The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress
Abstract
:1. Introduction
2. tRNA-Based Regulation of Cellular Processes
3. Ribonucleoside Modifications
3.1. tRNA Modification as Dynamic Marks
3.2. tRNA Modifications Prevent Translational Infidelity and Proteotoxic stress
3.3. tRNA Modification Enzymes as Essential Features of the Cell Stress Response
4. Mechanisms by which tRNA Modifications Function in the Cell Stress Response
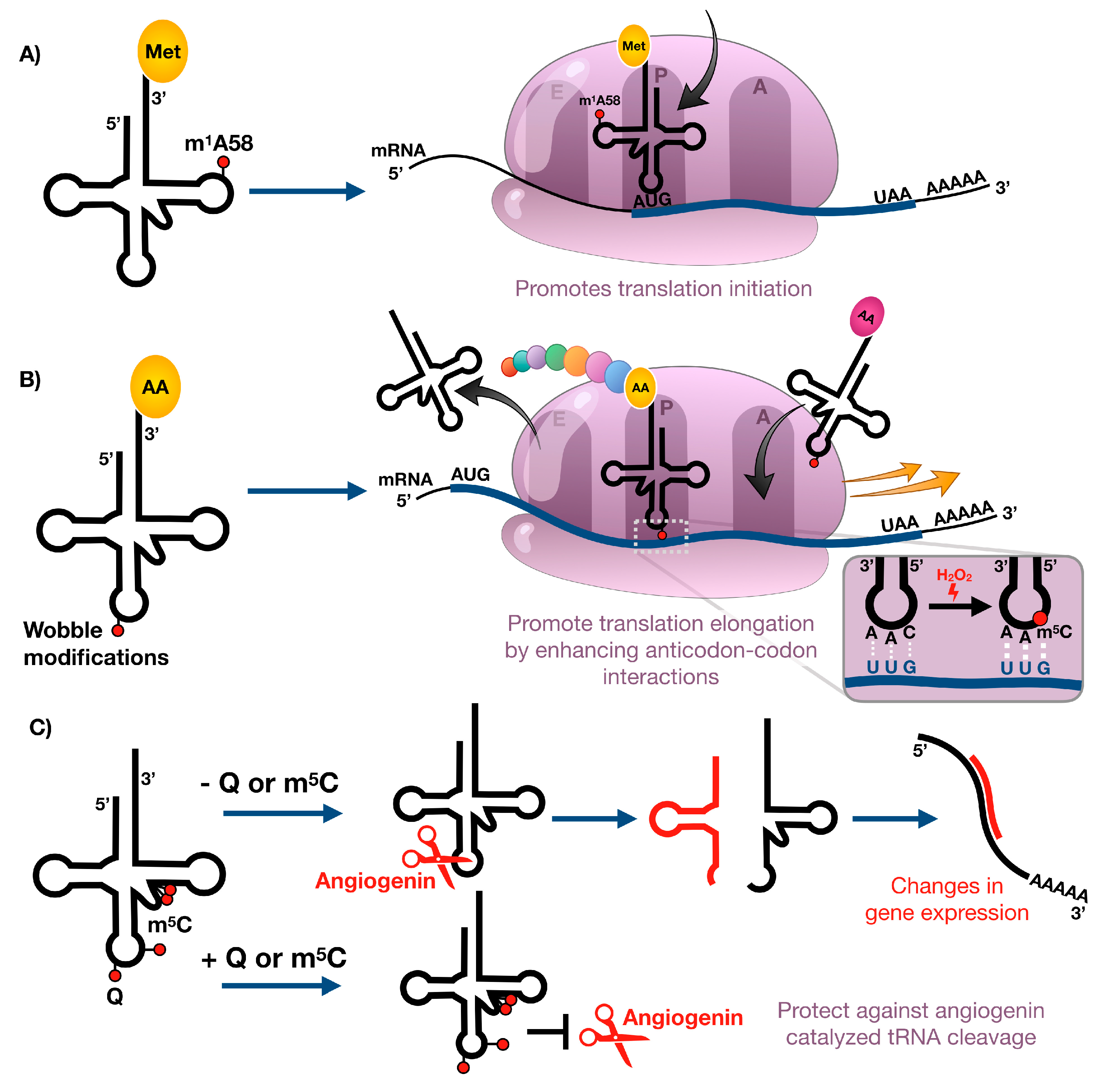
4.1. m1A Affects Translation Initiation during Cell Stress
4.2. Wobble tRNA Modifications Regulate Codon-Biased Translation of Stress Response Proteins
4.3. tRNA Modifications Restrict Stress-Induced tRNA Cleavage
4.4. tRNA Modifications Affect tRNA Maturation during Stress
5. RNA Modifications as Potential Biomarkers of Exposure and Disease Pathology
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Cell Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Kaufman, R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005, 74, 739–789. [Google Scholar] [CrossRef]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef]
- Zhang, X.; Cozen, A.E.; Liu, Y.; Chen, Q.; Lowe, T.M. Small RNA Modifications: Integral to Function and Disease. Trends Mol. Med. 2016, 22, 1025–1034. [Google Scholar] [CrossRef] [Green Version]
- Sarin, L.P.; Leidel, S.A. Modify or die?—RNA modification defects in metazoans. RNA Biol. 2015, 11, 1555–1567. [Google Scholar] [CrossRef]
- Mishima, E.; Inoue, C.; Saigusa, D.; Inoue, R.; Ito, K.; Suzuki, Y.; Jinno, D.; Tsukui, Y.; Akamatsu, Y.; Araki, M.; et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014, 25, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Yasukawa, T.; Ohta, S.; Akira, S.; Ishihara, K.; Watanabe, K.; Suzuki, T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. USA 2004, 101, 15070–15075. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, A.; Dyavaiah, M.; Joseph, F.; Rooney, J.P.; Chan, C.T.Y.; Dedon, P.C.; Begley, T.J. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 2012, 11, 3656–3665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.T.; Deng, W.; Li, F.; DeMott, M.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem. Res. Toxicol. 2015, 28, 978–988. [Google Scholar] [CrossRef]
- Endres, L.; Begley, U.; Clark, R.; Gu, C.; Dziergowska, A.; Małkiewicz, A.; Melendez, J.A.; Dedon, P.C.; Begley, T.J. Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage. PLoS ONE 2015, 10, e0131335. [Google Scholar] [CrossRef]
- Banerjee, R.; Chen, S.; Dare, K.; Gilreath, M.; Praetorius-Ibba, M.; Raina, M.; Reynolds, N.M.; Rogers, T.; Roy, H.; Yadavalli, S.S.; et al. tRNAs: Cellular barcodes for amino acids. FEBS Lett. 2010, 584, 387–395. [Google Scholar] [CrossRef]
- Francklyn, C.S.; Minajigi, A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010, 584, 366–375. [Google Scholar] [CrossRef]
- Harada, F.; Sawyer, R.C.; Dahlberg, J.E. A primer ribonucleic acid for initiation of in vitro Rous sarcarcoma virus deoxyribonucleic acid synthesis. J. Biol. Chem. 1975, 250, 3487–3497. [Google Scholar] [PubMed]
- Harada, F.; Peters, G.G.; Dahlberg, J.E. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 1979, 254, 10979–10985. [Google Scholar] [PubMed]
- Mak, J.; Kleiman, L. Primer tRNAs for reverse transcription. J. Virol. 1997, 71, 8087–8095. [Google Scholar]
- Isel, C.; Marquet, R.; Keith, G.; Ehresmann, C.; Ehresmann, B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993, 268, 25269–25272. [Google Scholar] [PubMed]
- Zhong, J.; Xiao, C.; Gu, W.; Du, G.; Sun, X.; He, Q.-Y.; Zhang, G. Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. PLoS Genet. 2015, 11, e1005302. [Google Scholar] [CrossRef]
- Wek, S.A.; Zhu, S.; Wek, R.C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 1995, 15, 4497–4506. [Google Scholar] [CrossRef]
- Dong, J.; Qiu, H.; Garcia-Barrio, M.; Anderson, J.; Hinnebusch, A.G. Uncharged tRNA Activates GCN2 by Displacing the Protein Kinase Moiety from a Bipartite tRNA-Binding Domain. Mol. Cell 2000, 6, 269–279. [Google Scholar] [CrossRef]
- Doyle, F.; Leonardi, A.; Endres, L.; Tenenbaum, S.A.; Dedon, P.C.; Begley, T.J. Gene- and genome-based analysis of significant codon patterns in yeast, rat and mice genomes with the CUT Codon UTilization tool. Methods 2016, 107, 98–109. [Google Scholar] [CrossRef]
- Mei, Y.; Yong, J.; Liu, H.; Shi, Y.; Meinkoth, J.; Dreyfuss, G.; Yang, X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 2010, 37, 668–678. [Google Scholar] [CrossRef]
- Mei, Y.; Yong, J.; Stonestrom, A.; Yang, X. tRNA and cytochrome c in cell death and beyond. Cell Cycle 2010, 9, 2936–2939. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Gebetsberger, J.; Zywicki, M.; Künzi, A.; Polacek, N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Davis, F.F.; Allen, F.W. Ribonucleic acids from yeast which contain a fifth nucleoside. J. Biol. Chem. 1957, 227, 907–915. [Google Scholar] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. tRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef]
- Bjork, G.R.; Wikstrom, P.M.; Bystrom, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989, 244, 986. [Google Scholar] [CrossRef] [PubMed]
- Shoffner, J.M.; Lott, M.T.; Lezza, A.M.S.; Seibel, P.; Ballinger, S.W.; Wallace, D.C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell 1990, 61, 931–937. [Google Scholar] [CrossRef]
- Yasukawa, T.; Suzuki, T.; Ishii, N.; Ohta, S.; Watanabe, K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001, 20, 4794–4802. [Google Scholar] [CrossRef]
- Yasukawa, T.; Suzuki, T.; Suzuki, T.; Ueda, T.; Ohta, S.; Watanabe, K. Modification Defect at Anticodon Wobble Nucleotide of Mitochondrial tRNAsLeu(UUR) with Pathogenic Mutations of Mitochondrial Myopathy, Encephalopathy, Lactic Acidosis, and Stroke-like Episodes. J. Biol. Chem. 2000, 275, 4251–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e816. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Fu, L.; Guerrero, C.R.; Zhong, N.; Amato, N.J.; Liu, Y.; Liu, S.; Cai, Q.; Ji, D.; Jin, S.G.; Niedernhofer, L.J.; et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014, 136, 11582–11585. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.M.; van Delft, P.; Mendil, L.; Bachman, M.; Smollett, K.; Werner, F.; Miska, E.A.; Balasubramanian, S. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem Eur. J. Chem. Biol. 2015, 16, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.M.; van Delft, P.; Tanpure, A.; Miska, E.A.; Balasubramanian, S. 2′-O-Methyl-5-hydroxymethylcytidine: A Second Oxidative Derivative of 5-Methylcytidine in RNA. J. Am. Chem. Soc. 2017, 139, 1766–1769. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Wang, F.; Ngoc, L.V.; Collignon, E.; Bonvin, E.; Deplus, R.; Calonne, E.; Hassabi, B.; Putmans, P.; Awe, S.; et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016, 351, 282. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Chan, C.T.; Dyavaiah, M.; Rooney, J.P.; Dedon, P.C.; Begley, T.J. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012, 9, 990–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [Green Version]
- Tukenmez, H.; Xu, H.; Esberg, A.; Bystrom, A.S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredes, J.A.; Carreto, L.; Simões, J.; Bezerra, A.R.; Gomes, A.C.; Santamaria, R.; Kapushesky, M.; Moura, G.R.; Santos, M.A.S. Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast. BMC Biol. 2012, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Lyko, F. Genome recoding by tRNA modifications. Open Biol. 2016, 6, 160287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2014, 16, 98. [Google Scholar] [CrossRef] [PubMed]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA Decay Can Result from Lack of Nonessential Modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Cload, S.T.; Liu, D.R.; Froland, W.A.; Schultz, P.G. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem. Biol. 1996, 3, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Esberg, A.; Huang, B.; Johansson, M.J.O.; Byström, A.S. Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Droogmans, L.; Roovers, M.; Bujnicki, J.M.; Tricot, C.; Hartsch, T.; Stalon, V.; Grosjean, H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003, 31, 2148–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, K.; Kunibayashi, T.; Tomikawa, C.; Ochi, A.; Kanai, T.; Hirata, A.; Iwashita, C.; Hori, H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011, 39, 2304–2318. [Google Scholar] [CrossRef]
- Rose, R.E.; Pazos, M.A., 2nd; Curcio, M.J.; Fabris, D. Global Epitranscriptomics Profiling of RNA Post-Transcriptional Modifications as an Effective Tool for Investigating the Epitranscriptomics of Stress Response. Mol. Cell. Proteom. 2016, 15, 932–944. [Google Scholar] [CrossRef]
- Basanta-Sanchez, M.; Temple, S.; Ansari, S.A.; D’Amico, A.; Agris, P.F. Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016, 44, e26. [Google Scholar] [CrossRef]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G.; Lomakin, I.B.; Pilipenko, E.V.; Shatsky, I.N.; Agol, V.I.; Hellen, C.U.T. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 2001, 98, 7029. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983, 47, 1–45. [Google Scholar]
- Basavappa, R.; Sigler, P.B. The 3 A crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 1991, 10, 3105–3111. [Google Scholar] [CrossRef]
- Anderson, J.; Phan, L.; Cuesta, R.; Carlson, B.A.; Pak, M.; Asano, K.; Björk, G.R.; Tamame, M.; Hinnebusch, A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998, 12, 3650–3662. [Google Scholar] [CrossRef] [Green Version]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Chan, C.T.Y.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Babu, I.R.; Su, D.; Yin, S.; Begley, T.J.; Dedon, P.C. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015, 11, e1005706. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; DiRenzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2008, 583, 437–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [Green Version]
- Motorin, Y.; Lyko, F.; Helm, M. 5-methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010, 38, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [Green Version]
- Haag, S.; Warda, A.S.; Kretschmer, J.; Günnigmann, M.A.; Höbartner, C.; Bohnsack, M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015, 21, 1532–1543. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science 2006, 311, 395. [Google Scholar] [CrossRef]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, F.; Nishimura, S. Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli. Universal presence of nucleoside O in the first position of the anticodons of these transfer ribonucleic acid. Biochemistry 1972, 11, 301–308. [Google Scholar] [CrossRef]
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Reyniers, J.P.; Pleasants, J.R.; Wostmann, B.S.; Katze, J.R.; Farkas, W.R. Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J. Biol. Chem. 1981, 256, 11591–11594. [Google Scholar]
- Katze, J.R.; Gunduz, U.; Smith, D.L.; Cheng, C.S.; McCloskey, J.A. Evidence that the nucleic acid base queuine is incorporated intact into tRNA by animal cells. Biochemistry 1984, 23, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Pathak, C.; Jaiswal, Y.K.; Vinayak, M. Queuine promotes antioxidant defence system by activating cellular antioxidant enzyme activities in cancer. Biosci. Rep. 2008, 28, 73. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Matuszek, Z.; Huang, Y.; Parisien, M.; Dai, Q.; Clark, W.; Schwartz, M.H.; Pan, T. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 2018, 24, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Nakanishi, K.; Macfarlane, R.D.; Torgerson, D.F.; Ohashi, Z.; McCloskey, J.A.; Gross, H.J.; Nishimura, S. The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J. Am. Chem. Soc. 1976, 98, 5044–5046. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Huang, B.; Esberg, A.; Johansson, M.J.O.; Byström, A.S. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 2005, 11, 1648–1654. [Google Scholar] [CrossRef]
- Ogawa, T.; Inoue, S.; Yajima, S.; Hidaka, M.; Masaki, H. Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res. 2006, 34, 6065–6073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Meidler, R.; Amitsur, M.; Kaufmann, G. Specific interaction between anticodon nuclease and the tRNALys wobble base11Edited by D. Draper. J. Mol. Biol. 2001, 305, 377–388. [Google Scholar] [CrossRef]
- Hopper, A.K.; Phizicky, E.M. tRNA transfers to the limelight. Genes Dev. 2003, 17, 162–180. [Google Scholar] [CrossRef] [Green Version]
- Hopper, A.K. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013, 194, 43–67. [Google Scholar] [CrossRef]
- Foretek, D.; Wu, J.; Hopper, A.K.; Boguta, M. Control of Saccharomyces cerevisiae pre-tRNA processing by environmental conditions. RNA 2016, 22, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Ohira, T.; Miyauchi, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T. Precise analysis of modification status at various stage of tRNA maturation in Saccharomyces cerevisiae. Nucleic Acids Symp. Ser. 2009, 53, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K.; De Robertis, E.M. RNA processing in microinjected Xenopus oocytes: Sequential addition of base modifications in a spliced transfer RNA. J. Mol. Biol. 1981, 145, 405–420. [Google Scholar] [CrossRef]
- Torres, A.G.; Piñeyro, D.; Rodríguez-Escribà, M.; Camacho, N.; Reina, O.; Saint-Léger, A.; Filonava, L.; Batlle, E.; Ribas de Pouplana, L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef] [PubMed]
- Gaston, K.W.; Rubio, M.A.T.; Spears, J.L.; Pastar, I.; Papavasiliou, F.N.; Alfonzo, J.D. C to U editing at position 32 of the anticodon loop precedes tRNA 5′ leader removal in trypanosomatids. Nucleic Acids Res. 2007, 35, 6740–6749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Miyauchi, K.; Harada, T.; Okita, R.; Takeshita, E.; Komaki, H.; Fujioka, K.; Yagasaki, H.; Goto, Y.-I.; Yanaka, K.; et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 2018, 9, 1875. [Google Scholar] [CrossRef] [PubMed]
- Baranowski, W.; Dirheimer, G.; Jakowicki, J.A.; Keith, G. Deficiency of Queuine, a Highly Modified Purine Base, in Transfer RNAs from Primary and Metastatic Ovarian Malignant Tumors in Women. Cancer Res. 1994, 54, 4468. [Google Scholar]
- Huang, B.-S.; Wu, R.-T.; Chien, K.-Y. Relationship of the Queuine Content of Transfer Ribonucleic Acids to Histopathological Grading and Survival in Human Lung Cancer. Cancer Res. 1992, 52, 4696. [Google Scholar]
- Dewe, J.M.; Fuller, B.L.; Lentini, J.M.; Kellner, S.M.; Fu, D. TRMT1-Catalyzed tRNA Modifications Are Required for Redox Homeostasis to Ensure Proper Cellular Proliferation and Oxidative Stress Survival. Mol. Cell. Biol. 2017, 37, e00214–e00217. [Google Scholar] [CrossRef]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770. [Google Scholar] [CrossRef]
- Wei, F.-Y.; Suzuki, T.; Watanabe, S.; Kimura, S.; Kaitsuka, T.; Fujimura, A.; Matsui, H.; Atta, M.; Michiue, H.; Fontecave, M.; et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Investig. 2011, 121, 3598–3608. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Nakamura, M.; Anai, S.; De Velasco, M.; Tanaka, M.; Tsujikawa, K.; Ouji, Y.; Konishi, N. A Novel Human AlkB Homologue, ALKBH8, Contributes to Human Bladder Cancer Progression. Cancer Res. 2009, 69, 3157. [Google Scholar] [CrossRef]
- Ueda, Y.; Ooshio, I.; Fusamae, Y.; Kitae, K.; Kawaguchi, M.; Jingushi, K.; Hase, H.; Harada, K.; Hirata, K.; Tsujikawa, K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017, 7, 42271. [Google Scholar] [CrossRef] [Green Version]
- Tasaki, M.; Shimada, K.; Kimura, H.; Tsujikawa, K.; Konishi, N. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br. J. Cancer 2011, 104, 700–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrenhofer-Murray, A.E. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules 2017, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009, 37, e12. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huber, S.M.; Leonardi, A.; Dedon, P.C.; Begley, T.J. The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics 2019, 7, 17. https://doi.org/10.3390/toxics7010017
Huber SM, Leonardi A, Dedon PC, Begley TJ. The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics. 2019; 7(1):17. https://doi.org/10.3390/toxics7010017
Chicago/Turabian StyleHuber, Sabrina M., Andrea Leonardi, Peter C. Dedon, and Thomas J. Begley. 2019. "The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress" Toxics 7, no. 1: 17. https://doi.org/10.3390/toxics7010017
APA StyleHuber, S. M., Leonardi, A., Dedon, P. C., & Begley, T. J. (2019). The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress. Toxics, 7(1), 17. https://doi.org/10.3390/toxics7010017





