miR–122–5p Promotes Cowshed Particulate Matter2.5-Induced Apoptosis in NR8383 by Targeting COL4A1
Abstract
:1. Introduction
2. Materials and Methods
2.1. PM2.5 Collection
2.2. Cell Culture and Treatments
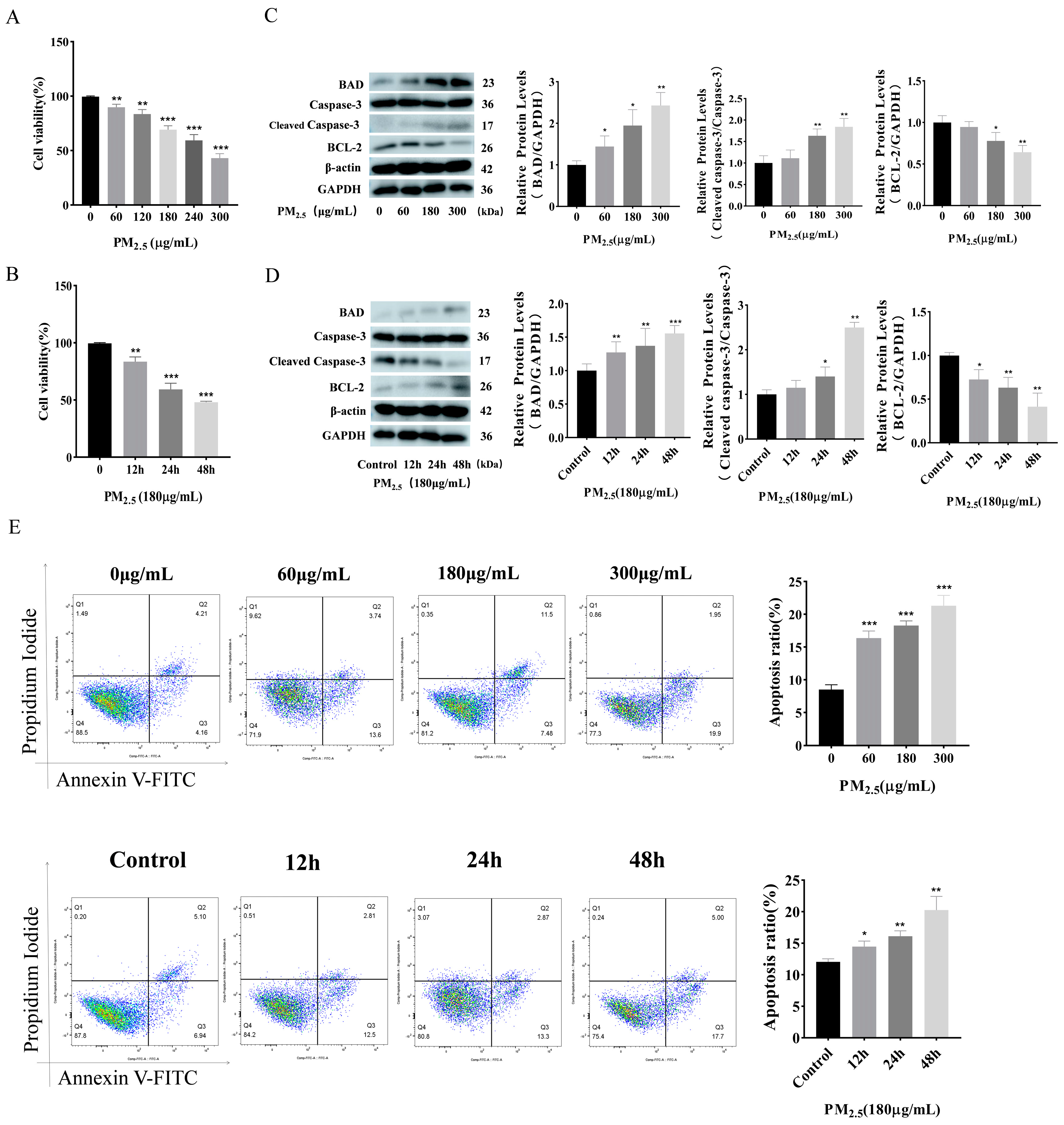
- The cells were exposed to escalating concentrations of cowshed PM2.5, ranging from 0 μg/mL to 300 μg/mL in increments of 60 μg/mL.
- A fixed concentration of PM2.5 (180 μg/mL) was used to treat cells for varying durations: 0, 12, 24, and 48 h.
2.3. Cell Viability (CCK-8)
2.4. Extraction and Real-Time Fluorescence Quantification of RNA
2.5. Plasmid Construction and Cell Transfection
2.6. Flow Cytometry
2.7. Hoechst 33342/PI Double Staining
2.8. Western Blotting
2.9. Statistical Analysis
3. Results
3.1. Cowshed PM2.5 Exposure Induces Apoptosis in NR8383 Cells
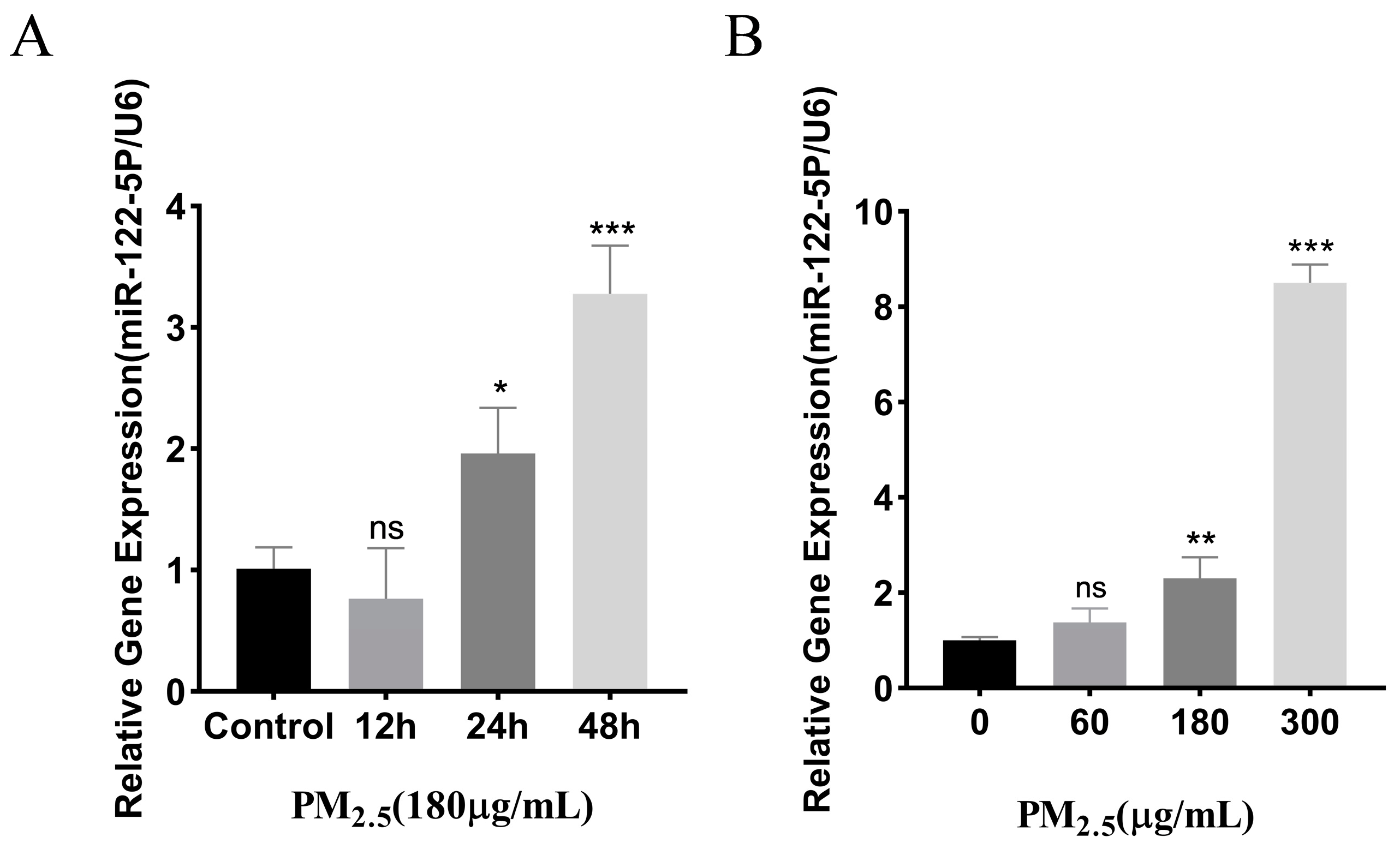
3.2. Cowshed PM2.5-Stimulated Upregulation of miR–122–5p
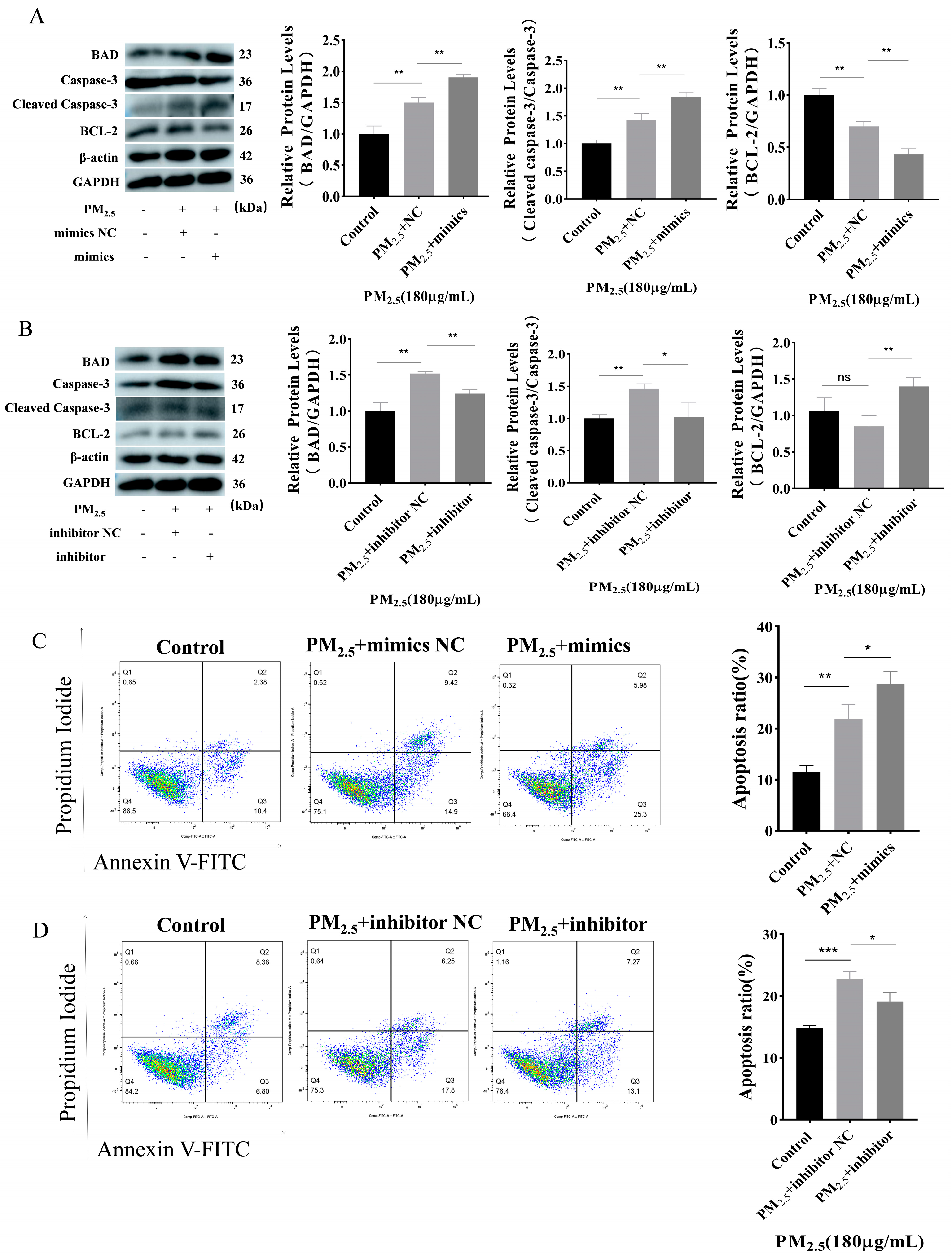
3.3. Promotion of Apoptosis by miR–122–5p
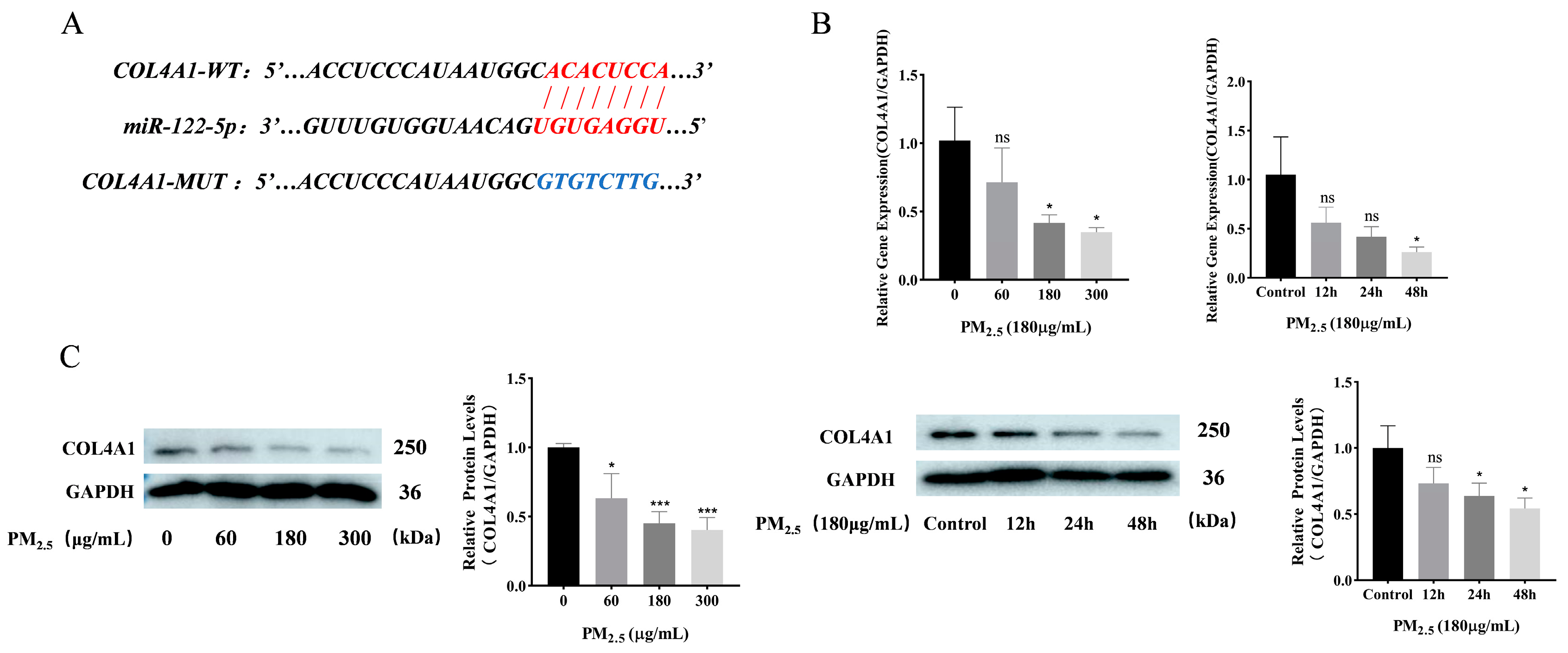
3.4. COL4A1 Is a Predicted Target of miR–122–5p and Downregulated in Cowshed PM2.5-Induced NR8383 Cells
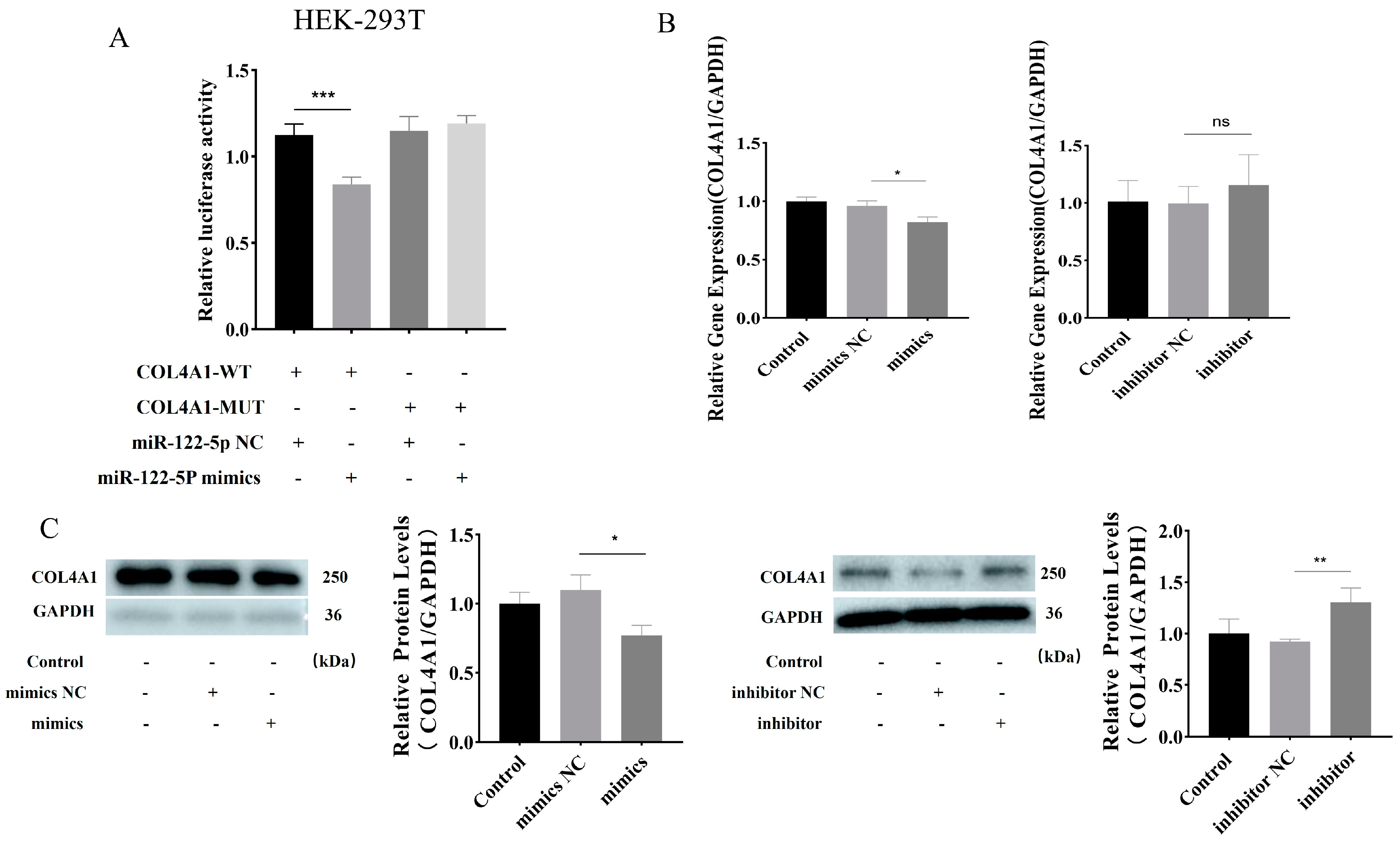
3.5. miR–122–5p Targeted COL4A1 and Showed Negative Regulatory Relationship
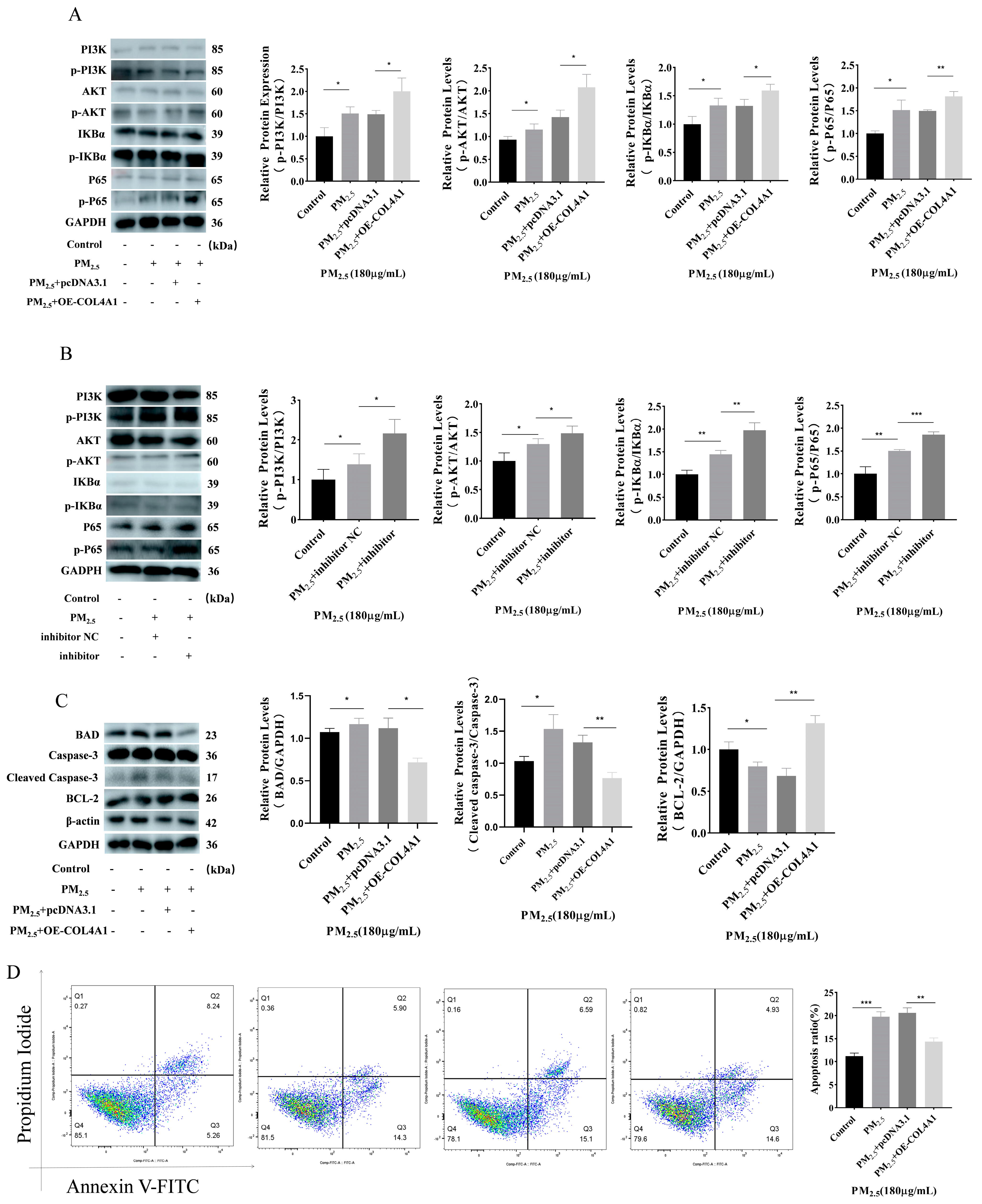
3.6. Overexpression of COL4A1 Enhances PI3K/AKT/NF–κΒ Signaling Pathway and Inhibits Cowshed PM2.5-Induced Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lary, D.J.; Faruque, F.S.; Malakar, N.; Moore, A.; Roscoe, B.; Adams, Z.L.; Eggelston, Y. Estimating the global abundance of ground level presence of particulate matter (PM2.5). Geospat. Health 2014, 8, S611–S630. [Google Scholar] [CrossRef] [PubMed]
- Alexeeff, S.E.; Deosaransingh, K.; Liao, N.S.; Van den Eeden, S.K.; Schwartz, J.; Sidney, S. Particulate Matter and Cardiovascular Risk in Adults with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2021, 204, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.Y.; Hu, D.Y.; Wang, X.F.; Chen, Y.H.; Wu, Y.; Pan, L.; Li, H.; Zhang, J.; Deng, F.; Guo, X.; et al. The modification of indoor PM2.5 exposure to chronic obstructive pulmonary disease in Chinese elderly people: A meet-in-metabolite analysis. Environ. Int. 2018, 121, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.M.; Hua, S.C.; Song, L. PM2.5 Exposure and Asthma Development: The Key Role of Oxidative Stress. Oxidative Med. Cell. Longev. 2022, 2022, 3618806. [Google Scholar] [CrossRef] [PubMed]
- Cogliati, M.G.; Paez, P.A.; Pianciola, L.A.; Caputo, M.A.; Mut, P.N. Bioaerosol Concentration in a Cattle Feedlot in Neuquén, Argentina. Atmosphere 2022, 13, 1761. [Google Scholar] [CrossRef]
- Lou, C.; Bai, Y.; Chai, T.; Yu, H.; Lin, T.; Hu, G.; Guan, Y.; Wu, B. Research progress on distribution and exposure risk of microbial aerosols in animal houses. Front. Vet. Sci. 2022, 9, 1015238. [Google Scholar] [CrossRef] [PubMed]
- Mebrahtu, T.F.; Santorelli, G.; Yang, T.C.; Wright, J.; Tate, J.; McEachan, R.R.C. The effects of exposure to NO2, PM2.5 and PM10 on health service attendances with respiratory illnesses: A time-series analysis. Environ. Pollut. 2023, 333, 122123. [Google Scholar] [CrossRef] [PubMed]
- Evren, E.; Ringqvist, E.; Willinger, T. Origin and ontogeny of lung macrophages: From mice to humans. Immunology 2020, 160, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.L.; Yang, Y.; Chu, C.Y.; Rung, S.A.; Wang, Z.Q.; Man, Y.; Lin, J.; Qu, Y. Macrophage response mediated by extracellular matrix: Recent progress. Biomed. Mater. 2023, 18, 012003. [Google Scholar] [CrossRef] [PubMed]
- Amirrah, I.N.; Lokanathan, Y.; Zulkiflee, I.; Wee, M.; Motta, A.; Fauzi, M.B. A Comprehensive Review on Collagen Type I Development of Biomaterials for Tissue Engineering: From Biosynthesis to Bioscaffold. Biomedicines 2022, 10, 2307. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K. Basement-membrane (type-iv) collagen. Matrix Biol. 1995, 14, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Yang, X.F.; Han, F.; Wang, H.D. Circ_0092291 attenuates angiotensin II-induced cell damages in human aortic vascular smooth muscle cells via mediating the miR-626/COL4A1 signal axis. J. Physiol. Biochem. 2022, 78, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Plancher, J.M.; Hufnagel, R.B.; Vagal, A.; Peariso, K.; Saal, H.M.; Broderick, J.P. Case of Small Vessel Disease Associated with COL4A1 Mutations following Trauma. Case Rep. Neurol. 2015, 7, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Iwashita, J.; Maeda, H.; Ishimura, M.; Murata, J. Type IV collagen reduces MUC5AC secretion in the lungs of ovalbumin-sensitized mice. Front. Pharmacol. 2022, 13, 851374. [Google Scholar] [CrossRef] [PubMed]
- Polette, M.; Thiblet, J.; Ploton, D.; Buisson, A.C.; Monboisse, J.C.; Tournier, J.M.; Birembaut, P. Distribution of α1(IV) and α3(IV) chains of type IV collagen in lung tumours. J. Pathol. 1999, 182, 185–191. [Google Scholar] [CrossRef]
- Dunn, W.; Trang, P.; Zhong, Q.; Yang, E.; van Belle, C.; Liu, F.Y. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell. Microbiol. 2005, 7, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.X.; Wang, S.N.; Zhang, Z.Y.; Lu, Y.; Yang, M.L.; Chen, P.; Chen, L.; Wang, M. MiRNA-6089 inhibits rheumatoid arthritis fibroblast-like synoviocytes proliferation and induces apoptosis by targeting CCR4. Arch. Physiol. Biochem. 2022, 128, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Li, P.Y.; Zhang, B.Y.; Han, B.; Su, X.; Wang, Q.; Wang, X.; Li, B.; Kang, H.; Zhou, L.; et al. miRNAs deregulation in serum of mice is associated with lung cancer related pathway deregulation induced by PM2.5. Environ. Pollut. 2019, 254, 112875. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, H.; Yu, J.; Li, Z.; Jia, G.; Ding, B.; Lv, C. Crocin suppresses breast cancer cell proliferation by down-regulating tumor promoter miR-122-5p and up-regulating tumor suppressors FOXP2 and SPRY2. Environ. Toxicol. 2023, 38, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Yanai, K.; Ishii, H.; Aomatsu, A.; Hirai, K.; Ookawara, S.; Ishibashi, K.; Morishita, Y. miR-122-5p Regulates Renal Fibrosis In Vivo. Int. J. Mol. Sci. 2022, 23, 15423. [Google Scholar] [CrossRef] [PubMed]
- Franke, T.F.; Hornik, C.P.; Segev, L.; Shostak, G.A.; Sugimoto, C. PI3K/Akt and apoptosis: Size matters. Oncogene 2003, 22, 8983–8998. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.W.; Ai, J.Y.; Fan, Y.; Gao, J.; Liu, W.W.; Feng, Q.; Liao, W.; Wu, L. NCAPG Promotes The Proliferation Of Hepatocellular Carcinoma Through PI3K/AKT Signaling. Oncotargets Ther. 2019, 12, 8537–8552. [Google Scholar] [CrossRef] [PubMed]
- Vara, J.Á.F.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Robbins, H.L.; Hague, A. The PI3K/Akt Pathway in Tumors of endocrine Tissues. Front. Endocrinol. 2016, 6, 174816. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.T.; Liu, D.J.; Shi, B.B.; Chen, M.; Weng, S.Y.; Guo, R.J.; Fu, C. Penehyclidine hydrochloride protects against lipopolysaccharide-induced acute lung injury by promoting the PI3K/Akt pathway. Int. J. Immunopathol. Pharmacol. 2023, 37, 03946320231192175. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Sun, Y.; Jia, Y.; Duan, X.; Ma, Z.; Zhang, X.; Wang, L.; Zhu, Y.; Gao, Y.; Basang, W. MicroRNA miR-212-5p Regulates the MEK/ERK Signaling Pathway by Targeting A-Raf proto-oncogene serine/threonine-protein kinase (ARAF) to Regulate Cowshed PM2.5-Induced NR8383 Apoptosis. Toxics 2023, 11, 981. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Sun, Y.; Du, X.; Zhang, X.; Ma, Z.; Gao, Y.; Liang, X. Lnc-Clic5 as a sponge for miR-212–5p to inhibit cow barn PM2.5-induced apoptosis in rat alveolar macrophages. Toxicology 2024, 504, 153797. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, Z.; Hao, P.; Ji, S.; Gao, Y. Characteristics and health impacts of bioaerosols in animal barns: A comprehensive study. Ecotoxicol. Environ. Saf. 2024, 278, 116381. [Google Scholar] [CrossRef]
- Jia, Y.N. Study on miR-212-5p Targeting LAMC2 and LAMA3 to Regulate Cell Apoptosis Induced by PM2.5 in Cowhouse; Jilin Agricultural University: Jinlin, China, 2023; Available online: https://kns.cnki.net/kcms2/article/abstract?v=JxCH2R2OgonxEmvdodzdIVK9LATq7Mza1UktlqVaU1AvnKYQ2z39D6RQiEvgwtaAZXTFx6Esnf5Sz9Zmj2hFqStlnt_Rx1mR-fda7mM6MOh49Il2zSiPK_ph3f-Xkaa5IeFDTXjizTxLsoZ8l32fMw==&uniplatform=NZKPT&language=CHS: (accessed on 1 May 2023).
- Ming, Y.; Zhou, X.A.; Liu, G.; Abudupataer, M.; Zhu, S.C.; Xiang, B.T.; Yin, X.; Lai, H.; Sun, Y.; Wang, C.; et al. PM2.5 exposure exacerbates mice thoracic aortic aneurysm and dissection by inducing smooth muscle cell apoptosis via the MAPK pathway. Chemosphere 2023, 313, 137500. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Xu, H.M.; Liu, H.W.; Zhang, Y.F. Unique regulatory roles of ncRNAs changed by PM2.5 in human diseases. Ecotoxicol. Environ. Saf. 2023, 255, 114812. [Google Scholar] [CrossRef] [PubMed]
- Li, D.W.; Chen, J.N.; Yun, C.; Li, X.Y.; Huang, Z.Z. MiR-122-5p regulates the pathogenesis of childhood obesity by targeting CPEB1. Obes. Res. Clin. Pract. 2022, 16, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Chen, Z.M.; Jiang, Z.; Huang, T.; Hu, J.; Luo, P.Q.; Zhang, H.; Huang, M.; Huang, L.; Chen, Y.; et al. MiR-122-5p suppresses the proliferation, migration, and invasion of gastric cancer cells by targeting LYN. Acta Biochim. Biophys. Sin. 2020, 52, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, X.Y. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. J. Cell. Physiol. 2020, 235, 2825–2835. [Google Scholar] [CrossRef] [PubMed]
- Song, W.L.; Zhang, T.N.; Yang, N.; Zhang, T.; Wen, R.; Liu, C.F. Inhibition of micro RNA miR-122-5p prevents lipopolysaccharide-induced myocardial injury by inhibiting oxidative stress, inflammation and apoptosis via targeting GIT1. Bioengineered 2021, 12, 1902–1915. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-Z.; Hu, Z.-L.; Liao, T.-Y.; Li, Y.; Pan, Y.-L. LncRNA SND1-IT1 facilitates TGF-β1-induced epithelial-to-mesenchymal transition via miR-124/COL4A1 axis in gastric cancer. Cell Death Discov. 2022, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, M.; Van Sinderen, M.; Rainczuk, K.; Dimitriadis, E. miR-29c overexpression and COL4A1 downregulation in infertile human endometrium reduces endometrial epithelial cell adhesive capacity in vitro implying roles in receptivity. Sci. Rep. 2019, 9, 8644. [Google Scholar] [CrossRef] [PubMed]
- Jalali, M.; Nikravesh, M.R.; Moeen, A.A.; Mohammadi, S.; Karimfar, M.H. Effects of Maternal Nicotine Exposure on Expression of Collagen Type IV and its Roles on Pulmonary Bronchogenesis and Alveolarization in Newborn Mice. Iran. J. Allergy Asthma Immunol. 2010, 9, 169–173. [Google Scholar] [PubMed]
- Sand, J.M.B.; Knox, A.J.; Lange, P.; Sun, S.; Kristensen, J.H.; Leeming, D.J.; Karsdal, M.A.; Bolton, C.E.; Johnson, S.R. Accelerated extracellular matrix turnover during exacerbations of COPD. Respir. Res. 2015, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Lan, K.P.; Lai, S.C. Angiostrongylus cantonensis: Induction of urokinase-type PA and degradation of type IV collagen in rat lung granulornatous fibrosis. Exp. Parasitol. 2008, 118, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Ge, G.X. Complexity of type IV collagens: From network assembly to function. Biol. Chem. 2019, 400, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Arseni, L.; Lombardi, A.; Orioli, D. From Structure to Phenotype: Impact of Collagen Alterations on Human Health. Int. J. Mol. Sci. 2018, 19, 1407. [Google Scholar] [CrossRef] [PubMed]
- Ecker, V.; Stumpf, M.; Brandmeier, L.; Neumayer, T.; Pfeuffer, L.; Engleitner, T.; Ringshausen, I.; Nelson, N.; Jücker, M.; Wanninger, S.; et al. Targeted PI3K/AKT-hyperactivation induces cell death in chronic lymphocytic leukemia. Nat. Commun. 2021, 12, 3526. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Shi, C.R.; Wei, C.T.; Wang, C.; Du, H.J.; Liu, R.; Wang, X.; Hong, Q.; Chen, X. Fufang Shenhua tablet inhibits renal fibrosis by inhibiting PI3K/AKT. Phytomedicine 2023, 116, 154873. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zheng, H.; Li, L.D.; Zhou, C.S.; Chen, X.; Zhou, X.Y.; Cao, Y. KIF3C Promotes Proliferation, Migration, and Invasion of Glioma Cells by Activating the PI3K/AKT Pathway and Inducing EMT. Biomed Res. Int. 2020, 2020, 6349312. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ling, Z.; Xie, K.; Wang, Y.; Miao, Z.; Ji, X.; Li, J.; Hou, W.; Tang, Q.; Yuan, X.; et al. The COL-4A1 polypeptide destroy endothelial cells through the TGF-β/PI3K/AKT pathway. Sci. Rep. 2021, 11, 15761. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, Y.; Gao, Y.; Li, X.; Tian, F.; Liu, F.; Du, R.; Li, P.; Wang, F.; Xu, S.; et al. MicroRNA-31 regulating apoptosis by mediating the phosphatidylinositol-3 kinase/protein kinase B signaling pathway in treatment of spinal cord injury. Brain Dev. 2019, 41, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Bouchier-Hayes, L.; Martin, S.J. CARDINAL roles in apoptosis and NFκB activation. Vitam. Horm. 2004, 67, 133–147. [Google Scholar] [PubMed]
| Gene | Primers’ Sequence (5′-3′) |
|---|---|
| COL4A1 | F: AGTTGGCTTTCCTGGTAGTC R: AAGGCCTGCTTGTCCTTT |
| GAPDH | F: CCTGCACCACCAACTGCTTA R: CATCACGCCACAGCTTCCA |
| U6 | F: CTCGCTTCGGCAGCACA R: AACGCTTCACGAATTTGCGT |
| miR–122–5p | F: CGCGTGGAGTGTGACAATGG R: AGTGCAGGGTCCGAGGTATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Sun, K.; Ma, Z.; Zhang, X.; Du, X.; Jia, Y.; Zhu, Y.; Inam, M.; Gao, Y.; Basang, W. miR–122–5p Promotes Cowshed Particulate Matter2.5-Induced Apoptosis in NR8383 by Targeting COL4A1. Toxics 2024, 12, 386. https://doi.org/10.3390/toxics12060386
Sun Y, Sun K, Ma Z, Zhang X, Du X, Jia Y, Zhu Y, Inam M, Gao Y, Basang W. miR–122–5p Promotes Cowshed Particulate Matter2.5-Induced Apoptosis in NR8383 by Targeting COL4A1. Toxics. 2024; 12(6):386. https://doi.org/10.3390/toxics12060386
Chicago/Turabian StyleSun, Yize, Ke Sun, Zhenhua Ma, Xiqing Zhang, Xiaohui Du, Yunna Jia, Yanbin Zhu, Muhammad Inam, Yunhang Gao, and Wangdui Basang. 2024. "miR–122–5p Promotes Cowshed Particulate Matter2.5-Induced Apoptosis in NR8383 by Targeting COL4A1" Toxics 12, no. 6: 386. https://doi.org/10.3390/toxics12060386







