Epigenetic Biomarkers Driven by Environmental Toxins Associated with Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the United States: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Sources
2.2. PRISMA Criteria
2.3. Meta Analysis
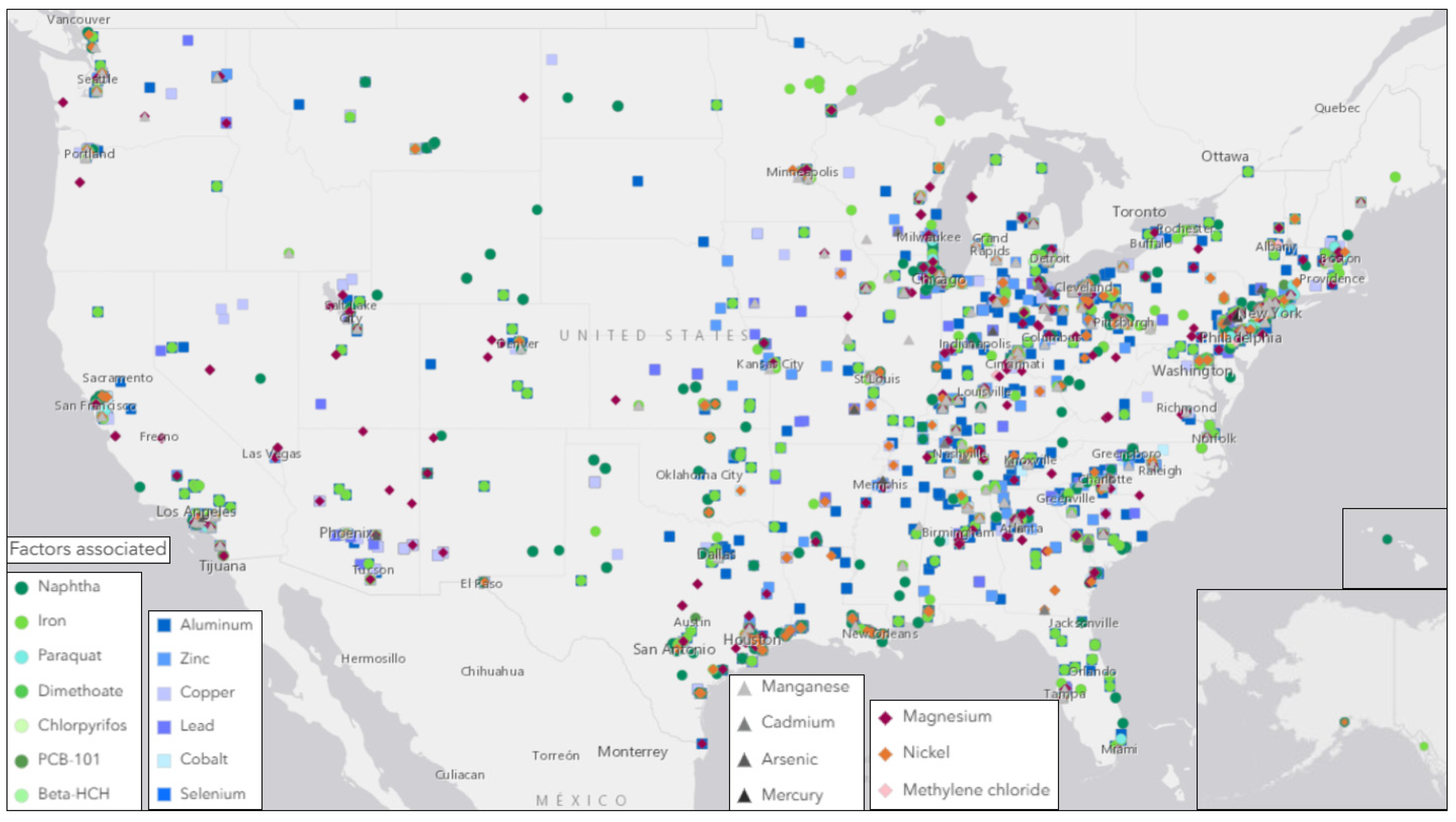
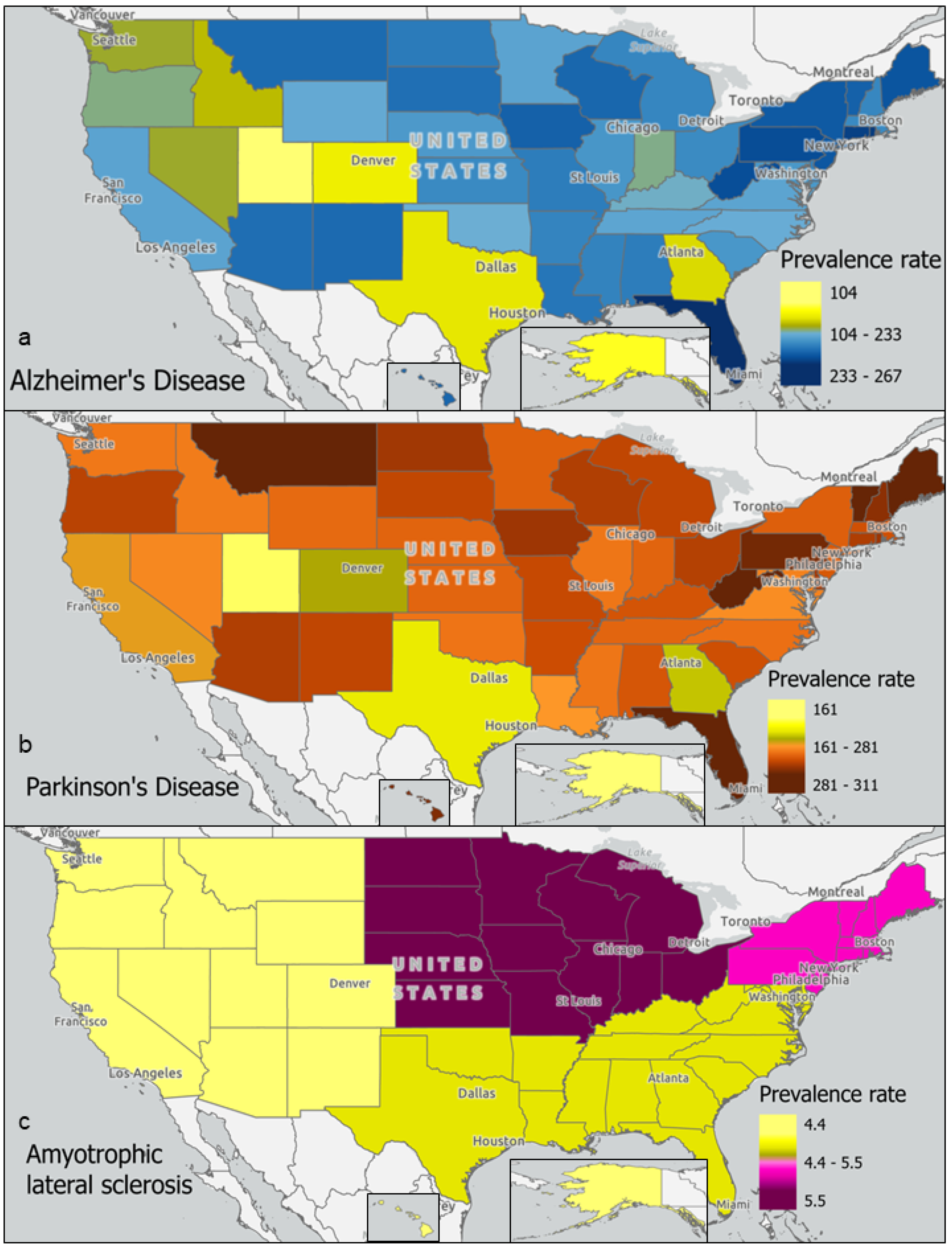
2.4. Geographic Analysis
2.5. Statistical Analysis
3. Results
3.1. Summary of the Systematic Literature Review
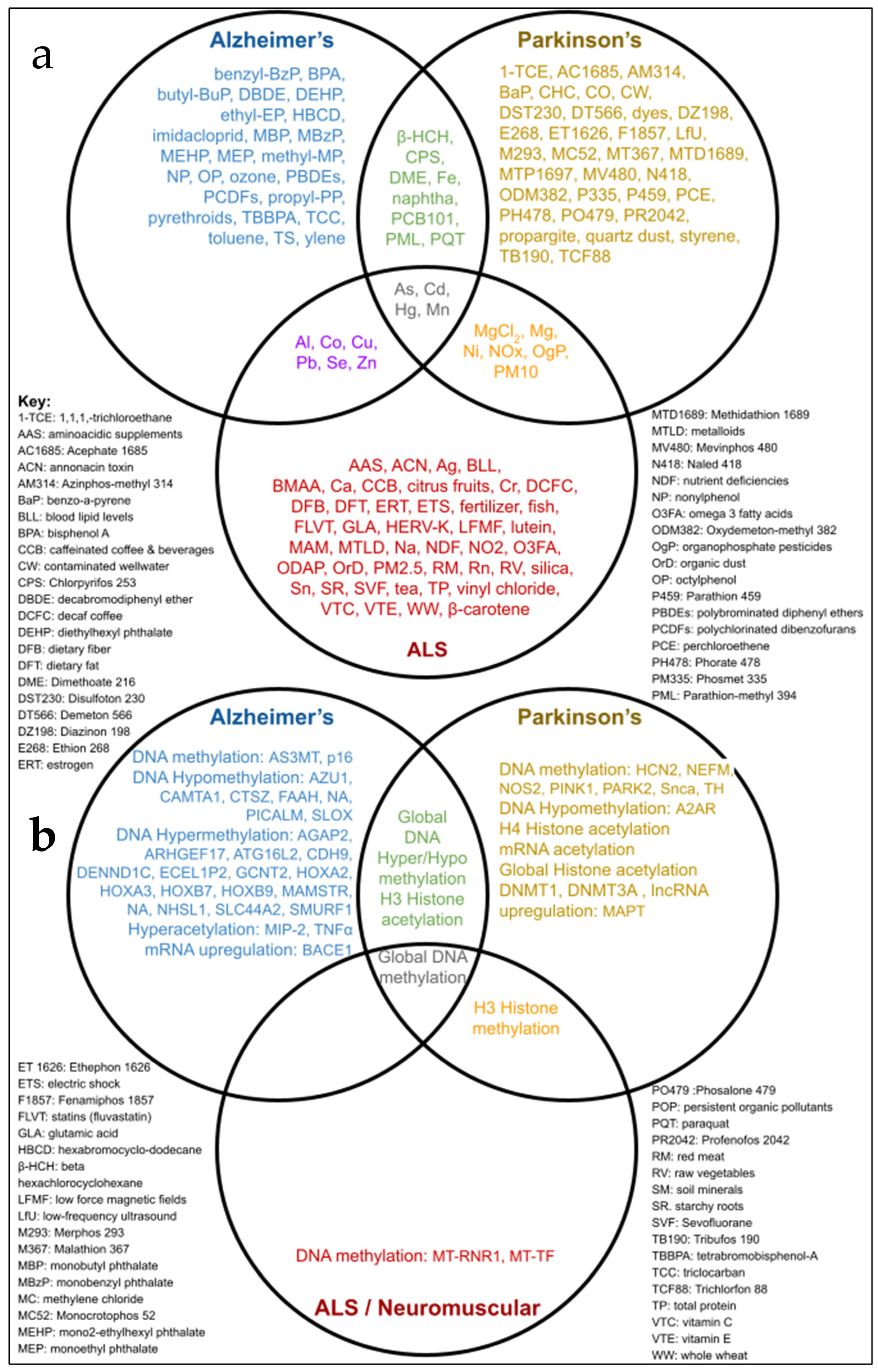
3.2. Environmental Toxins Common to AD, PD, and ALS
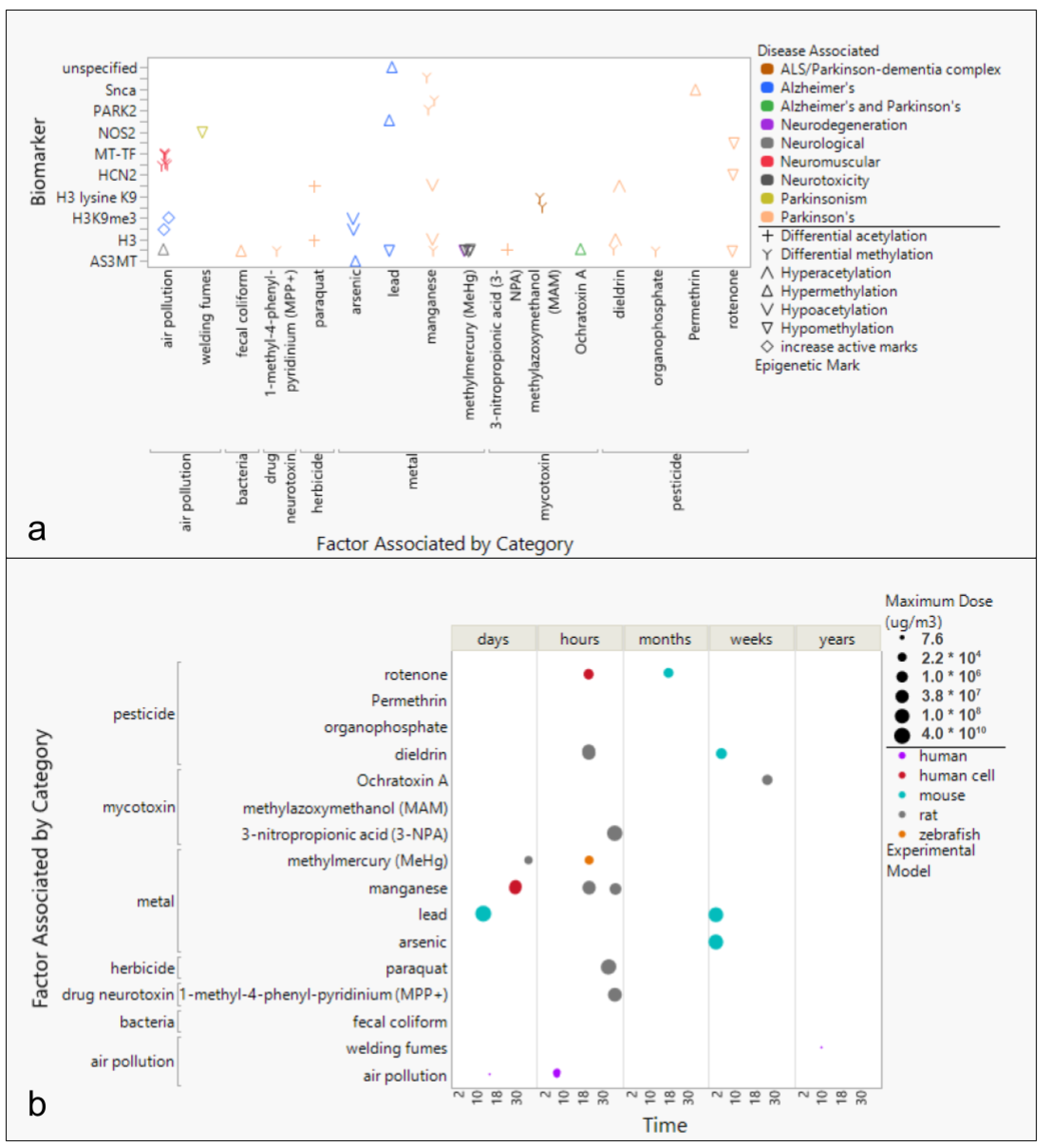
3.3. Environmental Factors Associated with Epigenetic Markers in AD, PD, and ALS
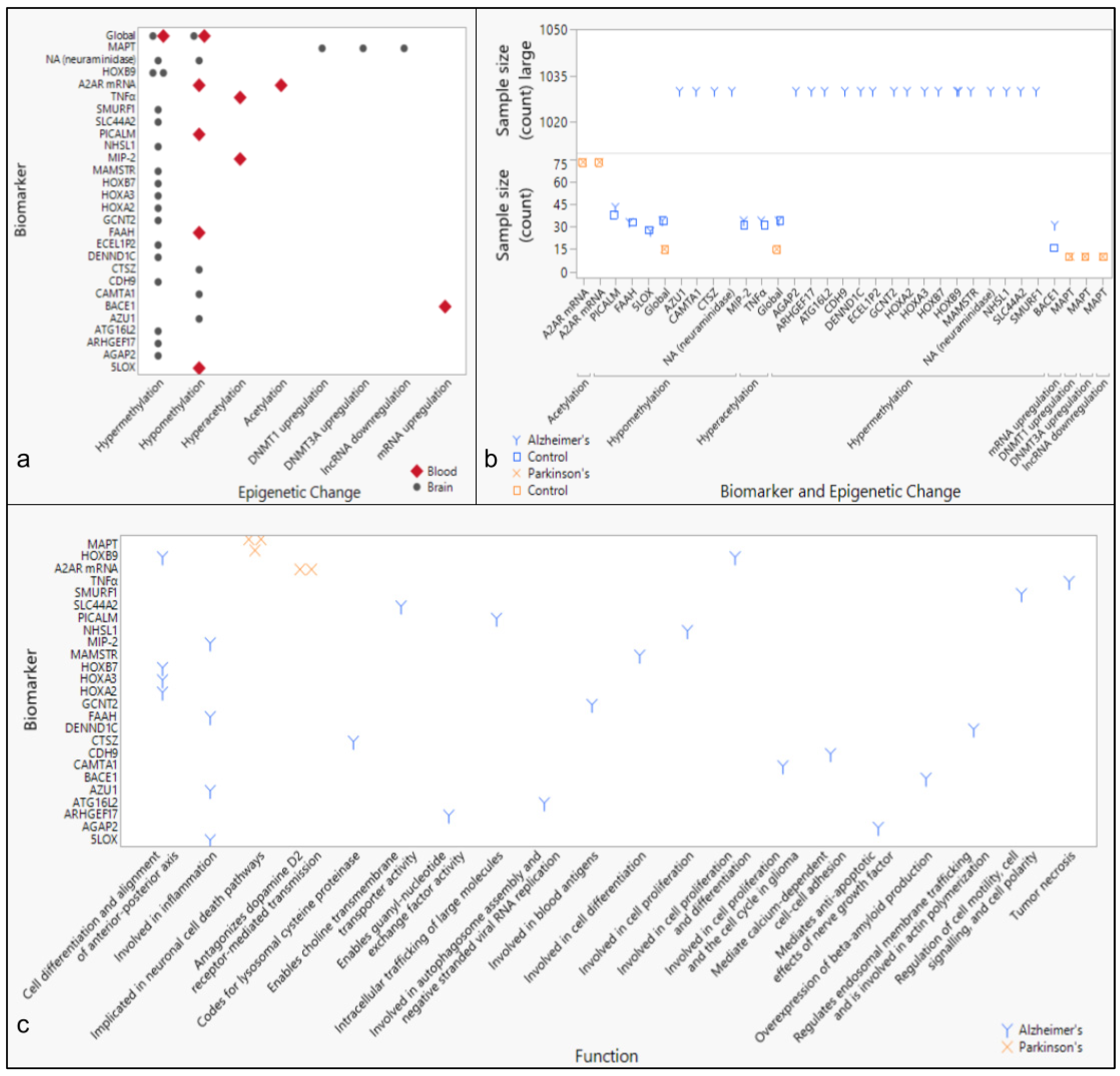
3.4. Epigenetic Markers of Neurodegenerative Diseases
4. Discussion
4.1. Neurodegenerative Disease Prevalence and Exposure to Environmental Toxins
4.2. Environmental Toxins Common to Neurodegenerative Diseases and Associated Epigenetic Changes
4.2.1. Epigenetic Changes Associated with Air Pollution
4.2.2. Epigenetic Changes Associated with Toxic Metals
4.2.3. Epigenetic Changes Associated with Pesticides and Herbicides
4.2.4. Epigenetic Changes Associated with Mycotoxins and Cyanotoxins
4.3. Differential DNA Methylation in Neurodegenerative Diseases
4.4. Considering a Lifetime of Exposures
4.5. Review Time Window
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, P.; Miah, M.R.; Aschner, M. Metals and Neurodegeneration. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Wakode, S.; Sharma, A.; Nair, N.; Dhobi, M.; Wani, M.A.; Potoo, F.H. Effect of environmental toxicants on neuronal functions. Environ. Sci. Pollut. Res. 2020, 27, 44906–44921. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.W. Hemispheres of Influence: Bridging the Disconnect between Environmental Neurotoxicology and Clinical Practice. Environ. Health Perspect. 2022, 130, 052001. [Google Scholar] [CrossRef] [PubMed]
- Tschanz, J.T.; Corcoran, C.D.; Schwartz, S.; Treiber, K.; Green, R.C.; Norton, M.C.; Mielke, M.M.; Piercy, K.; Steinberg, M.; Rabins, P.V.; et al. Progression of cognitive, functional, and neuropsychiatric symptom domains in a population cohort with alzheimer dementia: The cache county dementia progression study. Am. J. Geriatr. Psychiatry 2011, 19, 532–542. [Google Scholar] [CrossRef]
- Tsiouris, K.M.; Konitsiotis, S.; Koutsouris, D.D.; Fotiadis, D.I. Prognostic factors of Rapid symptoms progression in patients with newly diagnosed parkinson’s disease. Artif. Intell. Med. 2020, 103, 101807. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef]
- Zufiria, M.; Javier Gil-Bea, F.; Fernandez-Torron, R.; Jose Poza, J.; Luis Munoz-Blanco, J.; Rojas-Garcia, R.; Riancho, J.; Lopez de Munain, A. ALS: A bucket of genes, environment, metabolism and unknown ingredients. Prog. Neurobiol. 2016, 142, 104–129. [Google Scholar] [CrossRef]
- Rivera, C.M.; Ren, B. Mapping Human Epigenomes. Cell 2013, 155, 39–55. [Google Scholar] [CrossRef]
- Villota-Salazar, N.A.; Mendoza-Mendoza, A.; González-Prieto, J.M. Epigenetics: From the past to the present. Front. Life Sci. 2016, 9, 347–370. [Google Scholar] [CrossRef]
- Leda, K.; Elisavet, G.; Antrea, I.; Costas, H.; George, T.; Helen, T.; Sofia, K. P16 promoter methylation in Pb2+-exposed individuals. Clin. Toxicol. 2010, 48, 124–128. [Google Scholar]
- Song, C.; Kanthasamy, A.; Anantharam, V.; Sun, F.; Kanthasamy, A.G. Environmental Neurotoxic Pesticide Increases Histone Acetylation to Promote Apoptosis in Dopaminergic Neuronal Cells: Relevance to Epigenetic Mechanisms of Neurodegeneration. Mol. Pharmacol. 2010, 77, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, C.C.; Liu, X.Y.; Deng, X.N.; Tian, Q.; Du, Y.J. Epigenetic Modulation of Microglia Function and Phenotypes in Neurodegenerative Diseases. Neural Plast. 2021, 2021, 9912686. [Google Scholar] [CrossRef] [PubMed]
- Young, A.B. Four Decades of Neurodegenerative Disease Research: How Far We Have Come! J. Neurosci. 2009, 29, 12722–12728. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, L.F.J.R.; Matoso, R.D.O.; Rodrigues, M.V.; de Lima, T.O.L.; Nascimento, A.F.; Carvalho, F.C.; Moreira, D.R.; Fernandes, J.C.; de Paula, J.J.; Magno, L.A.V.; et al. Factors influencing possible delay in the diagnosis of Alzheimer’s disease: Findings from a tertiary Public University Hospital. Dement. Neuropsychol. 2011, 5, 328–331. [Google Scholar] [CrossRef]
- Breen, D.P.; Evans, J.R.; Farrell, K.; Brayne, C.; Barker, R.A. Determinants of delayed diagnosis in Parkinson’s disease. J. Neurol. 2013, 260, 1978–1981. [Google Scholar] [CrossRef]
- Longinetti, E.; Fang, F. Epidemiology of amyotrophic lateral sclerosis: An update of recent literature. Curr. Opin. Neurol. 2019, 32, 771–776. [Google Scholar] [CrossRef]
- Alzheimer’s Foundation. Alzheimer’s Disease Facts and Figures [Internet]; Alzheimer’s Association: Chicago, IL, USA, 2023; Available online: https://www.alz.org/alzheimers-dementia/facts-figures (accessed on 13 May 2022).
- Akushevich, I.; Yashkin, A.P.; Kravchenko, J.; Yashin, A.I. Analysis of Time Trends in Alzheimer’s Disease and Related Dementias Using Partitioning Approach. J. Alzheimer’s Dis. 2021, 82, 1277–1289. [Google Scholar] [CrossRef]
- de Lau, L.M.L.; Schipper, C.M.A.; Hofman, A.; Koudstaal, P.J.; Breteler, M.M.B. Prognosis of Parkinson Disease: Risk of Dementia and Mortality: The Rotterdam Study. Arch. Neurol. 2005, 62, 1265–1269. [Google Scholar] [CrossRef]
- Feigin, V.L.; Abajobir, A.A.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abera, S.F.; Abyu, G.Y.; Ahmed, M.B.; Aichour, A.N.; Aichour, I.; et al. Global, regional, and national burden of neurological disorders during 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017, 16, 877–897. [Google Scholar] [CrossRef]
- Marin, B.; Boumédiene, F.; Logroscino, G.; Couratier, P.; Babron, M.C.; Leutenegger, A.L.; Copetti, M.; Preux, P.-M.; Beghi, E. Variation in worldwide incidence of amyotrophic lateral sclerosis: A meta-analysis. Int. J. Epidemiol. 2016, 46, 57–74. [Google Scholar] [CrossRef]
- Newell, M.E.; Adhikari, S.; Halden, R.U. Systematic and state-of the science review of the role of environmental factors in Amyotrophic Lateral Sclerosis (ALS) or Lou Gehrig’s Disease. Sci. Total Environ. 2022, 817, 152504. [Google Scholar] [CrossRef] [PubMed]
- Testa, D.; Lovati, R.; Ferrarini, M.; Salmoiraghi, F.; Filippini, G. Survival of 793 patients with amyotrophic lateral sclerosis diagnosed over a 28-year period. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2004, 5, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, M.K. Riluzole and edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Aging (NIA). Parkinson’s Disease: Causes, Symptoms, and Treatments. National Institutes of Health [Internet]; National Institutes of Health, U.S. Department of Health & Human Services: Bethesda, MD, USA, 2022. Available online: https://www.nia.nih.gov/health/parkinsons-disease (accessed on 13 May 2022).
- Rahman, M.; White, E.M.; Mills, C.; Thomas, K.S.; Jutkowitz, E. Rural-urban differences in diagnostic incidence and prevalence of Alzheimer’s disease and related dementias. Alzheimer’s Dement. 2021, 17, 1213–1230. [Google Scholar] [CrossRef]
- Manivannan, B.; Yegambaram, M.; Supowit, S.; Beach, T.G.; Halden, R.U. Assessment of persistent, bioaccumulative and toxic organic environmental pollutants in liver and adipose tissue of alzheimer’s disease patients and age-matched controls. Curr. Alzheimer Res. 2019, 16, 1039–1049. [Google Scholar] [CrossRef]
- Aravindan, A.; Newell, M.E.; Halden, R.U. Literature review and meta-analysis of environmental toxins associated with increased risk of Parkinson’s disease. Sci. Total Environ. 2024, 931, 172838. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency (EPA). 2020 CDR Data [Internet]. Access CDR Data. Chemical Data Reporting; Updated 26 January 2023; US EPA: Washington, DC, USA, 2023. Available online: https://www.epa.gov/chemical-data-reporting/access-cdr-data#2020 (accessed on 13 May 2022).
- Marras, C.; Beck, J.C.; Bower, J.H.; Roberts, E.; Ritz, B.; Ross, G.W.; Abbott, R.D.; Savica, R.; Van Den Eeden, S.K.; Willis, A.W.; et al. Prevalence of Parkinson’s disease across North America. npj Park. Dis. 2018, 4, 21. [Google Scholar] [CrossRef]
- Mehta, P. Prevalence of Amyotrophic Lateral Sclerosis—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 1285–1289. [Google Scholar] [CrossRef]
- Searles Nielsen, S.; Checkoway, H.; Criswell, S.R.; Farin, F.M.; Stapleton, P.L.; Sheppard, L.; Racette, B.A. Inducible nitric oxide synthase gene methylation and parkinsonism in manganese-exposed welders. Park. Relat. Disord. 2015, 21, 355–360. [Google Scholar] [CrossRef]
- Calderón-Garcidueñas, L.; Herrera-Soto, A.; Jury, N.; Maher, B.A.; González-Maciel, A.; Reynoso-Robles, R.; Ruiz-Rudolph, P.; van Zundert, B.; Varela-Nallar, L. Reduced repressive epigenetic marks, increased DNA damage and Alzheimer’s disease hallmarks in the brain of humans and mice exposed to particulate urban air pollution. Environ. Res. 2020, 183, 109226. [Google Scholar] [CrossRef]
- Byun, H.M.; Panni, T.; Motta, V.; Hou, L.; Nordio, F.; Apostoli, P.; Bertazzi, P.A.; Baccarelli, A.A. Effects of airborne pollutants on mitochondrial DNA Methylation. Part. Fibre Toxicol. 2013, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Engström, K.S.; Hossain, M.B.; Lauss, M.; Ahmed, S.; Raqib, R.; Vahter, M.; Broberg, K. Efficient Arsenic Metabolism—The AS3MT Haplotype Is Associated with DNA Methylation and Expression of Multiple Genes Around AS3MT. PLoS ONE 2013, 8, e53732. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.R.; Trask, J.D. The virus of poliomyelitis in stools and sewage. J. Am. Med. Assoc. 1941, 116, 493–498. [Google Scholar] [CrossRef]
- Song, C.; Kanthasamy, A.; Jin, H.; Anantharam, V.; Kanthasamy, A.G. Paraquat induces epigenetic changes by promoting histone acetylation in cell culture models of dopaminergic degeneration. NeuroToxicology 2011, 32, 586–595. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, Z.; Wang, Q.; Zhang, J.; Wang, L.; Zhang, Q.; Li, H.; Wu, S. Manganese chloride induces histone acetylation changes in neuronal cells: Its role in manganese-induced damage. NeuroToxicology 2018, 65, 255–263. [Google Scholar] [CrossRef]
- Li, X.; Gao, J.; Huang, K.; Qi, X.; Dai, Q.; Mei, X.; Xu, W. Dynamic changes of global DNA methylation hypermethylation of cell adhesion-related genes in rat kidneys in response to ochratoxin, A. World Mycotoxin J. 2015, 8, 465–476. [Google Scholar] [CrossRef]
- Ranganayaki, S.; Govindaraj, P.; Gayathri, N.; Srinivas Bharath, M.M. Exposure to the neurotoxin 3-nitropropionic acid in neuronal cells induces unique histone acetylation pattern: Implications for neurodegeneration. Neurochem. Int. 2020, 140, 104846. [Google Scholar] [CrossRef]
- Maćkowiak, M.; Bator, E.; Latusz, J.; Mordalska, P.; Wedzony, K. Prenatal MAM administration affects histone H3 methylation in postnatal life in the rat medial prefrontal cortex. Eur. Neuropsychopharmacol. 2014, 24, 271–289. [Google Scholar] [CrossRef]
- Lozoya, O.A.; Xu, F.; Grenet, D.; Wang, T.; Grimm, S.A.; Godfrey, V.; Waidyanatha, S.; Woychik, R.P.; Santos, J.H. Single Nucleotide Resolution Analysis Reveals Pervasive, Long-Lasting DNA Methylation Changes by Developmental Exposure to a Mitochondrial Toxicant. Cell Rep. 2020, 32, 108131. [Google Scholar] [CrossRef]
- Cronican, A.A.; Fitz, N.F.; Carter, A.; Saleem, M.; Shiva, S.; Barchowsky, A.; Koldamova, R.; Schug, J.; Lefterov, I. Genome-Wide Alteration of Histone H3K9 Acetylation Pattern in Mouse Offspring Prenatally Exposed to Arsenic. PLoS ONE 2013, 8, e53478. [Google Scholar] [CrossRef]
- Alashwal, H.; Dosunmu, R.; Zawia, N.H. Integration of genome-wide expression and methylation data: Relevance to aging and Alzheimer’s disease. NeuroToxicology 2012, 33, 1450–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Silva, T.C.; Young, J.I.; Gomez, L.; Schmidt, M.A.; Hamilton-Nelson, K.L.; Kunkle, B.W.; Chen, X.; Martin, E.R.; Wang, L. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer’s disease. Nat. Commun. 2020, 11, 6114. [Google Scholar] [CrossRef] [PubMed]
- Kanthasamy, A.; Jin, H.; Anantharam, V.; Sondarva, G.; Rangasamy, V.; Rana, A.; Kanthasamy, A. Emerging neurotoxic mechanisms in environmental factors-induced neurodegeneration. NeuroToxicology 2012, 33, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Clinician Brief: Arsenic|Environmental Health and Medicine|ATSDR [Internet]. 2024. Available online: https://www.atsdr.cdc.gov/environmental-medicine/hcp/clinicianbriefarsenic/ (accessed on 30 January 2025).
- Wang, B.; Du, Y. Cadmium and its neurotoxic effects. Oxid. Med. Cell Longev. 2013, 2013, 898034. [Google Scholar] [CrossRef]
- Oliveira, C.S.; Nogara, P.A.; Ardisson-Araújo, D.M.P.; Aschner, M.; Rocha, J.B.T.; Dórea, J.G. Neurodevelopmental Effects of Mercury. Adv. Neurotoxicol. 2018, 2, 27. [Google Scholar]
- Dobson, A.W.; Erikson, K.M.; Aschner, M. Manganese neurotoxicity. Ann. N. Y. Acad. Sci. 2004, 1012, 115–128. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. National Ambient Air Quality Standards (NAAQS) for PM [Internet]. 2024. Available online: https://www.epa.gov/pm-pollution/national-ambient-air-quality-standards-naaqs-pm (accessed on 30 January 2025).
- Shukla, A.; Bunkar, N.; Kumar, R.; Bhargava, A.; Tiwari, R.; Chaudhury, K.; Goryacheva, I.Y.; Mishra, P.K. Air pollution associated epigenetic modifications: Transgenerational inheritance and underlying molecular mechanisms. Sci. Total Environ. 2019, 656, 760–777. [Google Scholar] [CrossRef]
- Dou, J.F.; Farooqui, Z.; Faulk, C.D.; Barks, A.K.; Jones, T.; Dolinoy, D.C.; Bakulski, K.M. Perinatal Lead (PB) Exposure and Cortical Neuron-Specific DNA Methylation in male Mice. Genes 2019, 10, 274. [Google Scholar] [CrossRef]
- Mythri, R.B.; Raghunath, N.R.; Narwade, S.C.; Pandareesh, M.D.R.; Sabitha, K.R.; Aiyaz, M.; Chand, B.; Sule, M.; Ghosh, K.; Kumar, S.; et al. Manganese- and 1-methyl-4-phenylpyridinium-induced neurotoxicity display differences in morphological, electrophysiological and genome-wide alterations: Implications for idiopathic Parkinson’s disease. J. Neurochem. 2017, 143, 334–358. [Google Scholar] [CrossRef]
- Tarale, P.; Sivanesan, S.; Daiwile, A.P.; Stöger, R.; Bafana, A.; Naoghare, P.K.; Parmar, D.; Chakrabarti, T.; Kannan, K. Global DNA methylation profiling of manganese-exposed human neuroblastoma SH-SY5Y cells reveals epigenetic alterations in Parkinson’s disease-associated genes. Arch. Toxicol. 2017, 91, 2629–2641. [Google Scholar] [CrossRef]
- Bose, R.; Onishchenko, N.; Edoff, K.; Janson Lang, A.M.; Ceccatelli, S. Inherited Effects of Low-Dose Exposure to Methylmercury in Neural Stem Cells. Toxicol. Sci. 2012, 130, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Head, J.; Nam, D.H.; Pilsner, J.R.; Carvan, M.J.; Chan, H.M.; Goetz, F.W.; Murphy, C.A.; Rouvinen-Watt, K.; Scheuhammer, A.M. Effects of methylmercury on epigenetic markers in three model species: Mink, chicken and yellow perch. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2013, 157, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.W.; Lee, D.H.; Won, H.R.; Kim, K.H.; Seong, Y.J.; Kwon, S.H. Influence of Toxicologically Relevant Metals on Human Epigenetic Regulation. Toxicol. Res. 2015, 31, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, L.; Nasuti, C.; Fedeli, D.; Galeazzi, R.; Laudadio, E.; Massaccesi, L.; López-Rodas, G.; Gabbianelli, R. Early impairment of epigenetic pattern in neurodegeneration: Additional mechanisms behind pyrethroid toxicity. Exp. Gerontol. 2019, 124, 110629. [Google Scholar] [CrossRef]
- Kochmanski, J.; Vanoeveren, S.E.; Patterson, J.R.; Bernstein, A.I. Developmental dieldrin exposure alters DNA methylation at genes related to dopaminergic neuron development and Parkinson’s disease in mouse midbrain. Toxicol. Sci. 2019, 169, 593–607. [Google Scholar] [CrossRef]
- Freeman, D.M.; Lou, D.; Li, Y.; Martos, S.N.; Wang, Z. The conserved DNMT1-dependent methylation regions in human cells are vulnerable to neurotoxicant rotenone exposure. Epigenet. Chromatin 2020, 13, 17. [Google Scholar] [CrossRef]
- Aroniadou-Anderjaska, V.; Figueiredo, T.H.; de Araujo Furtado, M.; Pidoplichko, V.I.; Braga, M.F.M. Mechanisms of Organophosphate Toxicity and the Role of Acetylcholinesterase Inhibition. Toxics 2023, 11, 866. [Google Scholar] [CrossRef]
- Collotta, M.; Bertazzi, P.A.; Bollati, V. Epigenetics and pesticides. Toxicology 2013, 307, 35–41. [Google Scholar] [CrossRef]
- Kamal, N.; Ali, S.S.Z.; Fazlullah, K.; Mohammed, B. Ochratoxin A–induced genotoxic and epigenetic mechanisms lead to Alzheimer disease: Its modulation with strategies. Environ. Sci. Pollut. Res. 2020, 27, 44673–44700. [Google Scholar]
- Khan, A.Q.; Thielen, L.; Le Pen, G.; Krebs, M.O.; Kebir, O.; Groh, A.; Deest, M.; Bleich, S.; Frieling, H.; Jahn, K. Methylation pattern and mRNA expression of synapse-relevant genes in the MAM model of schizophrenia in the time-course of adolescence. Schizophrenia 2022, 8, 110. [Google Scholar] [CrossRef]
- Han, J.; Wang, Q.C.; Zhu, C.C.; Liu, J.; Zhang, Y.; Cui, X.S.; Kim, N.H.; Sun, S.C. Deoxynivalenol exposure induces autophagy/apoptosis and epigenetic modification changes during porcine oocyte maturation. Toxicol. Appl. Pharmacol. 2016, 300, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, J.; Mu, P.; Lin, R.; Wen, J.; Deng, Y. Toxicokinetics and metabolism of deoxynivalenol in animals and humans. Arch. Toxicol. Arch. Toxikol. 2022, 96, 2639–2654. [Google Scholar] [CrossRef] [PubMed]
- Spencer, P.S.; Palmer, V.S.; Kisby, G.E. Western Pacific ALS-PDC: Evidence implicating cycad genotoxins. J. Neurol. Sci. 2020, 419, 117185. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, B.M.; Chung, C.; Thompson, B.; Kim, H.; Nakahara, A.; Anink, J.J.; Mills, J.D.; Phatnani, H.; Kwan, J.; Sareen, D.; et al. The cycad genotoxin methylazoxymethanol, linked to Guam ALS/PDC, induces transcriptional mutagenesis. Acta Neuropathol. Commun. 2024, 12, 30. [Google Scholar] [CrossRef]
- Chung, C.S.; Kou, Y.; Shemtov, S.J.; Verheijen, B.M.; Flores, I.; Love, K.; Del Dosso, A.; Thorwald, M.A.; Liu, Y.; Hicks, D.; et al. Transcript errors generate amyloid-like proteins in human cells. Nat. Commun. 2024, 15, 8676. [Google Scholar] [CrossRef]
- Lagrange, E.; Loriot, M.A.; Chaudhary, N.K.; Schultz, P.; Dirks, A.C.; Guissart, C.; James, T.Y.; Vernoux, J.P.; Camu, W.; Tripathi, A.; et al. Corrected speciation and gyromitrin content of false morels linked to ALS patients with mostly slow-acetylator phenotypes. eNeurologicalSci 2024, 35, 100502. [Google Scholar] [CrossRef]
- Gao, Z.; Fu, H.; Zhao, L.; Sun, Z.; Yang, Y.; Zhu, H. Aberrant DNA methylation associated with Alzheimer’s disease in the superior temporal gyrus. Exp. Ther. Med. 2017, 15, 103–108. [Google Scholar] [CrossRef]
- Henderson, A.R.; Wang, Q.; Meechoovet, B.; Siniard, A.L.; Naymik, M.; De Both, M.; Huentelman, M.J.; Caselli, R.J.; Driver-Dunckley, E.; Dunckley, T. DNA Methylation and Expression Profiles of Whole Blood in Parkinson’s Disease. Front. Genet. 2021, 12, 640266. [Google Scholar] [CrossRef]
- Morahan, J.M.; Yu, B.; Trent, R.J.; Pamphlett, R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2009, 10, 418–429. [Google Scholar] [CrossRef]
- Zhang, M.; Xi, Z.; Saez-Atienzar, S.; Chia, R.; Moreno, D.; Sato, C.; Montazer-Haghighi, M.; Traynor, B.J.; Zinman, L.; Rogaeva, E. Combined epigenetic/genetic study identified an ALS age of onset modifier. Acta Neuropathol. Commun. 2021, 9, 75. [Google Scholar] [CrossRef]
- Figueroa-Romero, C.; Hur, J.; Bender, D.E.; Delaney, C.E.; Cataldo, M.D.; Smith, A.L.; Yung, R.; Ruden, D.M.; Callaghan, B.C.; Feldman, E.L. Identification of Epigenetically Altered Genes in Sporadic Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e52672. [Google Scholar] [CrossRef] [PubMed]
- Mercorio, R.; Pergoli, L.; Galimberti, D.; Favero, C.; Carugno, M.; Valle, E.D.; Barretta, F.; Cortini, F.; Scarpini, E.; Valentina, V.B.; et al. PICALM Gene Methylation in Blood of Alzheimer’s Disease Patients Is Associated with Cognitive Decline. J. Alzheimer’s Dis. 2018, 65, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Francesco, A.D.; Arosio, B.; Gussago, C.; Dainese, E.; Mari, D.; D’Addario, C.; Maccarrone, M. Involvement of 5-Lipoxygenase in Alzheimer’s Disease: A Role for DNA Methylation. J. Alzheimer’s Dis. 2013, 37, 3–8. [Google Scholar] [CrossRef] [PubMed]
- D’Addario, C.; Di Francesco, A.; Arosio, B.; Gussago, C.; Dell’Osso, B.; Bari, M.; Galimberti, D.; Scarpini, E.; Altamura, A.C.; Mari, D.; et al. Epigenetic Regulation of Fatty Acid Amide Hydrolase in Alzheimer Disease. Estus S, editor. PLoS ONE 2012, 7, e39186. [Google Scholar]
- Coupland, K.G.; Kim, W.S.; Halliday, G.M.; Hallupp, M.; Dobson-Stone, C.; Kwok, J.B.J. Role of the Long Non-Coding RNA MAPT-AS1 in Regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson’s Disease. PLoS ONE 2016, 11, e0157924. [Google Scholar] [CrossRef]
- Falconi, A.; Bonito-Oliva, A.; Di Bartolomeo, M.; Massimini, M.; Fattapposta, F.; Locuratolo, N.; Dainese, E.; Pascale, E.; Fisone, G.; D’Addario, C. On the Role of Adenosine A2A Receptor Gene Transcriptional Regulation in Parkinson’s Disease. Front. Neurosci. 2019, 13, 683. [Google Scholar] [CrossRef]
- Logroscino, G.; Piccininni, M. Amyotrophic lateral sclerosis descriptive epidemiology: The origin of geographic difference. Neuroepidemiology 2019, 52, 93–103. [Google Scholar] [CrossRef]
- Masala, A.; Sanna, S.; Esposito, S.; Rassu, M.; Galioto, M.; Zinellu, A.; Carru, C.; Carrì, M.; Laccario, C.; Crosio, C. Epigenetic Changes Associated with the Expression of Amyotrophic Lateral Sclerosis (ALS) Causing Genes. Neuroscience 2018, 390, 1–11. [Google Scholar] [CrossRef]
- Xi, Z.; Zinman, L.; Moreno, D.; Schymick, J.; Liang, Y.; Sato, C.; Zheng, Y.; Ghani, M.; Dib, S.; Keth, J.; et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am. J. Hum. Genet. 2013, 92, 981–989. [Google Scholar] [CrossRef]
- Ying, K.; Paulson, S.; Eames, A.; Tyshkovskiy, A.; Li, S.; Perez-Guevara, M.; Emamifar, M.; Martínez, M.C.; Kwon, D.; Kosheleva, A.; et al. A Unified Framework for Systematic Curation and Evaluation of Aging Biomarkers. bioRxiv 2024. [Google Scholar] [CrossRef]
- Perez-Gracia, J.L.; Sanmamed, M.F.; Bosch, A.; Patiño-Garcia, A.; Schalper, K.A.; Segura, V.; Bellmunt, J.; Tabernero, J.; Sweeney, C.J.; Choueiri, T.K.; et al. Strategies to design clinical studies to identify predictive biomarkers in cancer research. Cancer Treat. Rev. 2017, 53, 79–97. [Google Scholar] [CrossRef]
- Upadhya, S.; Gingerich, D.; Lutz, M.W.; Chiba-Falek, O. Differential Gene Expression and DNA Methylation in the Risk of Depression in LOAD Patients. Biomolecules 2022, 12, 1679. [Google Scholar] [CrossRef]
- La Torre, A.; Vecchio, F.L.; Greco, A. Epigenetic Mechanisms of Aging and Aging-Associated Diseases. Cells 2023, 12, 1163. [Google Scholar] [CrossRef] [PubMed]
- Quiccione, M.S.; Tirozzi, A.; Cassioli, G.; Morelli, M.; Costanzo, S.; Pepe, A.; Bracone, F.; Magnacca, S.; Cerletti, C.; Licastro, D.; et al. Are Methylation Patterns in the KALRN Gene Associated with Cognitive and Depressive Symptoms? Findings from the Moli-sani Cohort. Int. J. Mol. Sci. 2024, 25, 10317. [Google Scholar] [CrossRef] [PubMed]
- Ijomone, O.M.; Ijomone, O.K.; Iroegbu, J.D.; Ifenatuoha, C.W.; Olung, N.F.; Aschner, M. Epigenetic influence of environmentally neurotoxic metals. NeuroToxicology 2020, 81, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Cookson, M.R.; Petrucelli, L.; La Spada, A.R. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018, 21, 1300–1309. [Google Scholar] [CrossRef]
- Ogonowski, N.S.; García-Marín, L.M.; Fernando, A.S.; Flores-Ocampo, V.; Rentería, M.E. Impact of genetic predisposition to late-onset neurodegenerative diseases on early life outcomes and brain structure. Transl. Psychiatry 2024, 14, 185. [Google Scholar] [CrossRef]
- Shaw, C.A.; Höglinger, G.U. Neurodegenerative diseases: Neurotoxins as sufficient etiologic agents? Neuromol. Med. 2008, 10, 1–9. [Google Scholar] [CrossRef]
- Angeles, A.K.; Janke, F.; Bauer, S.; Christopoulos, P.; Riediger, A.L.; Sültmann, H. Liquid biopsies beyond mutation calling: Genomic and epigenomic features of cell-free dna in cancer. Cancers 2021, 13, 5615. [Google Scholar] [CrossRef]
- Chen, L.; Shen, Q.; Xu, S.; Yu, H.; Pei, S.; Zhang, Y.; He, X.; Wang, Q.; Li, D. 5-Hydroxymethylcytosine Signatures in Circulating Cell-Free DNA as Diagnostic Biomarkers for Late-Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2022, 85, 569–581. [Google Scholar] [CrossRef]
- Warnecke, T.; Schäfer, K.H.; Claus, I.; Del Tredici, K.; Jost, W.H. Gastrointestinal involvement in Parkinson’s disease: Pathophysiology, diagnosis, and management. npj Park. Dis. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, W.D.; Papadopoulos, N.; Goodman, S.N.; Bjerregaard, N.C.; Laurberg, S.; Levin, B.; Juhl, H.; Arber, N.; Moi-nova, H.; et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 2009, 27, 858–863. [Google Scholar] [CrossRef] [PubMed]
- Markosian, C.; Mirzoyan, N. Wastewater-based epidemiology as a novel assessment approach for population-level metal exposure. Sci. Total Environ. 2019, 689, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Rousis, N.I.; Gracia-Lor, E.; Hernández, F.; Poretti, F.; Santos, M.M.; Zuccato, E.; Castiglioni, S. Wastewater-based epidemiology as a novel tool to evaluate human exposure to pesticides: Triazines and organophosphates as case studies. Sci. Total Environ. 2021, 793, 148618. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Newell, M.E.; Aravindan, A.; Babbrah, A.; Halden, R.U. Epigenetic Biomarkers Driven by Environmental Toxins Associated with Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the United States: A Systematic Review. Toxics 2025, 13, 114. https://doi.org/10.3390/toxics13020114
Newell ME, Aravindan A, Babbrah A, Halden RU. Epigenetic Biomarkers Driven by Environmental Toxins Associated with Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the United States: A Systematic Review. Toxics. 2025; 13(2):114. https://doi.org/10.3390/toxics13020114
Chicago/Turabian StyleNewell, Melanie Engstrom, Anumitha Aravindan, Ayesha Babbrah, and Rolf U. Halden. 2025. "Epigenetic Biomarkers Driven by Environmental Toxins Associated with Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the United States: A Systematic Review" Toxics 13, no. 2: 114. https://doi.org/10.3390/toxics13020114
APA StyleNewell, M. E., Aravindan, A., Babbrah, A., & Halden, R. U. (2025). Epigenetic Biomarkers Driven by Environmental Toxins Associated with Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis in the United States: A Systematic Review. Toxics, 13(2), 114. https://doi.org/10.3390/toxics13020114






