METTL3-Regulated lncRNA SNHG7 Drives MNNG-Induced Epithelial–Mesenchymal Transition in Gastric Precancerous Lesions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Lentiviral Packaging and Cell Transfection
2.3. RNA Total m6A Quantification
2.4. Construction of METTL3 Knockout Mice Model and Exposed by MNNG
2.5. Immunohistochemistry (IHC) Analysis
2.6. Cell Proliferation Assays
2.7. Migration and Invasion Assays
2.8. RNA Isolation and Quantitative Real-Time PCR (RT-qPCR)
2.9. Western Blot Analysis
2.10. m6A-Modified RNA Immunoprecipitation qPCR (m6A IP-qPCR)
2.11. RNA-Binding Protein Immunoprecipitation (RIP) Assay
2.12. Statistical Analyses
3. Results
3.1. Subsection
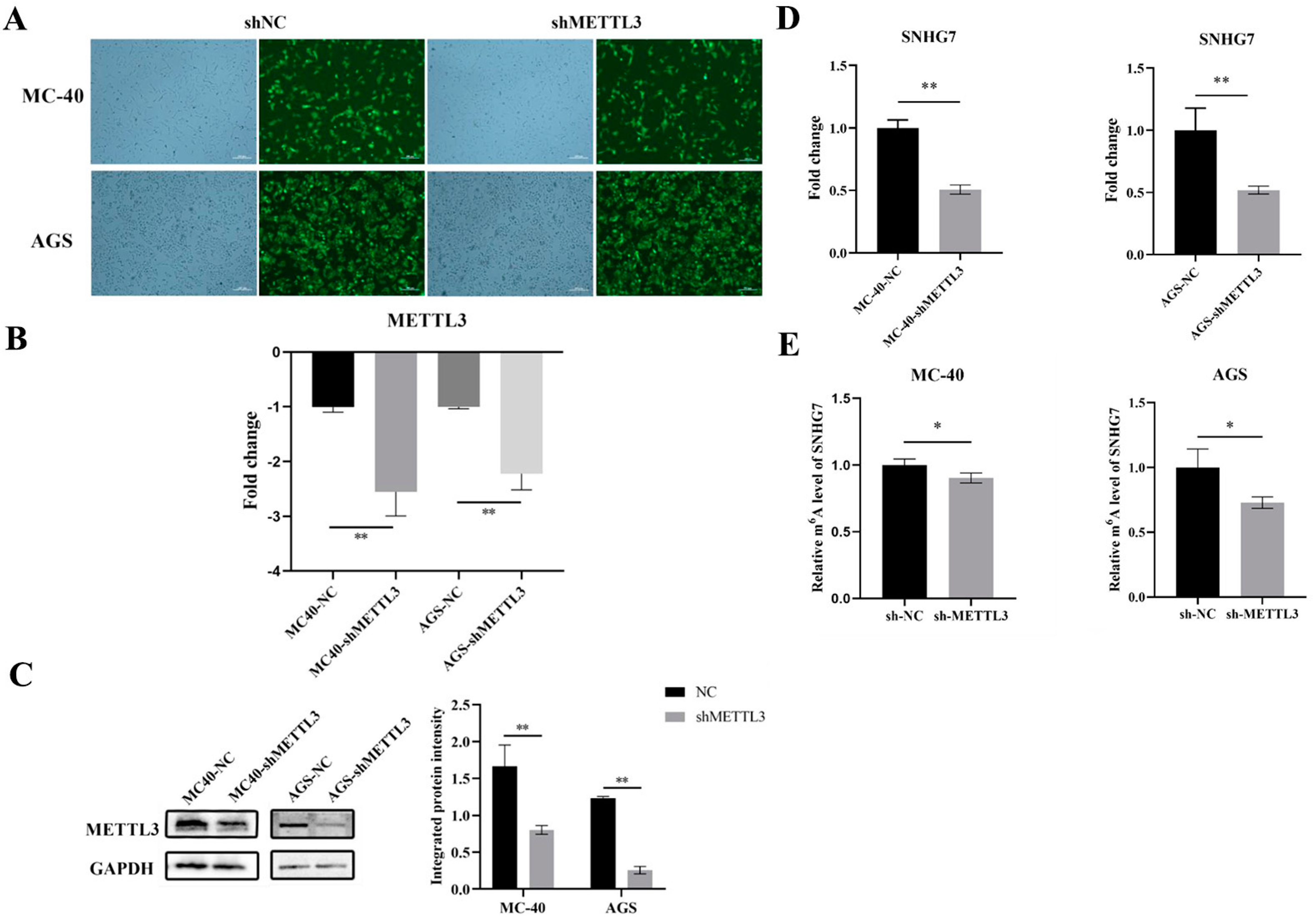
3.1.1. Knockdown of METTL3 Expression in MC-40 and GC Cells
3.1.2. Down-Regulation of METTL3 Inhibited lncRNA SNHG7 Expression
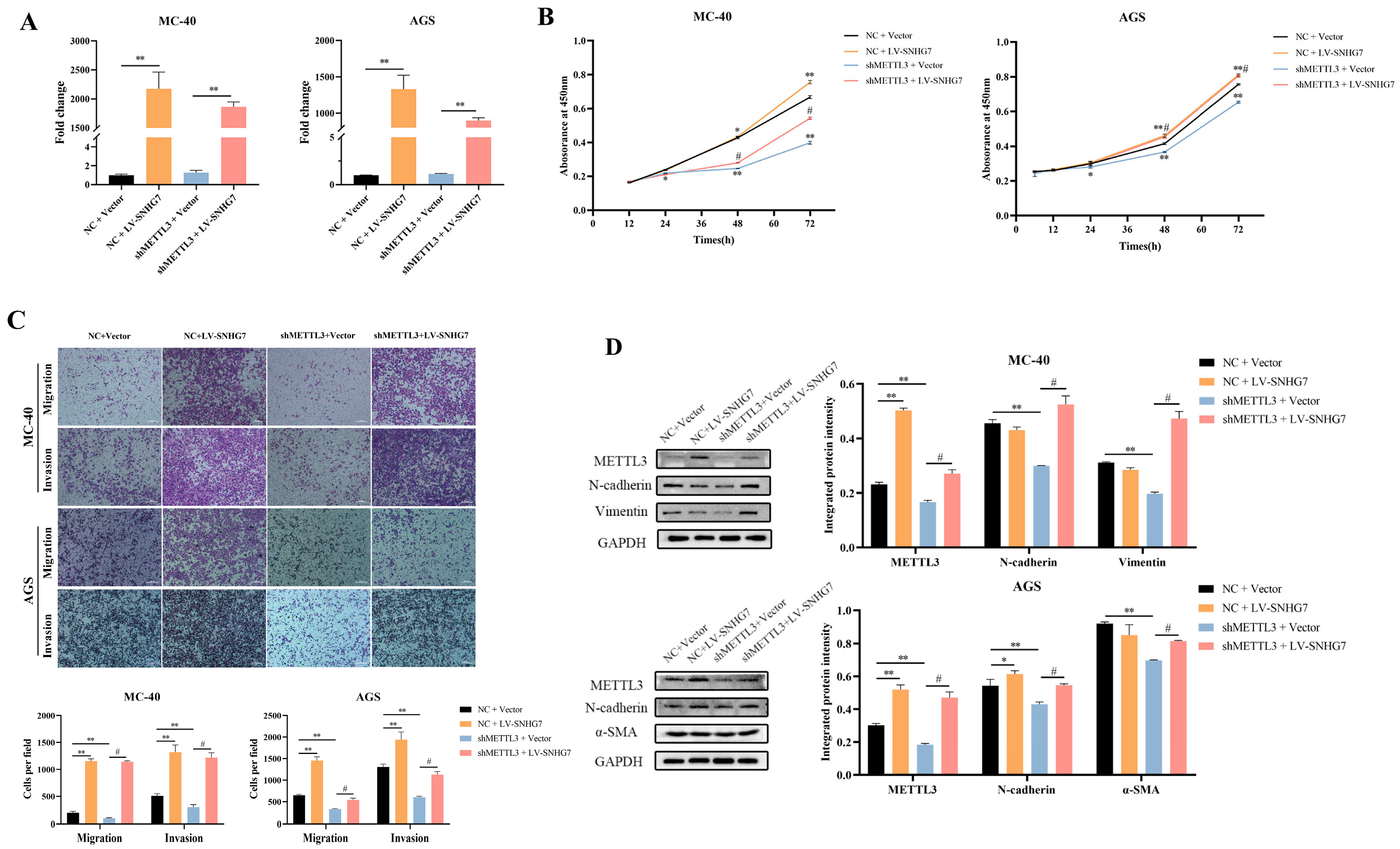
3.1.3. Overexpression of SNHG7 Reversed the Effect of Down-Regulation of METTL3 on the Proliferation, Migration, Invasion, and EMT of MC-40 and AGS Cells In Vitro
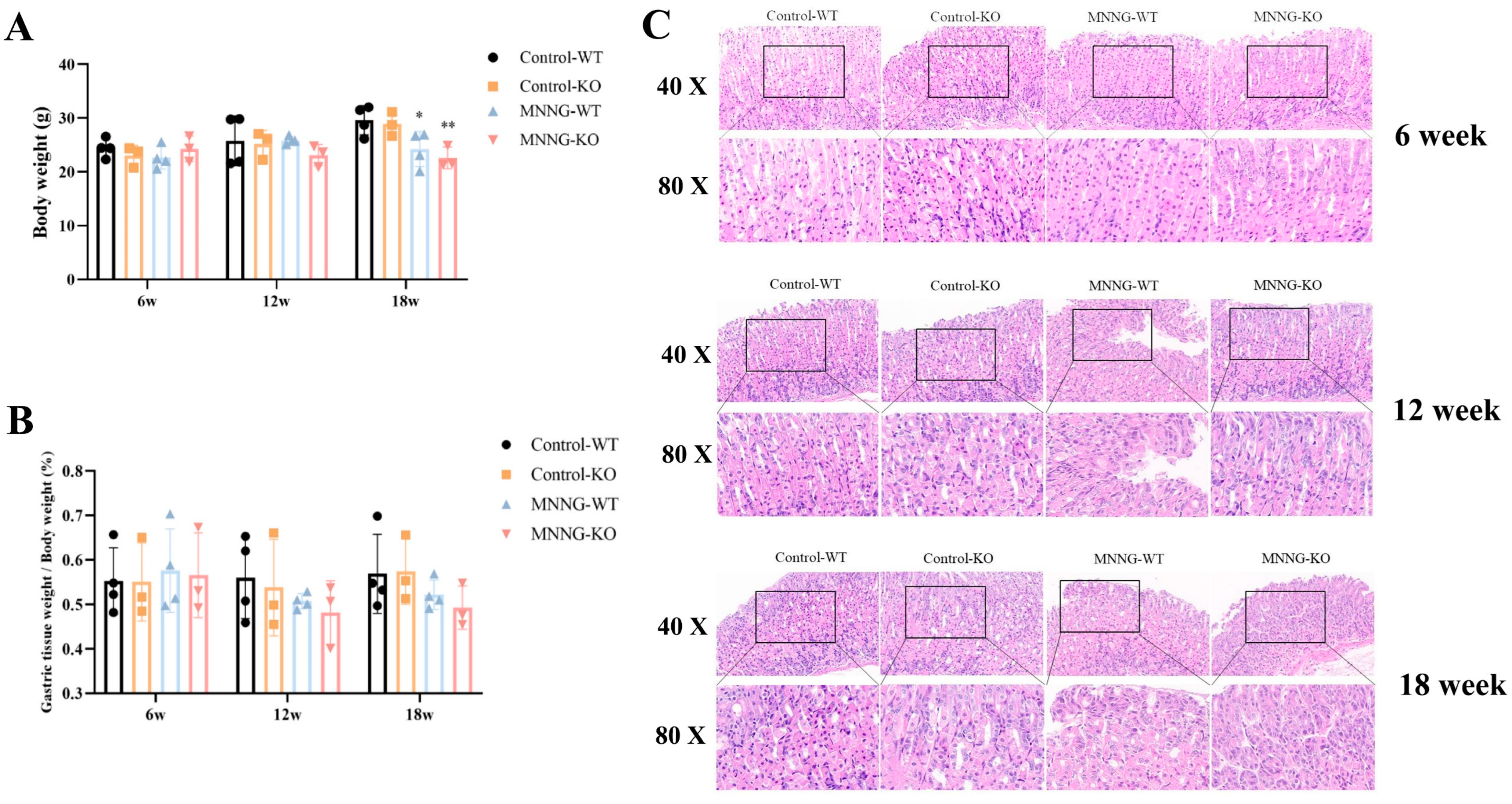
3.1.4. MNNG Exposure Exhibited a Significant Reduction in Gastric Weight, Body Weight, and Gastric Histopathological Changes in Mice
3.1.5. Chronic Exposure to MNNG Leads to METTL3 Up-Regulation and Increases m6A Levels in the Peripheral Plasma of Mice
3.1.6. Time-Dependent Increase in the Expression of lncRNA SNHG7 and Cell Proliferation Indicator Ki67 in METTL3-KO/WT Mice Exposed to MNNG
3.1.7. Knockout of METTL3 Suppressed EMT Induced by MNNG Exposure in the Mettl3-KO/WT Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chia, N.Y.; Tan, P. Molecular classification of gastric cancer. Ann. Oncol. 2016, 27, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Apicella, M.; Corso, S.; Giordano, S. Targeted therapies for gastric cancer: Failures and hopes from clinical trials. Oncotarget 2017, 8, 57654–57669. [Google Scholar] [CrossRef]
- Patel, T.H.; Cecchini, M. Targeted Therapies in Advanced Gastric Cancer. Curr. Treat. Options Oncol. 2020, 21, 70. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhou, M.; Xu, Y.; Gu, X.; Zou, M.; Abudushalamu, G.; Yao, Y.; Fan, X.; Wu, G. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol. Cancer 2023, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Zhang, C.; Zhao, Z.; Li, S.; Cai, H.; Chen, X.; Cai, D.; Liu, W.; Yan, Y.; Xie, K.; et al. The gastric mucosal protective effects of astragaloside IV in mnng-induced GPL rats. Biomed. Pharmacother. 2018, 104, 291–299. [Google Scholar] [CrossRef]
- Cai, T.; Zhang, C.; Zeng, X.; Zhao, Z.; Yan, Y.; Yu, X.; Wu, L.; Lin, L.; Pan, H. Protective effects of Weipixiao decoction against MNNG-induced gastric precancerous lesions in rats. Biomed. Pharmacother. 2019, 120, 109427. [Google Scholar] [CrossRef]
- Liu, T.; Yang, S.; Sui, J.; Xu, S.Y.; Cheng, Y.P.; Shen, B.; Zhang, Y.; Zhang, X.M.; Yin, L.H.; Pu, Y.P.; et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J. Cell Physiol. 2020, 235, 548–562. [Google Scholar] [CrossRef]
- Liu, C.; Yang, S.; Zhang, Y.; Wang, C.; Du, D.; Wang, X.; Liu, T.; Liang, G. Emerging Roles of N6-Methyladenosine Demethylases and Its Interaction with Environmental Toxicants in Digestive System Cancers. Cancer Manag. Res. 2021, 13, 7101–7114. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, T.; Xu, S.; Ren, Y.; Ge, Y.; Yin, L.; Pu, Y.; Liang, G. The role of N6-methyladenosine methylation in environmental exposure-induced health damage. Environ. Sci. Pollut. Res. Int. 2022, 29, 69153–69175. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Feng, Y.; Yang, S.; Ge, Y.; Zhang, T.; Li, J.; Li, C.; Ruan, Y.; Luo, B.; Liang, G. Depicting the Profile of METTL3-Mediated lncRNA m6A Modification Variants and Identified SNHG7 as a Prognostic Indicator of MNNG-Induced Gastric Cancer. Toxics 2023, 11, 944. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Feng, Y.L.; Wang, R.Y.; Yang, S.; Ge, Y.L.; Zhang, T.Y.; Li, J.; Li, C.Y.; Ruan, Y.; Luo, B.; et al. Long-term MNNG exposure promotes gastric carcinogenesis by activating METTL3/m6A/miR1184 axis-mediated epithelial-mesenchymal transition. Sci. Total Environ. 2024, 913, 169752. [Google Scholar] [CrossRef]
- Liu, H.T.; Liu, S.; Liu, L.; Ma, R.R.; Gao, P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res. 2018, 78, 5877–5890. [Google Scholar] [CrossRef]
- Hui, Y.; Yang, Y.; Li, D.; Wang, J.; Di, M.; Zhang, S.; Wang, S. LncRNA FEZF1-AS1 Modulates Cancer Stem Cell Properties of Human Gastric Cancer Through miR-363-3p/HMGA2. Cell Transplant. 2020, 29, 963689720925059. [Google Scholar] [CrossRef]
- Wang, S.; Liu, F.; Deng, J.; Cai, X.; Han, J.; Liu, Q. Long Noncoding RNA ROR Regulates Proliferation, Invasion, and Stemness of Gastric Cancer Stem Cell. Cell Reprogram 2016, 18, 319–326. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.; He, Y.; Deng, T.; Liu, R.; Wang, W.; Zhu, K.; Bai, M.; Ning, T.; Yang, H.; et al. Chemotoxicity-induced exosomal lncFERO regulates ferroptosis and stemness in gastric cancer stem cells. Cell Death Dis. 2021, 12, 1116. [Google Scholar] [CrossRef]
- Wong, Y.L.; LeBon, L.; Basso, A.M.; Kohlhaas, K.L.; Nikkel, A.L.; Robb, H.M.; Donnelly-Roberts, D.L.; Prakash, J.; Swensen, A.M.; Rubinstein, N.D.; et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife 2019, 8, e42940. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Lv, X.X.; Wang, Z.Q.; Zhou, Y.M.; Jiang, B.; Wang, S.N.; Chen, X.D. The significance of the microlymphangiogenesis, microangiogenesis, and combined detection of programmed cell death-1 protein (PD-1)/ki67 in gastric cancer tissues. J. Cancer Res. Clin. Oncol. 2023, 149, 9129–9137. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Gregory, R.I. RNAmod: An integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019, 47, W548–W555. [Google Scholar] [CrossRef]
- Ma, J.Z.; Yang, F.; Zhou, C.C.; Liu, F.; Yuan, J.H.; Wang, F.; Wang, T.T.; Xu, Q.G.; Zhou, W.P.; Sun, S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Gong, C.; Liu, H.; Song, R.; Zhong, T.; Lou, M.; Wang, T.; Qi, H.; Shen, J.; Zhu, L.; Shao, J. ATR-CHK1-E2F3 signaling transactivates human ribonucleotide reductase small subunit M2 for DNA repair induced by the chemical carcinogen MNNG. Biochim. Biophys. Acta 2016, 1859, 612–626. [Google Scholar] [CrossRef]
- Takahashi, E.; Okamoto, K.; Arimoto, S.; Yamanaka, H.; Negishi, T. Involvement of the drug efflux protein TolC in mutagenicity induced by MNNG or Trp-P-2. Mutat. Res. 2006, 605, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Gunes-Bayir, A.; Guler, E.M.; Bilgin, M.G.; Ergun, I.S.; Kocyigit, A.; Dadak, A. Anti-Inflammatory and Antioxidant Effects of Carvacrol on N-Methyl-N’-Nitro-N-Nitrosoguanidine (MNNG) Induced Gastric Carcinogenesis in Wistar Rats. Nutrients 2022, 14, 2848. [Google Scholar] [CrossRef]
- Yu, C.; Su, Z.; Li, Y.; Li, Y.; Liu, K.; Chu, F.; Liu, T.; Chen, R.; Ding, X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed. Pharmacother. 2020, 126, 110036. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Xian, Q.; Wang, Q.; Wu, C.; Yan, H.; Li, X.; Lu, L.; Wu, C.; Zhu, D.; Xu, X.; et al. m6A Methyltransferase 3 Promotes the Proliferation and Migration of Gastric Cancer Cells through the m6A Modification of YAP1. J. Oncol. 2021, 2021, 8875424. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Y.; Shu, Y.; He, J.; Gao, W. Interaction between N6-methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol. Cancer 2020, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef]
- Ban, Y.; Tan, P.; Cai, J.; Li, J.; Hu, M.; Zhou, Y.; Mei, Y.; Tan, Y.; Li, X.; Zeng, Z.; et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol. Oncol. 2020, 14, 1282–1296. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, X.; Li, D.; Jiang, X. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging 2020, 12, 24967–24982. [Google Scholar] [CrossRef] [PubMed]
- Huo, F.C.; Zhu, Z.M.; Zhu, W.T.; Du, Q.Y.; Liang, J.; Mou, J. METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene 2021, 40, 2968–2981. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Du, J.; Yang, L.; Wang, X.; Di, W.; Hu, B.; An, J.; Kong, L.; et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2021, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Song, Y.; Huang, L.L.; Tian, Y.J.; Wang, X.L.; Hua, L.Y. m6A transferase METTL3 regulates endothelial-mesenchymal transition in diabetic retinopathy via lncRNA SNHG7/KHSRP/MKL1 axis. Genomics 2022, 114, 110498. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yuan, J.F.; Wang, Y.Z. METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-Myc axis. Exp. Cell Res. 2022, 416, 113149. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Ma, J.; Pan, Y.; Hu, J.; Liu, B.; Jia, L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018, 9, 722. [Google Scholar] [CrossRef]
- Yang, X.; Sun, L.; Wang, L.; Yao, B.; Mo, H.; Yang, W. LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed. Pharmacother. 2019, 118, 109386. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Song, Y.; Tang, L.; Sun, D.H.; Ji, D.G. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7788–7797. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, S.; Song, X.; Gui, J.; Li, Y.; Li, M. ELK1/lncRNA-SNHG7/miR-2682-5p feedback loop enhances bladder cancer cell growth. Life Sci. 2020, 262, 118386. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Fan, J.; Ma, Y.; Zhou, Y.; Qin, K.; Shi, M.; Yang, J. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, Y.; Zhang, Y.; Cheng, L.; Zhou, X.; Chen, K. SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle 2020, 19, 142–152. [Google Scholar] [CrossRef]
- Wang, M.W.; Liu, J.; Liu, Q.; Xu, Q.H.; Li, T.F.; Jin, S.; Xia, T.S. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4613–4622. [Google Scholar]
- He, Y.; Liu, H.H.; Zhou, X.L.; He, T.T.; Zhang, A.Z.; Wang, X.; Wei, S.Z.; Li, H.T.; Chen, L.S.; Chang, L.; et al. Rutaecarpine Ameliorates Murine N-Methyl-N’-Nitro-N-Nitrosoguanidine-Induced Chronic Atrophic Gastritis by Sonic Hedgehog Pathway. Molecules 2023, 28, 6294. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Lu, W.; Liu, J.; Ren, H.; Zhao, X.; Yang, W. miR-520f-3p blocks MNNG-induced gastric precancerous lesions via the KLF7/NFkappaB pathway. Toxicol. Lett. 2024, 392, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhu, X.; Wang, C.; Yang, S.; Qiao, Y.; Qiao, R.; Zhang, J. Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer. Sci. Rep. 2019, 9, 5166. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ran, L.; Zhang, W.; Leng, X.; Wang, K.; Liu, G.; Song, J.; Wang, Y.; Zhang, X.; Wang, Y.; et al. FOXS1 is regulated by GLI1 and miR-125a-5p and promotes cell proliferation and EMT in gastric cancer. Sci. Rep. 2019, 9, 5281. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Liu, C.; Pan, Y.; Han, Y.; Yang, L.; Xia, J.; Xu, F. SHP2 deficiency promotes Staphylococcus aureus pneumonia following influenza infection. Cell Prolif. 2020, 53, e12721. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Wang, Z.; Suo, R.; Ma, R.; Zhang, J. miR-181-5p/KLHL5 Promoted Proliferation and Migration of Gastric Cancer Through Activating METTL3-Mediated m6A Process. Mol. Biotechnol. 2023, 1–11. [Google Scholar] [CrossRef]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef]
- Ni, W.; Zhang, Y.; Zhan, Z.; Ye, F.; Liang, Y.; Huang, J.; Chen, K.; Chen, L.; Ding, Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J. Hematol. Oncol. 2017, 10, 91. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Jiang, D.M.; Hu, S.S.; Zhao, L.; Wang, L.; Yang, M.H.; Ai, M.L.; Jiang, H.J.; Han, Y.; Ding, Y.Q.; et al. SATB2-AS1 Suppresses Colorectal Carcinoma Aggressiveness by Inhibiting SATB2-Dependent Snail Transcription and Epithelial-Mesenchymal Transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer | Sequence |
|---|---|---|
| Ki67 | Forward primer | 5′-ATCATTGACCGCTCCTTTAGGT-3′ |
| Reverse primer | 5′-GCTCGCCTTGATGGTTCCT-3′ | |
| METTL3 | Forward primer | 5′-CTGGGCACTTGGATTTAAGGAA-3′ |
| Reverse primer | 5′-TGAGAGGTGGTGTAGCAACTT-3′ | |
| SNHG7 | Forward primer | 5′-ATGCTGACCATGCAACCCTT-3′ |
| Reverse primer | 5′-GACATTTTGCAGAGCCGTGG-3′ | |
| GAPDH | Forward primer | 5′-AGGTCGGTGTGAACGGATTTG-3′ |
| Reverse primer | 5′-GGGGTCGTTGATGGCAACA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jian, J.; Feng, Y.; Wang, R.; Li, C.; Zhang, L.; Ruan, Y.; Luo, B.; Liang, G.; Liu, T. METTL3-Regulated lncRNA SNHG7 Drives MNNG-Induced Epithelial–Mesenchymal Transition in Gastric Precancerous Lesions. Toxics 2024, 12, 573. https://doi.org/10.3390/toxics12080573
Jian J, Feng Y, Wang R, Li C, Zhang L, Ruan Y, Luo B, Liang G, Liu T. METTL3-Regulated lncRNA SNHG7 Drives MNNG-Induced Epithelial–Mesenchymal Transition in Gastric Precancerous Lesions. Toxics. 2024; 12(8):573. https://doi.org/10.3390/toxics12080573
Chicago/Turabian StyleJian, Jiabei, Yanlu Feng, Ruiying Wang, Chengyun Li, Lin Zhang, Ye Ruan, Bin Luo, Geyu Liang, and Tong Liu. 2024. "METTL3-Regulated lncRNA SNHG7 Drives MNNG-Induced Epithelial–Mesenchymal Transition in Gastric Precancerous Lesions" Toxics 12, no. 8: 573. https://doi.org/10.3390/toxics12080573
APA StyleJian, J., Feng, Y., Wang, R., Li, C., Zhang, L., Ruan, Y., Luo, B., Liang, G., & Liu, T. (2024). METTL3-Regulated lncRNA SNHG7 Drives MNNG-Induced Epithelial–Mesenchymal Transition in Gastric Precancerous Lesions. Toxics, 12(8), 573. https://doi.org/10.3390/toxics12080573








