triazine herbicide metribuzin by the strain Bacillus sp. N1. J. Environ. Sci. Health Part B 2014, 49, 79–86. [Google Scholar] [CrossRef]
- Gaona, L.; Bedmar, F.; Gianelli, V.; Faberi, A.; Angelini, H. Estimating the risk of groundwater contamination and environmental impact of pesticides in an agricultural basin in Argentina. Int. J. Environ. Sci. Technol. 2019, 16, 6657–6670. [Google Scholar] [CrossRef]
- Henriksen, T.; Svensmark, B.; Juhler, R.K. Analysis of metribuzin and transformation products in soil by pressurized liquid extraction and liquid chromatographic–tandem mass spectrometry. J. Chromatogr. A 2002, 957, 79–87. [Google Scholar] [CrossRef]
- Li, G.; Li, D.; Rao, H.; Liu, X. Potential neurotoxicity, immunotoxicity, and carcinogenicity induced by metribuzin and tebuconazole exposure in earthworms (Eisenia fetida) revealed by transcriptome analysis. Sci. Total Environ. 2022, 807, 150760. [Google Scholar] [CrossRef]
- Lone, A.; Raverkar, K.; Navneet Pareek, N.P.; Ramesh Chandra, R.C. Response of soil microbial communities to the selective herbicides: A microcosm approach. J. Pure Appl. Microbiol. 2014, 8, 1559–1567. [Google Scholar]
- Arora, S.; Arora, S.; Sahni, D.; Sehgal, M.; Srivastava, D.; Singh, A. Pesticides use and its effect on soil bacteria and fungal populations, microbial biomass carbon and enzymatic activity. Curr. Sci. 2019, 116, 643–649. [Google Scholar] [CrossRef]
- Gao, Y.; Truong, Y.B.; Cacioli, P.; Butler, P.; Kyratzis, I.L. Bioremediation of pesticide contaminated water using an organophosphate degrading enzyme immobilized on nonwoven polyester textiles. Enzym. Microb. Technol. 2014, 54, 38–44. [Google Scholar] [CrossRef]
- Wahla, A.Q.; Iqbal, S.; Anwar, S.; Firdous, S.; Mueller, J.A. Optimizing the metribuzin degrading potential of a novel bacterial consortium based on Taguchi design of experiment. J. Hazard. Mater. 2019, 366, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mutua, G.K.; Ngigi, A.N.; Getenga, Z.M. Degradation characteristics of metribuzin in soils within the Nzoia River Drainage Basin, Kenya. Toxicol. Environ. Chem. 2016, 98, 800–813. [Google Scholar]
- Khoury, R.; Coste, C.M.; Kawar, N.S. Degradation of metribuzin in two soil types of Lebanon. J. Environ. Sci. Health Part B 2006, 41, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, S.; Yuan, J.; Yang, P.; Liu, Y.; Stewart, K. Rapid biodegradation of atrazine by Ensifer sp. strain and its degradation genes. Int. Biodeterior. Biodegrad. 2017, 116, 133–140. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Zhang, K.; Wang, X.; Ma, C.; Tang, H.; Xu, P. Atrazine degradation by a simple consortium of Klebsiella sp. A1 and Comamonas sp. A2 in nitrogen enriched medium. Biodegradation 2010, 21, 97–105. [Google Scholar] [CrossRef]
- Mirzavand, S.; Aeini, M.; Najafgholi, H.M.; Rezamahalleh, H.M. Identification and Characterization of Bacterial Strains Capable of Degrading Atrazine and Metribuzin Herbicides in Sugarcane Fields. Sugar Tech 2024, 26, 95–105. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, J. Biodegradation of atrazine and nicosulfuron by Streptomyces nigra LM01: Performance, degradative pathway, and possible genes involved. J. Hazard. Mater. 2024, 471, 134336. [Google Scholar] [CrossRef]
- Herrera-Gallardo, B.E.; Guzmán-Gil, R.; Colín-Luna, J.A.; García-Martínez, J.C.; León-Santiesteban, H.H.; González-Brambila, O.M.; González-Brambila, M.M. Atrazine biodegradation in soil by Aspergillus niger. Can. J. Chem. Eng. 2021, 99, 932–946. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Wong, M.; Geng, B. Atrazine biodegradation in water by co-immobilized Citricoccus sp. strain TT3 with Chlorella vulgaris under a harsh environment. Algal Res. 2023, 70, 102994. [Google Scholar] [CrossRef]
- Gopal, M.; Dutta, D.; Jha, S.; Kalra, S.; Bandyopadhyay, S.; Das, S. Biodegradation of imidacloprid and metribuzin by Burkholderia cepacia strain CH9. Pestic. Res. J. 2011, 23, 36–40. [Google Scholar]
- Sharma, R.; Saroop, S. Role of microbes in pesticide bioremediation: Recent advances and biotechnological implications. Pestic. A Chang. Environ. 2024, 223–250. [Google Scholar]
- Wahla, A.Q.; Anwar, S.; Mueller, J.A.; Arslan, M.; Iqbal, S. Immobilization of metribuzin degrading bacterial consortium MB3R on biochar enhances bioremediation of potato vegetated soil and restores bacterial community structure. J. Hazard. Mater. 2020, 390, 121493. [Google Scholar] [CrossRef]
- Pan, Z.; Wu, Y.; Zhai, Q.; Tang, Y.; Liu, X.; Xu, X.; Liang, S.; Zhang, H. Immobilization of bacterial mixture of Klebsiella variicola FH-1 and Arthrobacter sp. NJ-1 enhances the bioremediation of atrazine-polluted soil en-vironments. Front. Microbiol. 2023, 14, 1056264. [Google Scholar] [CrossRef] [PubMed]
- Phillips, T.; Boopathy, R. Biotransformation of metribuzin under various electron acceptor conditions. Environ. Qual. Manag. 2022, 32, 223–231. [Google Scholar] [CrossRef]
- Kadam, S.R.; Pejaver, M.K. Biodegradation and Kinetic Study of Hazardous Metribuzin Herbicide Using a Novel Soil Bacterial Isolate Olivibacter oleidegradans Strain SP01 in Aqueous Solution. Nat. Environ. Pollut. Technol. 2023, 22, 1009–1015. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Costa, J.S.D.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Aouar, L.; Boukelloul, I.; Benadjila, A. Identification of antagonistic Streptomyces strains isolated from Algerian Saharan soils and their plant growth promoting properties. Biodiversitas J. Biol. Divers. 2020, 21. [Google Scholar] [CrossRef]
- McGinnis, M.R.; D’Amato, R.F.; Land, G.A. Pictorial Handbook of Medically Important Fungi and Aerobic Actinomycetes; Praeger: New York, NY, USA, 1982. [Google Scholar]
- Djebbah, F.Z.; Belyagoubi, L.; Abdelouahid, D.E.; Kherbouche, F.; Al-Dhabi, N.A.; Arasu, M.V.; Ravindran, B. Isolation and characterization of novel Streptomyces strain from Algeria and its in-vitro antimicrobial properties against microbial pathogens. J. Infect. Public Health 2021, 14, 1671–1678. [Google Scholar] [CrossRef]
- Boudemagh, A.; Bensouici, K. The effect of thermic pretreatment and antibiotics on the selective isolation of the culturable actinomycetes from the Algerian desert soil. Sci. Technologie. C Biotechnol. 2014, 39, 25–32. [Google Scholar]
- Hocinat, A.; Boudemagh, A. Biodegradation of commercial Ortiva fungicide by isolated actinomycetes from the activated sludge. Desalination Water Treat. 2016, 57, 6091–6097. [Google Scholar] [CrossRef]
- Briceño, G.; Fuentes, M.S.; Palma, G.; Jorquera, M.; Amoroso, M.; Diez, M. Chlorpyrifos biodegradation and 3, 5, 6-trichloro-2-pyridinol production by actinobacteria isolated from soil. Int. Biodeterior. Biodegrad. 2012, 73, 1–7. [Google Scholar] [CrossRef]
- Silini, S.; Ali-Khodja, H.; Boudemagh, A.; Terrouche, A.; Bouziane, M. Isolation and preliminary identification of actinomycetes isolated from a wastewater treatment plant and capable of growing on methyl ethyl ketone as a sole source of carbon and energy. Desalination Water Treat. 2016, 57, 12108–12117. [Google Scholar] [CrossRef]
- Shirling, E.T.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Williams, S.; Goodfellow, M.; Wellington, E.; Vickers, J.; Alderson, G.; Sneath, P.; Sackin, M.; Mortimer, A. A probability matrix for identification of some streptomycetes. Microbiology 1983, 129, 1815–1830. [Google Scholar] [CrossRef]
- Tatsinkou Fossi, B.; Tavea, F.; Ndjouenkeu, R. Production and partial characterization of thermostable alpha amylase from ascomycete yeast strain isolated from starchy soil. Afr. J. Biotechnol. 2005, 4, 14–18. [Google Scholar]
- Li, Q.; Chen, X.; Jiang, Y.; Jiang, C. Cultural, physiological, and biochemical identification of actinobacteria. Actinobacteria-Basics Biotechnol. Appl. 2016, 87–111. [Google Scholar]
- Minotto, E.; Milagre, L.P.; Oliveira, M.T.; Van Der Sand, S.T. Enzyme characterization of endophytic actinobacteria isolated from tomato plants. J. Adv. Sci. Res. 2014, 5, 16–23. [Google Scholar]
- Abraham, J.; Gajendiran, A. Biodegradation of fipronil and its metabolite fipronil sulfone by Streptomyces rochei strain AJAG7 and its use in bioremediation of contaminated soil. Pestic. Biochem. Physiol. 2019, 155, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Albero, B.; Fernández, M.D.; García-Gómez, C.; Pérez, R.A. Rapid determination of metribuzin and three major transformation products in soil and plant by Gas Chromatography–Tandem Mass Spectrometry. Separations 2022, 9, 386. [Google Scholar] [CrossRef]
- Korkmaz, V.; Yildirim, N.; Erguven, G.O.; Durmus, B.; Nuhoglu, Y. The bioremediation of glyphosate in soil media by some newly isolated bacteria: The COD, TOC removal efficiency and mortality assessment for Daphnia magna. Environ. Technol. Innov. 2021, 22, 101535. [Google Scholar] [CrossRef]
- Griffiths, P.R. Fourier transform infrared spectrometry. Science 1983, 222, 297–302. [Google Scholar] [CrossRef]
- Milosevic, M. Internal reflection and ATR spectroscopy. Appl. Spectrosc. Rev. 2004, 39, 365–384. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Liu, L.; Gao, C.; Ru, S.; Yang, L. Occurrence, ecological risk, and advanced removal methods of herbicides in waters: A timely review. Environ. Sci. Pollut. Res. 2024, 31, 3297–3319. [Google Scholar] [CrossRef]
- Rossi, F.; Carles, L.; Donnadieu, F.; Batisson, I.; Artigas, J. Glyphosate-degrading behavior of five bacterial strains isolated from stream biofilms. J. Hazard. Mater. 2021, 420, 126651. [Google Scholar] [CrossRef] [PubMed]
- Naccarato, A.; Vommaro, M.L.; Amico, D.; Sprovieri, F.; Pirrone, N.; Tagarelli, A.; Giglio, A. Triazine herbicide and NPK fertilizer exposure: Accumulation of heavy metals and rare earth elements, effects on cuticle melanization, and immunocompetence in the model species Tenebrio molitor. Toxics 2023, 11, 499. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, C.W.; Muia, A.W.; Ngigi, A.N.; Moturi, W.N. Isolation of metribuzin degrading soil bacteria and assessment of their growth in response to selected physical chemical conditions. J. Biol. Agric. Healthc. 2017, 7, 2224–3208. [Google Scholar]
- Gouma, S.; Papadaki, A.A.; Markakis, G.; Magan, N.; Goumas, D. Studies on pesticides mixture degradation by white rot fungi. J. Ecol. Eng. 2019, 20, 16–26. [Google Scholar] [CrossRef]
- Pizzutti, I.R.; Kok, A.d.; Silva, R.C.d.; Rohers, G.N. Comparison between three chromatographic (GC-ECD, GC-PFPD and GC-ITD-MS) methods and a UV-Vis spectrophotometric method for the determination of dithiocarbamates in lettuce. J. Braz. Chem. Soc. 2017, 28, 775–781. [Google Scholar] [CrossRef]
- Zameer, M.; Tahir, U.; Khalid, S.; Zahra, N.; Sarwar, A.; Aziz, T.; Saidal, A.; Alhomrani, M.; Alamri, A.S.; Dablool, A.S. Isolation and characterization of indigenous bacterial assemblage for biodegradation of persistent herbicides in the soil. Acta Biochim. Pol. 2023, 70, 325–334. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, H.; Chen, F.; Song, M. Colonization of Paracoccus sp. QCT6 and enhancement of metribuzin degradation in maize rhizosphere soil. Curr. Microbiol. 2018, 75, 156–162. [Google Scholar] [CrossRef]
- Zhu, J.; Fu, L.; Meng, Z.; Jin, C. Characteristics of an atrazine degrading bacterium and the construction of a microbial agent for effective atrazine degradation. Water Environ. J. 2021, 35, 7–17. [Google Scholar] [CrossRef]



| Strains | Metribuzin Concentrations (mg/L) | |||||
|---|---|---|---|---|---|---|
| 1 | 10 | 25 | 50 | 100 | 200 | |
| CH | +++ | ++ | +++ | +++ | + | + |
| C1 | +++ | +++ | +++ | +++ | + | + |
| B1 | − | − | − | − | − | − |
| B2 | +++ | ++ | +++ | ++ | + | + |
| C2 | +++ | +++ | ++ | − | − | − |
| C3 | +++ | ++ | ++ | +++ | − | − |
| C4 | +++ | + | +++ | − | − | − |
| C | ++ | ++ | + | − | − | − |
| CB | +++ | ++ | ++ | − | − | − |
| 3G | +++ | + | + | − | − | − |
| GH | − | − | − | − | − | − |
| GL | − | − | − | − | − | − |
| CH | B2 | C1 | C3 | |
| Temperature | ||||
| 25 °C | + | + | + | + |
| 30 °C | ++ | ++ | ++ | ++ |
| 37 °C | + | + | + | + |
| 40 °C | −/+ | −/+ | −/+ | −/+ |
| pH | ||||
| 2 | + | + | + | + |
| 5 | + | + | + | + |
| 7 | ++ | ++ | ++ | ++ |
| 9 | + | + | + | + |
| 12 | + | + | + | + |
| NaCl | ||||
| 2% | ++ | ++ | ++ | ++ |
| 5% | + | − | − | − |
| 9% | − | − | − | − |
| 15% | − | − | − | − |
| Enzymatic activity | ||||
| Starch hydrolysis | + | + | + | + |
| Casein hydrolysis | + | − | + | − |
| Gelatin Hydrolysis | − | + | + | + |
| Catalase | + | + | + | + |
| Carbon source | ||||
| D-Fructose | − | + | + | + |
| D-Glucose | + | + | + | + |
| D-Galactose | + | + | + | + |
| Nitrogen source | ||||
| Proline | − | − | + | + |
| Arginine | − | + | + | + |
| Threonine | − | − | − | + |
| Histidine | + | − | + | − |
| Asparagine | − | + | + | + |
| Tyrosine | + | − | + | − |
| Methionine | + | + | − | + |
| Bacteria | Close Strain | Accession. N |
|---|---|---|
| CH | Streptomyces toxytricini | PP413749 |
| B2 | Streptomyces stelliscabiei | PP413746 |
| C1 | Streptomyces heliomycini | PP413747 |
| C3 | Streptomyces heliomycini | PP413748 |
| Strains | k | t1/2 | R2 |
|---|---|---|---|
| CH | 0.0054 | 3.7 | 0.96 |
| B2 | 0.0044 | 4.54 | 0.83 |
| C1 | 0.0066 | 3.03 | 0.99 |
| C3 | 0.0051 | 3.92 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rebai, H.; Sholkamy, E.N.; Abdelhamid, M.A.A.; Prakasam Thanka, P.; Aly Hassan, A.; Pack, S.P.; Ki, M.-R.; Boudemagh, A. Soil Actinobacteria Exhibit Metabolic Capabilities for Degrading the Toxic and Persistent Herbicide Metribuzin. Toxics 2024, 12, 709. https://doi.org/10.3390/toxics12100709
Rebai H, Sholkamy EN, Abdelhamid MAA, Prakasam Thanka P, Aly Hassan A, Pack SP, Ki M-R, Boudemagh A. Soil Actinobacteria Exhibit Metabolic Capabilities for Degrading the Toxic and Persistent Herbicide Metribuzin. Toxics. 2024; 12(10):709. https://doi.org/10.3390/toxics12100709
Chicago/Turabian StyleRebai, Hadjer, Essam Nageh Sholkamy, Mohamed A. A. Abdelhamid, Pratheesh Prakasam Thanka, Ashraf Aly Hassan, Seung Pil Pack, Mi-Ran Ki, and Allaoueddine Boudemagh. 2024. "Soil Actinobacteria Exhibit Metabolic Capabilities for Degrading the Toxic and Persistent Herbicide Metribuzin" Toxics 12, no. 10: 709. https://doi.org/10.3390/toxics12100709
APA StyleRebai, H., Sholkamy, E. N., Abdelhamid, M. A. A., Prakasam Thanka, P., Aly Hassan, A., Pack, S. P., Ki, M.-R., & Boudemagh, A. (2024). Soil Actinobacteria Exhibit Metabolic Capabilities for Degrading the Toxic and Persistent Herbicide Metribuzin. Toxics, 12(10), 709. https://doi.org/10.3390/toxics12100709












