Transcriptome-Wide Analysis of N6-Methyladenosine-Modified Long Noncoding RNAs in Particulate Matter-Induced Lung Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals Experiment
2.2. MeRIP Sequencing and Identification of Differentially Methylated lncRNAs
2.3. LncRNA Sequencing
2.4. Real-Time qPCR (RT-qPCR)
2.5. MeRIP-qPCR
2.6. Data Analysis
3. Results
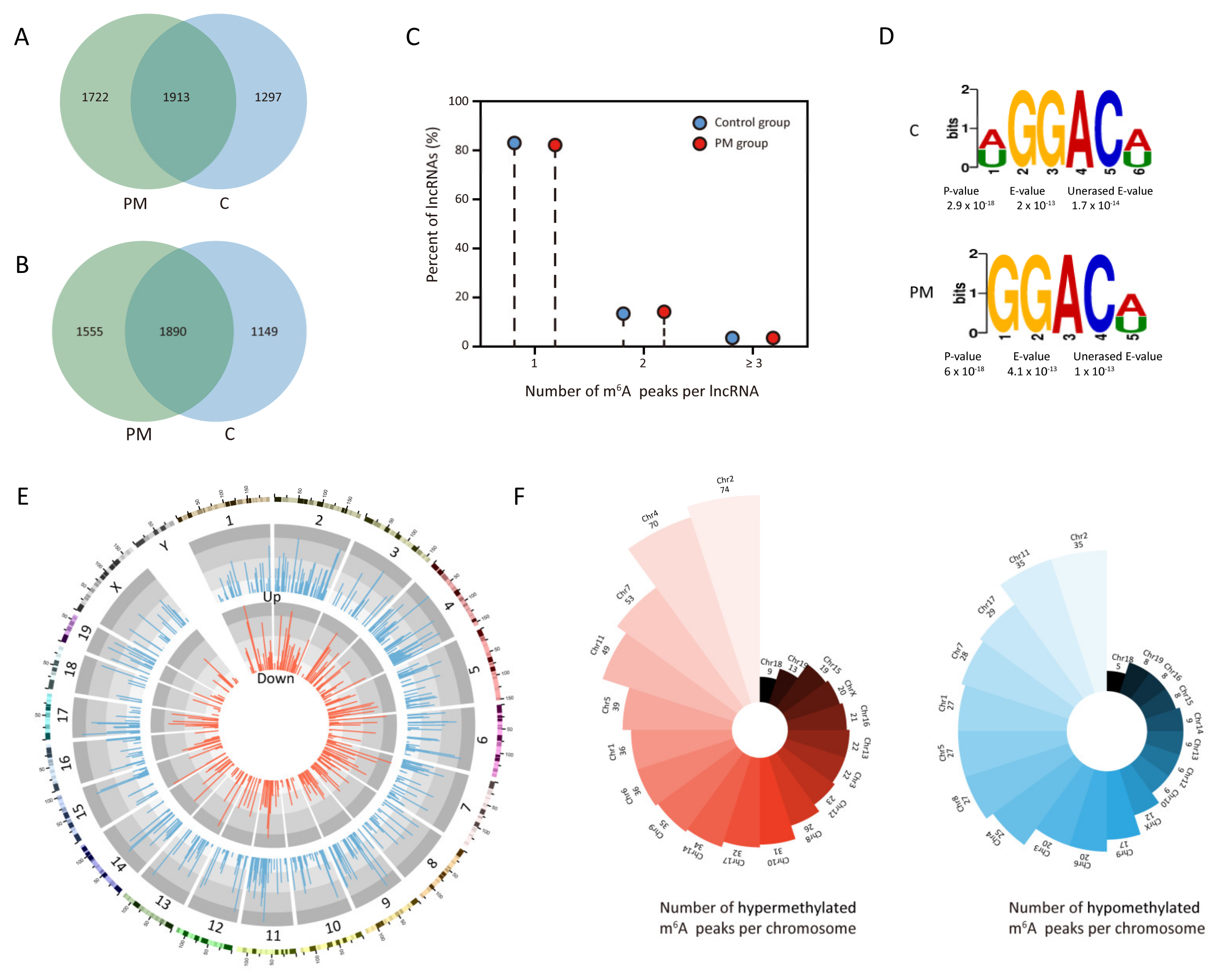
3.1. Characteristics of m6A Peaks on lncRNAs in Lung Tissues of Mice Exposed to PM
3.2. Function Enrichment and Pathway Analysis of m6A-Modified lncRNAs
3.3. Differential Expression and Functional Analysis of lncRNAs in Lung Tissues of Mice Exposed to PM
3.4. Conjoint Analysis of MeRIP-Seq and lncRNA-Seq Data
3.5. Construction of m6A-Modified lncRNAs-mRNAs Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, C.; Chen, R.; Sera, F.; Vicedo-Cabrera, A.M.; Guo, Y.; Tong, S.; Coelho, M.; Saldiva, P.H.N.; Lavigne, E.; Matus, P.; et al. Ambient Particulate Air Pollution and Daily Mortality in 652 Cities. N. Engl. J. Med. 2019, 381, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, J.; Yang, L.; Xu, Y.; Zhang, X.; Bai, C.; Kang, J.; Ran, P.; Shen, H.; Wen, F.; et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): A national cross-sectional study. Lancet 2018, 391, 1706–1717. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Yang, T.; Xu, J.; Yang, L.; Zhao, J.; Zhang, X.; Bai, C.; Kang, J.; Ran, P.; Shen, H.; et al. Prevalence, risk factors, and management of asthma in China: A national cross-sectional study. Lancet 2019, 394, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, J.; Wang, L.; Chen, C.; Yang, D.; Jin, M.; Bai, C.; Song, Y. Urban particulate matter triggers lung inflammation via the ROS-MAPK-NF-κB signaling pathway. J. Thorac. Dis. 2017, 9, 4398. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Song, J.; Zeng, Y.; Xia, S.; Chen, C.; Jin, M.; Song, Y. The circular RNA circTXNRD1 promoted ambient particulate matter-induced inflammation in human bronchial epithelial cells by regulating miR-892a/COX-2 axis. Chemosphere 2022, 286, 131614. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Ye, L.; Chen, C.; She, J.; Song, Y. MiR-29b-3p promotes particulate matter-induced inflammatory responses by regulating the C1QTNF6/AMPK pathway. Aging 2020, 12, 1141–1158. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Zhu, M.; Chen, S.; Chen, C.; Miao, C.; He, H.; Song, Y. Osteopontin potentiates PM-induced IL-1α and IL-1β production via the ERK/JNK signaling pathway. Ecotoxicol. Environ. Saf. 2019, 171, 467–474. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Wang, L.; Chen, C.; Song, Y. Amphiregulin potentiates airway inflammation and mucus hypersecretion induced by urban particulate matter via the EGFR-PI3Kalpha-AKT/ERK pathway. Cell. Signal. 2019, 53, 122–131. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m(6) A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- He, X.; Zhang, L.; Liu, S.; Wang, J.; Liu, Y.; Xiong, A.; Jiang, M.; Luo, L.; Ying, X.; Li, G. Methyltransferase-like 3 leads to lung injury by up-regulation of interleukin 24 through N6-methyladenosine-dependent mRNA stability and translation efficiency in mice exposed to fine particulate matter 2.5. Environ. Pollut. 2022, 308, 119607. [Google Scholar] [CrossRef]
- Song, J.; Zeng, Y.; Zhu, M.; Zhu, G.; Chen, C.; Jin, M.; Wang, J.; Song, Y. Comprehensive analysis of transcriptome-wide m(6)A methylome in the lung tissues of mice with acute particulate matter exposure. Ecotoxicol. Environ. Saf. 2022, 241, 113810. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Pei, Y.H.; Chen, J.; Wu, X.; He, Y.; Qin, W.; He, S.Y.; Chang, N.; Jiang, H.; Zhou, J.; Yu, P.; et al. LncRNA PEAMIR inhibits apoptosis and inflammatory response in PM2.5 exposure aggravated myocardial ischemia/reperfusion injury as a competing endogenous RNA of miR-29b-3p. Nanotoxicology 2020, 14, 638–653. [Google Scholar] [CrossRef]
- He, F.; Wang, N.; Yu, X.; Zheng, Y.; Liu, Q.; Chen, Q.; Pu, J.; Li, N.; Zou, W.; Li, B.; et al. GATA3/long noncoding RNA MHC-R regulates the immune activity of dendritic cells in chronic obstructive pulmonary disease induced by air pollution particulate matter. J. Hazard. Mater. 2022, 438, 129459. [Google Scholar] [CrossRef]
- Li, S.X.; Yan, W.; Liu, J.P.; Zhao, Y.J.; Chen, L. Long noncoding RNA SNHG4 remits lipopolysaccharide-engendered inflammatory lung damage by inhibiting METTL3—Mediated m(6)A level of STAT2 mRNA. Mol. Immunol. 2021, 139, 10–22. [Google Scholar] [CrossRef]
- Qian, X.; Yang, J.; Qiu, Q.; Li, X.; Jiang, C.; Li, J.; Dong, L.; Ying, K.; Lu, B.; Chen, E.; et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J. Hematol. Oncol. 2021, 14, 112. [Google Scholar] [CrossRef]
- Chen, Z.H.; Wu, Y.F.; Wang, P.L.; Wu, Y.P.; Li, Z.Y.; Zhao, Y.; Zhou, J.S.; Zhu, C.; Cao, C.; Mao, Y.Y.; et al. Autophagy is essential for ultrafine particle-induced inflammation and mucus hyperproduction in airway epithelium. Autophagy 2016, 12, 297–311. [Google Scholar] [CrossRef]
- Zeng, Y.; Bai, X.; Zhu, G.; Zhu, M.; Peng, W.; Song, J.; Cai, H.; Ye, L.; Chen, C.; Song, Y.; et al. m(6)A-mediated HDAC9 upregulation promotes particulate matter-induced airway inflammation via epigenetic control of DUSP9-MAPK axis and acts as an inhaled nanotherapeutic target. J. Hazard. Mater. 2024, 477, 135093. [Google Scholar] [CrossRef]
- Teng, F.; Tang, W.; Wuniqiemu, T.; Qin, J.; Zhou, Y.; Huang, X.; Wang, S.; Zhu, X.; Tang, Z.; Yi, L.; et al. N(6)-Methyladenosine Methylomic Landscape of Lung Tissues in Murine Acute Allergic Asthma. Front. Immunol. 2021, 12, 740571. [Google Scholar] [CrossRef]
- Qu, M.; Chen, Z.; Qiu, Z.; Nan, K.; Wang, Y.; Shi, Y.; Shao, Y.; Zhong, Z.; Zhu, S.; Guo, K.; et al. Neutrophil extracellular traps-triggered impaired autophagic flux via METTL3 underlies sepsis-associated acute lung injury. Cell Death Discov. 2022, 8, 375. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, P.; Xie, Y.; Fan, L.; You, X.; Yang, S.; Yao, Y.; Chen, W.; Ma, J. Insights into the mechanism underlying crystalline silica-induced pulmonary fibrosis via transcriptome-wide m(6)A methylation profile. Ecotoxicol. Environ. Saf. 2022, 247, 114215. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.; Lim, E.L.; Weeden, C.E.; Lee, C.; Augustine, M.; Chen, K.; Kuan, F.C.; Marongiu, F.; Evans, E.J., Jr.; Moore, D.A.; et al. Lung adenocarcinoma promotion by air pollutants. Nature 2023, 616, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhu, G.; Zhu, M.; Song, J.; Cai, H.; Song, Y.; Wang, J.; Jin, M. Edaravone Attenuated Particulate Matter-Induced Lung Inflammation by Inhibiting ROS-NF-kappaB Signaling Pathway. Oxidative Med. Cell. Longev. 2022, 2022, 6908884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fang, X.Y.; Wu, J.; Fan, Y.G.; Leng, R.X.; Liu, B.; Lv, X.J.; Yan, Y.L.; Mao, C.; Ye, D.A.-O. Association of Combined Exposure to Ambient Air Pollutants, Genetic Risk, and Incident Rheumatoid Arthritis: A Prospective Cohort Study in the UK Biobank. Environ. Health Perspect. 2023, 131, 37008. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, Y.; Lin, Y.; Zhong, Y.; Yu, H.; Huang, Y.; Yang, J.; Cai, Y.; Liu, F.; Li, Y.; et al. PM2.5 induces pulmonary microvascular injury in COPD via METTL16-mediated m6A modification. Environ. Pollut. 2022, 303, 119115. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Ho, S.C.; Sun, W.L.; Feng, P.H.; Lin, C.W.; Chen, K.Y.; Chuang, H.C.; Tseng, C.H.; Chen, T.T.; Wu, S.A.-O. Lnc-IL7R alleviates PM(2.5)-mediated cellular senescence and apoptosis through EZH2 recruitment in chronic obstructive pulmonary disease. Cell Biol. Toxicol. 2022, 38, 1097–1120. [Google Scholar] [CrossRef]
- Li, B.; Huang, N.; Wei, S.; Xv, J.; Meng, Q.; Aschner, M.; Li, X.; Chen, R. lncRNA TUG1 as a ceRNA promotes PM exposure-induced airway hyper-reactivity. J. Hazard. Mater. 2021, 416, 125878. [Google Scholar] [CrossRef]
- Wu, X.; Ma, C.; Ma, Q.; Zhuang, P.; Deng, G.A.-O. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model. Pathogens 2022, 11, 532. [Google Scholar] [CrossRef]
- Sun, L.; Fu, J.; Lin, S.H.; Sun, J.L.; Xia, L.; Lin, C.H.; Liu, L.; Zhang, C.; Yang, L.; Xue, P.; et al. Particulate matter of 2.5 μm or less in diameter disturbs the balance of T(H)17/regulatory T cells by targeting glutamate oxaloacetate transaminase 1 and hypoxia-inducible factor 1α in an asthma model. J. Allergy Clin. Immunol. 2020, 145, 402–414. [Google Scholar] [CrossRef]
- Xia, M.; Harb, H.; Saffari, A.; Sioutas, C.; Chatila, T.A. A Jagged 1-Notch 4 molecular switch mediates airway inflammation induced by ultrafine particles. J. Allergy Clin. Immunol. 2018, 142, 1243–1256.e17. [Google Scholar] [CrossRef] [PubMed]
- Guohua, F.; Tieyuan, Z.; Xinping, M.; Juan, X. Melatonin protects against PM2.5-induced lung injury by inhibiting ferroptosis of lung epithelial cells in a Nrf2-dependent manner. Ecotoxicol. Environ. Saf. 2021, 223, 112588. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, A.; Woo, H.; Raju, S.; Belz, D.C.; Putcha, N.; Williams, M.S.; McCormack, M.C.; Kohler, K.; Hansel, N.N. Indoor particulate matter concentrations and air cleaner intervention association with biomarkers in former smokers with COPD. Environ. Res. 2024, 243, 117874. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Huff, R.D.; Rider, C.F.; Yuen, A.C.Y.; Carlsten, C. Controlled human exposures to diesel exhaust or particle-depleted diesel exhaust with allergen modulates transcriptomic responses in the lung. Sci. Total Environ. 2024, 945, 173688. [Google Scholar] [CrossRef]




| lncRNAs | Regulation | Chromosome | txStart | txEnd | Peak Length | Fold Change | p Value |
|---|---|---|---|---|---|---|---|
| ENSMUST00000135583 | up | chr15 | 79427219 | 79427400 | 181 | 170.6 | 5.93134 × 10−13 |
| uc008juf.1 | up | chr2 | 60369992 | 60370540 | 548 | 157.52 | 9.23891 × 10−13 |
| ENSMUST00000154751 | up | chr5 | 30071281 | 30071580 | 299 | 157 | 1.29438 × 10−9 |
| ENSMUST00000172465 | up | chr14 | 25978576 | 25978680 | 104 | 143.5 | 2.36309 × 10−9 |
| ENSMUST00000175738 | up | chr7 | 15265181 | 15265620 | 439 | 143.5 | 2.98183 × 10−9 |
| NR_046191 | up | chr5 | 38345415 | 38345493 | 78 | 140.1 | 1.8444 × 10−9 |
| NR_038088 | up | chr10 | 79697343 | 79697350 | 7 | 136.7 | 1.81193 × 10−9 |
| ENSMUST00000120341 | up | chr4 | 122205841 | 122205903 | 62 | 136.7 | 4.27563 × 10−12 |
| ENSMUST00000144723 | up | chr6 | 83153701 | 83154300 | 599 | 133.3 | 1.5141 × 10−9 |
| NR_029464 | up | chr12 | 111942281 | 111942700 | 419 | 119.7 | 1.989 × 10−11 |
| ENSMUST00000140004 | down | chr7 | 28086221 | 28086880 | 659 | 305.3 | 6.90569 × 10−13 |
| ENSMUST00000144451 | down | chr8 | 93952946 | 93953024 | 78 | 209.3 | 4.4487 × 10−10 |
| NR_015540 | down | chr1 | 89676821 | 89677080 | 259 | 202.3 | 1.51477 × 10−13 |
| NR_045969 | down | chr7 | 98178421 | 98178533 | 112 | 141.7 | 3.48445 × 10−10 |
| ENSMUST00000152848 | down | chr12 | 56696301 | 56697200 | 899 | 137.9 | 2.57292 × 10−9 |
| ENSMUST00000136726 | down | chr14 | 30002362 | 30002820 | 458 | 132.2 | 5.80821 × 10−12 |
| ENSMUST00000148311 | down | chr3 | 93288941 | 93289320 | 379 | 131.9 | 2.82892 × 10−10 |
| NR_045480 | down | chr6 | 126287781 | 126288340 | 559 | 123 | 1.70799 × 10−9 |
| uc012ejf.1 | down | chr6 | 29799438 | 29799584 | 146 | 119.3 | 3.08992 × 10−11 |
| ENSMUST00000159769 | down | chr14 | 101828967 | 101829058 | 91 | 116.2 | 8.16182 × 10−9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Y.; Zhu, G.; Peng, W.; Cai, H.; Lu, C.; Ye, L.; Jin, M.; Wang, J. Transcriptome-Wide Analysis of N6-Methyladenosine-Modified Long Noncoding RNAs in Particulate Matter-Induced Lung Injury. Toxics 2025, 13, 98. https://doi.org/10.3390/toxics13020098
Zeng Y, Zhu G, Peng W, Cai H, Lu C, Ye L, Jin M, Wang J. Transcriptome-Wide Analysis of N6-Methyladenosine-Modified Long Noncoding RNAs in Particulate Matter-Induced Lung Injury. Toxics. 2025; 13(2):98. https://doi.org/10.3390/toxics13020098
Chicago/Turabian StyleZeng, Yingying, Guiping Zhu, Wenjun Peng, Hui Cai, Chong Lu, Ling Ye, Meiling Jin, and Jian Wang. 2025. "Transcriptome-Wide Analysis of N6-Methyladenosine-Modified Long Noncoding RNAs in Particulate Matter-Induced Lung Injury" Toxics 13, no. 2: 98. https://doi.org/10.3390/toxics13020098
APA StyleZeng, Y., Zhu, G., Peng, W., Cai, H., Lu, C., Ye, L., Jin, M., & Wang, J. (2025). Transcriptome-Wide Analysis of N6-Methyladenosine-Modified Long Noncoding RNAs in Particulate Matter-Induced Lung Injury. Toxics, 13(2), 98. https://doi.org/10.3390/toxics13020098






