Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions
Abstract
:1. Introduction
2. Exposome Measurement Approaches
2.1. Chemical Approaches
2.1.1. Low-Resolution Mass Spectrometry
2.1.2. High-Resolution Mass Spectrometry
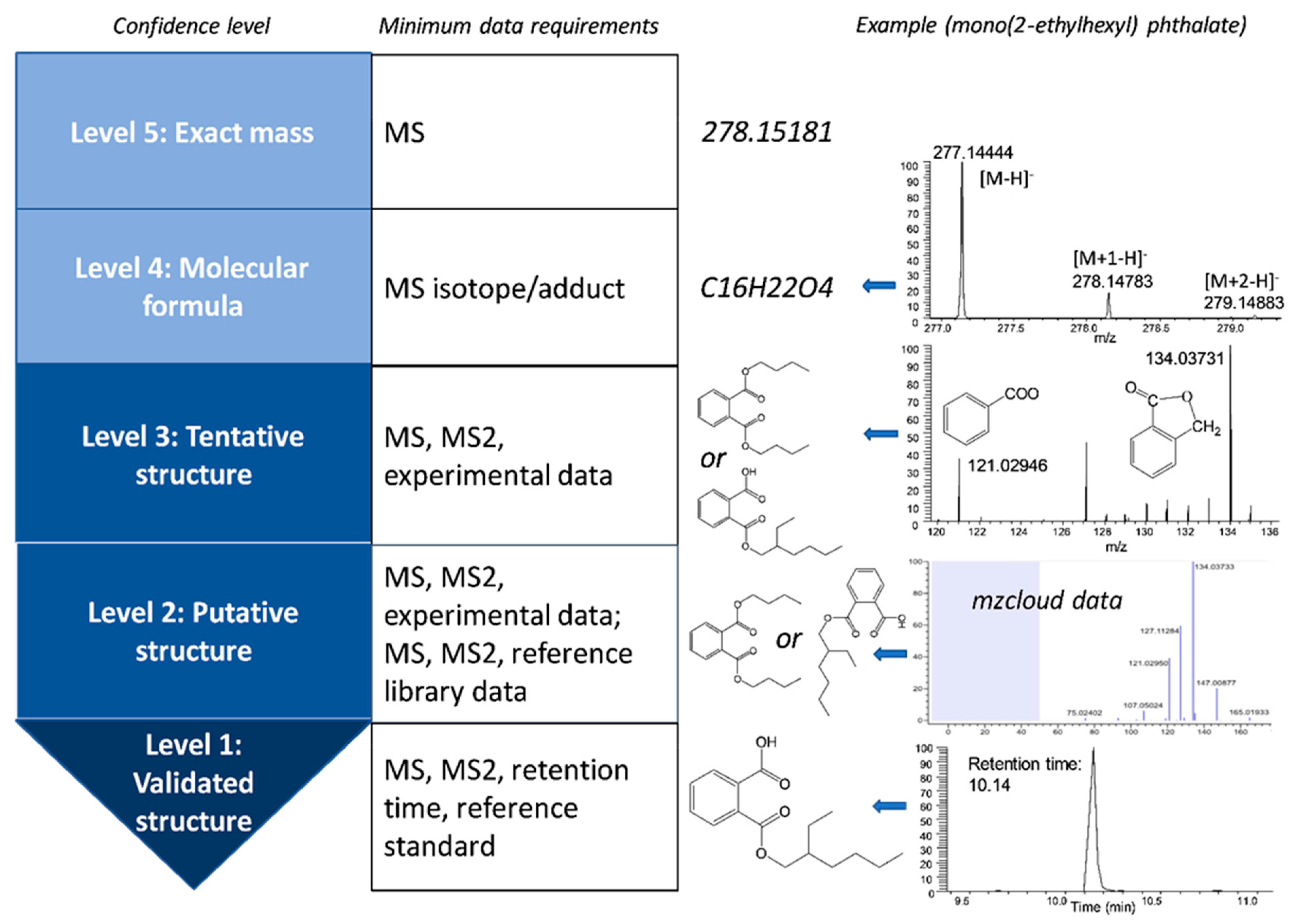
- (i)
- grouping all the features (ions or peaks) from the same compound based on the full scan MS chromatogram and determining monoisotopic or neutral molecular mass. An enormous number of ions are present in the chromatogram, but not every ion represents an individual compound. One compound may form different adducts (e.g., protonated and deprotonated ions, [M + Na]+, and [M + NH4]+), neutral losses (e.g., [M + H-H2O]+), isotopes (e.g., M+1 and M+2 isotope of precursor ion), and even in-source fragments. A variety of strategies have been proposed to group peaks including comparing expected theoretical distances between known ion adduct masses with experimental distances. One recent study suggested to extract MS pseudospectra based on peak shape and peak abundance based on the assumption that all the peaks with different m/z ratios from the same compounds ideally have a similar peak shape and strong linear relation in relative abundance across samples [51].
- (ii)
- acquiring a list of potential chemical candidates by searching the monoisotopic mass or the molecular formula assigned against the databases. It has been reported that monoisotopic mass-based searching resulted in a higher percentage of chemicals in the number one rank position than chemical formula-based searching [54]. Available chemical substance databases such as PubChem and ChemSpider have been reviewed [52]. Recently, a few more databases have been developed for search including CompTox Chemistry Dashboard, Exposome-Explorer, and Toxic Exposome Database (T3DB) [55,56,57].
- (iii)
- ranking the candidate list based on other information of the unknown chemical, including MS/MS spectral information, retention time, biochemical pathway and environmental chemistry knowledge. Fragments information can differentiate molecules with the same neutral mass in most cases. There are databases with experimental or in silico MS/MS spectral information available for reference, such as the Human Metabolome Database (HMDB) and METLIN. Currently available mass spectral databases have been reviewed elsewhere [52,58]. Retention time information can be obtained through quantitative structure-retention relationships (QSRR) models when reference standards are not available [59,60]. Biochemical pathway and environmental chemistry knowledge can also be used to narrow down putative identified compounds. In metabolomics, many bioinformatics tools are using biochemical pathways to filter and rank lists of candidates such as XCMS, xMSannotator, and mummichog [61,62,63]. It is expected that environmental chemistry knowledge will be incorporated into bioinformatic tools to facilitate the identification of xenobiotics.
2.2. Biological Approaches
2.3. Other Approaches
3. Measurement-Based Exposome Studies
3.1. Top-Down Exposome Approach
3.2. Bottom-Up Exposome Approach
4. Publicly Accessible Data-Based Exposome Studies
5. Challenges in Exposome Research
5.1. Challenges in Measuring the Exposome
5.2. Challenges in Associating Exposome with Diseases
6. Future Directions
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rappaport, S.M.; Smith, M.T. Epidemiology. Environment and disease risks. Science 2010, 330, 460–461. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.C. Balancing life-style and genomics research for disease prevention. Science 2002, 296, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Polderman, T.J.; Benyamin, B.; de Leeuw, C.A.; Sullivan, P.F.; van Bochoven, A.; Visscher, P.M.; Posthuma, D. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 2015, 47, 702–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappaport, S.M. Genetic Factors Are Not the Major Causes of Chronic Diseases. PLoS ONE 2016, 11, e0154387. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, M.M.; Miller, G.W. The Exposome Paradigm in Human Health: Lessons from the Emory Exposome Summer Course. Environ. Health Perspect. 2017, 125, 064502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Balshaw, D.M.; Kwok, R.K.; Thompson, C.L.; Collman, G.W.; Birnbaum, L.S. The Exposome: Embracing the Complexity for Discovery in Environmental Health. Environ. Health Perspect. 2016, 124, A137–A140. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Athersuch, T.J.; Collman, G.W.; Dhungana, S.; Grant, D.F.; Jones, D.P.; Patel, C.J.; Vasiliou, V. Yale school of public health symposium on lifetime exposures and human health: The exposome; summary and future reflections. Hum. Genom. 2017, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Dennis, K.K.; Marder, E.; Balshaw, D.M.; Cui, Y.; Lynes, M.A.; Patti, G.J.; Rappaport, S.M.; Shaughnessy, D.T.; Vrijheid, M.; Barr, D.B. Biomonitoring in the Era of the Exposome. Environ. Health Perspect. 2017, 125, 502–510. [Google Scholar] [CrossRef]
- Al-Chalabi, A.; Pearce, N. Commentary: Mapping the Human Exposome: Without It, How Can We Find Environmental Risk Factors for ALS? Epidemiology 2015, 26, 821–823. [Google Scholar] [CrossRef]
- Buck Louis, G.M.; Smarr, M.M.; Patel, C.J. The Exposome Research Paradigm: An Opportunity to Understand the Environmental Basis for Human Health and Disease. Curr. Environ. Health Rep. 2017, 4, 89–98. [Google Scholar] [CrossRef]
- Lioy, P.J.; Rappaport, S.M. Exposure science and the exposome: An opportunity for coherence in the environmental health sciences. Environ. Health Perspect. 2011, 119, A466–A467. [Google Scholar] [CrossRef]
- Rappaport, S.M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011, 21, 5–9. [Google Scholar] [CrossRef]
- Stingone, J.A.; Buck Louis, G.M.; Nakayama, S.F.; Vermeulen, R.C.; Kwok, R.K.; Cui, Y.; Balshaw, D.M.; Teitelbaum, S.L. Toward Greater Implementation of the Exposome Research Paradigm within Environmental Epidemiology. Annu. Rev. Public Health 2017, 38, 315–327. [Google Scholar] [CrossRef]
- Buck Louis, G.M.; Yeung, E.; Sundaram, R.; Laughon, S.K.; Zhang, C. The exposome—Exciting opportunities for discoveries in reproductive and perinatal epidemiology. Paediatr. Perinat. Epidemiol. 2013, 27, 229–236. [Google Scholar] [CrossRef]
- Escher, B.I.; Hackermuller, J.; Polte, T.; Scholz, S.; Aigner, A.; Altenburger, R.; Bohme, A.; Bopp, S.K.; Brack, W.; Busch, W.; et al. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 2017, 99, 97–106. [Google Scholar] [CrossRef]
- Siroux, V.; Agier, L.; Slama, R. The exposome concept: A challenge and a potential driver for environmental health research. Eur. Respir. Rev. 2016, 25, 124–129. [Google Scholar] [CrossRef]
- Wild, C.P. The exposome: From concept to utility. Int. J. Epidemiol. 2012, 41, 24–32. [Google Scholar] [CrossRef]
- Miller, G.W.; Jones, D.P. The nature of nurture: Refining the definition of the exposome. Toxicol. Sci. 2014, 137, 1–2. [Google Scholar] [CrossRef]
- Turner, M.C.; Nieuwenhuijsen, M.; Anderson, K.; Balshaw, D.; Cui, Y.; Dunton, G.; Hoppin, J.A.; Koutrakis, P.; Jerrett, M. Assessing the Exposome with External Measures: Commentary on the State of the Science and Research Recommendations. Annu. Rev. Public Health 2017, 38, 215–239. [Google Scholar] [CrossRef] [Green Version]
- van Tongeren, M.; Cherrie, J.W. An integrated approach to the exposome. Environ. Health Perspect. 2012, 120, A103–A104, author reply A104. [Google Scholar] [CrossRef]
- Rappaport, S.M.; Barupal, D.K.; Wishart, D.; Vineis, P.; Scalbert, A. The blood exposome and its role in discovering causes of disease. Environ. Health Perspect. 2014, 122, 769–774. [Google Scholar] [CrossRef]
- Simon, E.; van Velzen, M.; Brandsma, S.H.; Lie, E.; Loken, K.; de Boer, J.; Bytingsvik, J.; Jenssen, B.M.; Aars, J.; Hamers, T.; et al. Effect-directed analysis to explore the polar bear exposome: Identification of thyroid hormone disrupting compounds in plasma. Environ. Sci. Technol. 2013, 47, 8902–8912. [Google Scholar] [CrossRef]
- Soltow, Q.A.; Strobel, F.H.; Mansfield, K.G.; Wachtman, L.; Park, Y.; Jones, D.P. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013, 9, S132–S143. [Google Scholar] [CrossRef]
- Andra, S.S.; Austin, C.; Patel, D.; Dolios, G.; Awawda, M.; Arora, M. Trends in the application of high-resolution mass spectrometry for human biomonitoring: An analytical primer to studying the environmental chemical space of the human exposome. Environ. Int. 2017, 100, 32–61. [Google Scholar] [CrossRef] [Green Version]
- Asimakopoulos, A.G.; Xue, J.; De Carvalho, B.P.; Iyer, A.; Abualnaja, K.O.; Yaghmoor, S.S.; Kumosani, T.A.; Kannan, K. Urinary biomarkers of exposure to 57 xenobiotics and its association with oxidative stress in a population in Jeddah, Saudi Arabia. Environ. Res. 2016, 150, 573–581. [Google Scholar] [CrossRef]
- Xue, J.; Wu, Q.; Sakthivel, S.; Pavithran, P.V.; Vasukutty, J.R.; Kannan, K. Urinary levels of endocrine-disrupting chemicals, including bisphenols, bisphenol A diglycidyl ethers, benzophenones, parabens, and triclosan in obese and non-obese Indian children. Environ. Res. 2015, 137, 120–128. [Google Scholar] [CrossRef]
- Lenters, V.; Portengen, L.; Smit, L.A.; Jonsson, B.A.; Giwercman, A.; Rylander, L.; Lindh, C.H.; Spano, M.; Pedersen, H.S.; Ludwicki, J.K.; et al. Phthalates, perfluoroalkyl acids, metals and organochlorines and reproductive function: A multipollutant assessment in Greenlandic, Polish and Ukrainian men. Occup. Environ. Med. 2015, 72, 385–393. [Google Scholar] [CrossRef]
- Shoemaker, J.; Dietrich, W. Single Laboratory Validated Method for Determination of Cylindrospermopsin and Anatoxin-a in Ambient Water by Liquid Chromatography/Tandem Mass Spectrometry (LC/MS/MS); US EPA Office of Research and Development: Washington, DC, USA, 2017.
- Schlittenbauer, L.; Seiwert, B.; Reemtsma, T. A false positive finding in liquid chromatography/triple quadrupole mass spectrometry analysis by a non-isobaric matrix component: The case of benzotriazole in urine for human biomonitoring. Rapid. Commun. Mass Spectrom. 2016, 30, 1560–1566. [Google Scholar] [CrossRef]
- CDC, Center for Disease Control and Prevention. National Biomonitoring Program. Available online: https://www.cdc.gov/biomonitoring/about.html (accessed on 1 August 2017).
- Chung, M.K.; Kannan, K.; Louis, G.M.; Patel, C.J. Toward Capturing the Exposome: Exposure Biomarker Variability and Co-Exposure Patterns in the Shared Environment. Environ. Sci. Technol. 2018, 52, 8801–8810. [Google Scholar] [CrossRef]
- Improved LC/MS/MS Pesticide Multiresidue Analysis Using Triggered MRM and Online Dilution. Available online: https://www.agilent.com/cs/library/applications/5991-7193EN.pdf (accessed on 18 August 2019).
- Dresen, S.; Ferreiros, N.; Gnann, H.; Zimmermann, R.; Weinmann, W. Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal. Bioanal. Chem. 2010, 396, 2425–2434. [Google Scholar] [CrossRef]
- Mueller, C.A.; Weinmann, W.; Dresen, S.; Schreiber, A.; Gergov, M. Development of a multi-target screening analysis for 301 drugs using a QTrap liquid chromatography/tandem mass spectrometry system and automated library searching. Rapid Commun. Mass Spectrom. 2005, 19, 1332–1338. [Google Scholar] [CrossRef]
- Lin, L.; Lin, H.; Zhang, M.; Dong, X.; Yin, X.; Qu, C.; Ni, J. Types, principle, and characteristics of tandem high-resolution mass spectrometry and its applications. RSC Adv. 2015, 5, 107623–107636. [Google Scholar] [CrossRef]
- Marshall, A.G.; Hendrickson, C.L. High-resolution mass spectrometers. Annu. Rev. Anal. Chem. 2008, 1, 579–599. [Google Scholar] [CrossRef]
- Krauss, M.; Singer, H.; Hollender, J. LC-high resolution MS in environmental analysis: From target screening to the identification of unknowns. Anal. Bioanal. Chem. 2010, 397, 943–951. [Google Scholar] [CrossRef]
- Hernández, F.; Ibáñez, M.; Bade, R.; Bijlsma, L.; Sancho, J.V. Investigation of pharmaceuticals and illicit drugs in waters by liquid chromatography-high-resolution mass spectrometry. TrAC Trends Anal. Chem. 2014, 63, 140–157. [Google Scholar]
- Romero-González, R.; Frenich, A.G. Applications in High Resolution Mass Spectrometry: Food Safety and Pesticide; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Dickel, T.; Plass, W.R.; Lippert, W.; Lang, J.; Yavor, M.I.; Geissel, H.; Scheidenberger, C. Isobar Separation in a Multiple-Reflection Time-of-Flight Mass Spectrometer by Mass-Selective Re-Trapping. J. Am. Soc. Mass Spectrom. 2017, 28, 1079–1090. [Google Scholar] [CrossRef]
- Lacorte, S.; Fernandez-Alba, A.R. Time of flight mass spectrometry applied to the liquid chromatographic analysis of pesticides in water and food. Mass Spectrom. Rev. 2006, 25, 866–880. [Google Scholar] [CrossRef]
- Sancho, J.V.; Pozo, O.J.; Ibanez, M.; Hernandez, F. Potential of liquid chromatography/time-of-flight mass spectrometry for the determination of pesticides and transformation products in water. Anal. Bioanal. Chem. 2006, 386, 987–997. [Google Scholar] [CrossRef]
- Zhang, N.R.; Yu, S.; Tiller, P.; Yeh, S.; Mahan, E.; Emary, W.B. Quantitation of small molecules using high-resolution accurate mass spectrometers—A different approach for analysis of biological samples. Rapid Commun. Mass Spectrom. 2009, 23, 1085–1094. [Google Scholar] [CrossRef]
- Henry, H.; Sobhi, H.R.; Scheibner, O.; Bromirski, M.; Nimkar, S.B.; Rochat, B. Comparison between a high-resolution single-stage Orbitrap and a triple quadrupole mass spectrometer for quantitative analyses of drugs. Rapid Commun. Mass Spectrom. 2012, 26, 499–509. [Google Scholar] [CrossRef]
- Reinholds, I.; Pugajeva, I.; Bartkevičs, V. Comparison of Tandem Quadrupole Mass Spectrometry and Orbitrap High Resolution Mass Spectrometry for Analysis of Pharmaceutical Residues in Biota Samples. Mat. Sci. Appl. Chem. 2016, 33, 5–10. [Google Scholar] [CrossRef]
- Kern, S.; Fenner, K.; Singer, H.P.; Schwarzenbach, R.P.; Hollender, J. Identification of transformation products of organic contaminants in natural waters by computer-aided prediction and high-resolution mass spectrometry. Environ. Sci. Technol. 2009, 43, 7039–7046. [Google Scholar] [CrossRef]
- Hernandez, F.; Ibanez, M.; Gracia-Lor, E.; Sancho, J.V. Retrospective LC-QTOF-MS analysis searching for pharmaceutical metabolites in urban wastewater. J. Sep. Sci 2011, 34, 3517–3526. [Google Scholar] [CrossRef]
- Cappiello, A.; Famiglini, G.; Palma, P.; Termopoli, V.; Lavezzi, A.M.; Matturri, L. Determination of selected endocrine disrupting compounds in human fetal and newborn tissues by GC-MS. Anal. Bioanal. Chem. 2014, 406, 2779–2788. [Google Scholar] [CrossRef]
- Gerona, R.R.; Schwartz, J.M.; Pan, J.; Friesen, M.M.; Lin, T.; Woodruff, T.J. Suspect screening of maternal serum to identify new environmental chemical biomonitoring targets using liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Expo. Sci. Environ. Epidemiol. 2018, 28, 101–108. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Benton, H.P.; Siuzdak, G. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem 2018, 90, 480–489. [Google Scholar] [CrossRef]
- Yi, L.; Dong, N.; Yun, Y.; Deng, B.; Ren, D.; Liu, S.; Liang, Y. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal. Chim Acta 2016, 914, 17–34. [Google Scholar] [CrossRef]
- Gorrochategui, E.; Jaumot, J.; Lacorte, S.; Tauler, R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow. TrAC Trends Anal. Chem. 2016, 82, 425–442. [Google Scholar] [CrossRef]
- McEachran, A.D.; Sobus, J.R.; Williams, A.J. Identifying known unknowns using the US EPA’s CompTox Chemistry Dashboard. Anal. Bioanal. Chem. 2017, 409, 1729–1735. [Google Scholar] [CrossRef]
- Williams, A.J.; Grulke, C.M.; Edwards, J.; McEachran, A.D.; Mansouri, K.; Baker, N.C.; Patlewicz, G.; Shah, I.; Wambaugh, J.F.; Judson, R.S.; et al. The CompTox Chemistry Dashboard: A community data resource for environmental chemistry. J. Cheminformatics 2017, 9, 61. [Google Scholar] [CrossRef]
- Neveu, V.; Moussy, A.; Rouaix, H.; Wedekind, R.; Pon, A.; Knox, C.; Wishart, D.S.; Scalbert, A. Exposome-Explorer: A manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017, 45, D979–D984. [Google Scholar] [CrossRef]
- Wishart, D.; Arndt, D.; Pon, A.; Sajed, T.; Guo, A.C.; Djoumbou, Y.; Knox, C.; Wilson, M.; Liang, Y.; Grant, J.; et al. T3DB: The toxic exposome database. Nucleic Acids Res. 2015, 43, D928–D934. [Google Scholar] [CrossRef]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2016, 9999, 1–20. [Google Scholar] [CrossRef]
- Randazzo, G.M.; Tonoli, D.; Strajhar, P.; Xenarios, I.; Odermatt, A.; Boccard, J.; Rudaz, S. Enhanced metabolite annotation via dynamic retention time prediction: Steroidogenesis alterations as a case study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1071, 11–18. [Google Scholar] [CrossRef]
- Randazzo, G.M.; Tonoli, D.; Hambye, S.; Guillarme, D.; Jeanneret, F.; Nurisso, A.; Goracci, L.; Boccard, J.; Rudaz, S. Prediction of retention time in reversed-phase liquid chromatography as a tool for steroid identification. Anal. Chim. Acta 2016, 916, 8–16. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef]
- Forsberg, E.M.; Huan, T.; Rinehart, D.; Benton, H.P.; Warth, B.; Hilmers, B.; Siuzdak, G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat. Protoc. 2018, 13, 633–651. [Google Scholar] [CrossRef]
- Spicer, R.; Salek, R.M.; Moreno, P.; Canueto, D.; Steinbeck, C. Navigating freely-available software tools for metabolomics analysis. Metabolomics 2017, 13, 106. [Google Scholar] [CrossRef]
- Misra, B.B.; Fahrmann, J.F.; Grapov, D. Review of emerging metabolomic tools and resources: 2015-2016. Electrophoresis 2017, 38, 2257–2274. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Metz, T.O.; Baker, E.S.; Schymanski, E.L.; Renslow, R.S.; Thomas, D.G.; Causon, T.J.; Webb, I.K.; Hann, S.; Smith, R.D.; Teeguarden, J.G. Integrating ion mobility spectrometry into mass spectrometry-based exposome measurements: What can it add and how far can it go? Bioanalysis 2017, 9, 81–98. [Google Scholar] [CrossRef]
- Hernandez, F.; Portoles, T.; Pitarch, E.; Lopez, F.J. Searching for anthropogenic contaminants in human breast adipose tissues using gas chromatography-time-of-flight mass spectrometry. J. Mass Spectrom. 2009, 44, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Broecker, S.; Herre, S.; Wust, B.; Zweigenbaum, J.; Pragst, F. Development and practical application of a library of CID accurate mass spectra of more than 2,500 toxic compounds for systematic toxicological analysis by LC-QTOF-MS with data-dependent acquisition. Anal. Bioanal. Chem. 2011, 400, 101–117. [Google Scholar] [CrossRef]
- Roca, M.; Leon, N.; Pastor, A.; Yusa, V. Comprehensive analytical strategy for biomonitoring of pesticides in urine by liquid chromatography-orbitrap high resolution masss pectrometry. J. Chromatogr. A 2014, 1374, 66–76. [Google Scholar] [CrossRef]
- Li, X.; Shen, B.; Jiang, Z.; Huang, Y.; Zhuo, X. Rapid screening of drugs of abuse in human urine by high-performance liquid chromatography coupled with high resolution and high mass accuracy hybrid linear ion trap-Orbitrap mass spectrometry. J. Chromatogr. A 2013, 1302, 95–104. [Google Scholar] [CrossRef]
- Helfer, A.G.; Michely, J.A.; Weber, A.A.; Meyer, M.R.; Maurer, H.H. Orbitrap technology for comprehensive metabolite-based liquid chromatographic-high resolution-tandem mass spectrometric urine drug screening—Exemplified for cardiovascular drugs. Anal. Chim. Acta 2015, 891, 221–233. [Google Scholar] [CrossRef]
- Plassmann, M.M.; Brack, W.; Krauss, M. Extending analysis of environmental pollutants in human urine towards screening for suspected compounds. J. Chromatogr. A 2015, 1394, 18–25. [Google Scholar] [CrossRef]
- Senyuva, H.Z.; Gokmen, V.; Sarikaya, E.A. Future perspectives in Orbitrap-high-resolution mass spectrometry in food analysis: A review. Food Addit. Contam. Part. A Chem. Anal. Control. Expo. Risk Assess. 2015, 32, 1568–1606. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Singer, H.P.; Longree, P.; Loos, M.; Ruff, M.; Stravs, M.A.; Ripolles Vidal, C.; Hollender, J. Strategies to characterize polar organic contamination in wastewater: Exploring the capability of high resolution mass spectrometry. Environ. Sci. Technol. 2014, 48, 1811–1818. [Google Scholar] [CrossRef]
- Wild, C.P.; Scalbert, A.; Herceg, Z. Measuring the exposome: A powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 2013, 54, 480–499. [Google Scholar] [CrossRef]
- Niedzwiecki, M.M.; Walker, D.I.; Vermeulen, R.; Chadeau-Hyam, M.; Jones, D.P.; Miller, G.W. The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 107–127. [Google Scholar] [CrossRef]
- Rappaport, S.M.; Li, H.; Grigoryan, H.; Funk, W.E.; Williams, E.R. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol. Lett. 2012, 213, 83–90. [Google Scholar] [CrossRef]
- Dogruer, G.; Weijs, L.; Tang, J.Y.; Hollert, H.; Kock, M.; Bell, I.; Madden Hof, C.A.; Gaus, C. Effect-based approach for screening of chemical mixtures in whole blood of green turtles from the Great Barrier Reef. Sci. Total Environ. 2018, 612, 321–329. [Google Scholar] [CrossRef]
- Tang, J.Y.; McCarty, S.; Glenn, E.; Neale, P.A.; Warne, M.S.; Escher, B.I. Mixture effects of organic micropollutants present in water: Towards the development of effect-based water quality trigger values for baseline toxicity. Water Res. 2013, 47, 3300–3314. [Google Scholar] [CrossRef]
- Brack, W.; Ait-Aissa, S.; Burgess, R.M.; Busch, W.; Creusot, N.; Di Paolo, C.; Escher, B.I.; Mark Hewitt, L.; Hilscherova, K.; Hollender, J.; et al. Effect-directed analysis supporting monitoring of aquatic environments—An in-depth overview. Sci. Total Environ. 2016, 544, 1073–1118. [Google Scholar] [CrossRef]
- Escher, B.I.; Dutt, M.; Maylin, E.; Tang, J.Y.M.; Toze, S.; Wolf, C.R.; Lang, M. Water quality assessment using the AREc32 reporter gene assay indicative of the oxidative stress response pathway. J. Environ. Monitor. 2012, 14, 2877–2885. [Google Scholar] [CrossRef]
- Tang, J.Y.; Aryal, R.; Deletic, A.; Gernjak, W.; Glenn, E.; McCarthy, D.; Escher, B.I. Toxicity characterization of urban stormwater with bioanalytical tools. Water Res. 2013, 47, 5594–5606. [Google Scholar] [CrossRef]
- Brack, W. Effect-directed analysis: A promising tool for the identification of organic toxicants in complex mixtures? Anal. Bioanal. Chem. 2003, 377, 397–407. [Google Scholar] [CrossRef]
- Tian, Z.; Gold, A.; Nakamura, J.; Zhang, Z.; Vila, J.; Singleton, D.R.; Collins, L.B.; Aitken, M.D. Nontarget Analysis Reveals a Bacterial Metabolite of Pyrene Implicated in the Genotoxicity of Contaminated Soil after Bioremediation. Environ. Sci. Technol. 2017, 51, 7091–7100. [Google Scholar] [CrossRef]
- Simon, E.; Lamoree, M.H.; Hamers, T.; de Boer, J. Challenges in effect-directed analysis with a focus on biological samples. Trends Anal. Chem. 2015, 67, 179–191. [Google Scholar] [CrossRef]
- Van Breda, S.G.; Wilms, L.C.; Gaj, S.; Jennen, D.G.; Briede, J.J.; Kleinjans, J.C.; de Kok, T.M. The exposome concept in a human nutrigenomics study: Evaluating the impact of exposure to a complex mixture of phytochemicals using transcriptomics signatures. Mutagenesis 2015, 30, 723–731. [Google Scholar] [CrossRef]
- Pleil, J.D.; Stiegel, M.A. Evolution of environmental exposure science: Using breath-borne biomarkers for “discovery” of the human exposome. Anal. Chem. 2013, 85, 9984–9990. [Google Scholar] [CrossRef]
- Asante-Duah, K. Public Health Risk Assessment for Human Exposure to Chemicals; Springer: Washington, DC, USA, 2017. [Google Scholar]
- Robinson, O.; Basagana, X.; Agier, L.; de Castro, M.; Hernandez-Ferrer, C.; Gonzalez, J.R.; Grimalt, J.O.; Nieuwenhuijsen, M.; Sunyer, J.; Slama, R.; et al. The Pregnancy Exposome: Multiple Environmental Exposures in the INMA-Sabadell Birth Cohort. Environ. Sci. Technol. 2015, 49, 10632–10641. [Google Scholar] [CrossRef] [Green Version]
- Go, Y.M.; Walker, D.I.; Liang, Y.; Uppal, K.; Soltow, Q.A.; Tran, V.; Strobel, F.; Quyyumi, A.A.; Ziegler, T.R.; Pennell, K.D.; et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol. Sci. 2015, 148, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Neujahr, D.C.; Uppal, K.; Force, S.D.; Fernandez, F.; Lawrence, C.; Pickens, A.; Bag, R.; Lockard, C.; Kirk, A.D.; Tran, V.; et al. Bile acid aspiration associated with lung chemical profile linked to other biomarkers of injury after lung transplantation. Am. J. Transplant. 2014, 14, 841–848. [Google Scholar] [CrossRef]
- Park, Y.H.; Lee, K.; Soltow, Q.A.; Strobel, F.H.; Brigham, K.L.; Parker, R.E.; Wilson, M.E.; Sutliff, R.L.; Mansfield, K.G.; Wachtman, L.M.; et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012, 295, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Osborn, M.P.; Park, Y.; Parks, M.B.; Burgess, L.G.; Uppal, K.; Lee, K.; Jones, D.P.; Brantley, M.A., Jr. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS ONE 2013, 8, e72737. [Google Scholar] [CrossRef]
- Cribbs, S.K.; Park, Y.; Guidot, D.M.; Martin, G.S.; Brown, L.A.; Lennox, J.; Jones, D.P. Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res. Hum. Retroviruses 2014, 30, 579–585. [Google Scholar] [CrossRef]
- Roede, J.R.; Uppal, K.; Park, Y.; Lee, K.; Tran, V.; Walker, D.; Strobel, F.H.; Rhodes, S.L.; Ritz, B.; Jones, D.P. Serum metabolomics of slow vs. rapid motor progression Parkinson’s disease: A pilot study. PLoS ONE 2013, 8, e77629. [Google Scholar] [CrossRef]
- Go, Y.M.; Walker, D.I.; Soltow, Q.A.; Uppal, K.; Wachtman, L.M.; Strobel, F.H.; Pennell, K.; Promislow, D.E.; Jones, D.P. Metabolome-wide association study of phenylalanine in plasma of common marmosets. Amino Acids 2015, 47, 589–601. [Google Scholar] [CrossRef]
- Frediani, J.K.; Jones, D.P.; Tukvadze, N.; Uppal, K.; Sanikidze, E.; Kipiani, M.; Tran, V.T.; Hebbar, G.; Walker, D.I.; Kempker, R.R.; et al. Plasma metabolomics in human pulmonary tuberculosis disease: A pilot study. PLoS ONE 2014, 9, e108854. [Google Scholar] [CrossRef]
- Warth, B.; Spangler, S.; Fang, M.; Johnson, C.H.; Forsberg, E.M.; Granados, A.; Martin, R.L.; Domingo-Almenara, X.; Huan, T.; Rinehart, D.; et al. Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing. Anal. Chem 2017, 89, 11505–11513. [Google Scholar] [CrossRef]
- Cecchi, L.; D’Amato, G.; Annesi-Maesano, I. External exposome and allergic respiratory and skin diseases. J. Allergy Clin. Immunol. 2018, 141, 846–857. [Google Scholar] [CrossRef] [Green Version]
- Gehring, U.; Wijga, A.H.; Brauer, M.; Fischer, P.; de Jongste, J.C.; Kerkhof, M.; Oldenwening, M.; Smit, H.A.; Brunekreef, B. Traffic-related air pollution and the development of asthma and allergies during the first 8 years of life. Am. J. Respir. Crit. Care Med. 2010, 181, 596–603. [Google Scholar] [CrossRef]
- Kramer, U.; Sugiri, D.; Ranft, U.; Krutmann, J.; von Berg, A.; Berdel, D.; Behrendt, H.; Kuhlbusch, T.; Hochadel, M.; Wichmann, H.E.; et al. Eczema, respiratory allergies, and traffic-related air pollution in birth cohorts from small-town areas. J. Dermatol. Sci. 2009, 56, 99–105. [Google Scholar] [CrossRef]
- Huang, C.C.; Wen, H.J.; Chen, P.C.; Chiang, T.L.; Lin, S.J.; Guo, Y.L. Prenatal air pollutant exposure and occurrence of atopic dermatitis. Br. J. Dermatol. 2015, 173, 981–988. [Google Scholar] [CrossRef]
- Southam, A.D.; Lange, A.; Al-Salhi, R.; Hill, E.M.; Tyler, C.R.; Viant, M.R. Distinguishing between the metabolome and xenobiotic exposome in environmental field samples analysed by direct-infusion mass spectrometry based metabolomics and lipidomics. Metabolomics 2014, 10, 1050–1058. [Google Scholar] [CrossRef] [Green Version]
- Patel, C.J.; Bhattacharya, J.; Butte, A.J. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE 2010, 5, e10746. [Google Scholar] [CrossRef]
- Tzoulaki, I.; Patel, C.J.; Okamura, T.; Chan, Q.; Brown, I.J.; Miura, K.; Ueshima, H.; Zhao, L.; Van Horn, L.; Daviglus, M.L.; et al. A nutrient-wide association study on blood pressure. Circulation 2012, 126, 2456–2464. [Google Scholar] [CrossRef]
- Hall, M.A.; Dudek, S.M.; Goodloe, R.; Crawford, D.C.; Pendergrass, S.A.; Peissig, P.; Brilliant, M.; McCarty, C.A.; Ritchie, M.D. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. Pac. Symp. Biocomput. 2014, 200–211. [Google Scholar]
- Patel, C.J.; Rehkopf, D.H.; Leppert, J.T.; Bortz, W.M.; Cullen, M.R.; Chertow, G.M.; Ioannidis, J.P. Systematic evaluation of environmental and behavioural factors associated with all-cause mortality in the United States national health and nutrition examination survey. Int. J. Epidemiol. 2013, 42, 1795–1810. [Google Scholar] [CrossRef]
- Patel, C.J.; Manrai, A.K.; Corona, E.; Kohane, I.S. Systematic correlation of environmental exposure and physiological and self-reported behaviour factors with leukocyte telomere length. Int. J. Epidemiol. 2017, 46, 44–56. [Google Scholar] [CrossRef]
- Juarez, P.D.; Hood, D.B.; Rogers, G.L.; Baktash, S.H.; Saxton, A.M.; Matthews-Juarez, P.; Im, W.; Cifuentes, M.P.; Phillips, C.A.; Lichtveld, M.Y.; et al. A novel approach to analyzing lung cancer mortality disparities: Using the exposome and a graph-theoretical toolchain. Environ. Dis. 2017, 2, 33–44. [Google Scholar]
- Agier, L.; Portengen, L.; Chadeau-Hyam, M.; Basagana, X.; Giorgis-Allemand, L.; Siroux, V.; Robinson, O.; Vlaanderen, J.; Gonzalez, J.R.; Nieuwenhuijsen, M.J.; et al. A Systematic Comparison of Linear Regression-Based Statistical Methods to Assess Exposome-Health Associations. Environ. Health Perspect. 2016, 124, 1848–1856. [Google Scholar] [CrossRef]
- Barrera-Gomez, J.; Agier, L.; Portengen, L.; Chadeau-Hyam, M.; Giorgis-Allemand, L.; Siroux, V.; Robinson, O.; Vlaanderen, J.; Gonzalez, J.R.; Nieuwenhuijsen, M.; et al. A systematic comparison of statistical methods to detect interactions in exposome-health associations. Environ. Health 2017, 16, 74. [Google Scholar] [CrossRef]
- Patel, C.J.; Ioannidis, J.P. Placing epidemiological results in the context of multiplicity and typical correlations of exposures. J. Epidemiol. Community Health 2014, 68, 1096–1100. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Bessonneau, V.; Pawliszyn, J.; Rappaport, S.M. The Saliva Exposome for Monitoring of Individuals’ Health Trajectories. Environ. Health Perspect. 2017, 125, 077014. [Google Scholar] [CrossRef]
- Slama, R.; Vrijheid, M. Some challenges of studies aiming to relate the Exposome to human health. Occup. Environ. Med. 2015, 72, 383–384. [Google Scholar] [CrossRef]
- Vineis, P.; Chadeau-Hyam, M.; Gmuender, H.; Gulliver, J.; Herceg, Z.; Kleinjans, J.; Kogevinas, M.; Kyrtopoulos, S.; Nieuwenhuijsen, M.; Phillips, D.H.; et al. The exposome in practice: Design of the EXPOsOMICS project. Int. J. Hyg. Environ. Health 2017, 220, 142–151. [Google Scholar] [CrossRef]
- Vrijheid, M.; Slama, R.; Robinson, O.; Chatzi, L.; Coen, M.; van den Hazel, P.; Thomsen, C.; Wright, J.; Athersuch, T.J.; Avellana, N.; et al. The human early-life exposome (HELIX): Project rationale and design. Environ. Health Perspect. 2014, 122, 535–544. [Google Scholar] [CrossRef]
- Pragst, F.; Broecker, S.; Hastedt, M.; Herre, S.; Andresen-Streichert, H.; Sachs, H.; Tsokos, M. Methadone and illegal drugs in hair from children with parents in maintenance treatment or suspected for drug abuse in a German community. Ther. Drug Monit. 2013, 35, 737–752. [Google Scholar] [CrossRef]
- Andra, S.S.; Austin, C.; Wright, R.O.; Arora, M. Reconstructing pre-natal and early childhood exposure to multi-class organic chemicals using teeth: Towards a retrospective temporal exposome. Environ. Int. 2015, 83, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Andra, S.S.; Austin, C.; Arora, M. The tooth exposome in children’s health research. Curr. Opin. Pediatr. 2016, 28, 221–227. [Google Scholar] [CrossRef]
- Liu, K.H.; Walker, D.I.; Uppal, K.; Tran, V.; Rohrbeck, P.; Mallon, T.M.; Jones, D.P. High-Resolution Metabolomics Assessment of Military Personnel: Evaluating Analytical Strategies for Chemical Detection. J. Occup. Environ. Med. 2016, 58, S53–S61. [Google Scholar] [CrossRef]
- Jia, S.; Xu, T.; Huan, T.; Chong, M.; Liu, M.; Fang, W.; Fang, M. Chemical Isotope Labeling Exposome (CIL-EXPOSOME): One High-Throughput Platform for Human Urinary Global Exposome Characterization. Environ. Sci. Technol. 2019, 53, 5445–5453. [Google Scholar] [CrossRef]
- Walker, D.I.; Mallon, C.T.; Hopke, P.K.; Uppal, K.; Go, Y.M.; Rohrbeck, P.; Pennell, K.D.; Jones, D.P. Deployment-Associated Exposure Surveillance With High-Resolution Metabolomics. J. Occup. Environ. Med. 2016, 58, S12–S21. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.R.; Lange, B.M. Open-access metabolomics databases for natural product research: Present capabilities and future potential. Front. Bioeng. Biotechnol. 2015, 3, 22. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, J.; Lai, Y.; Liu, C.-W.; Ru, H. Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions. Toxics 2019, 7, 41. https://doi.org/10.3390/toxics7030041
Xue J, Lai Y, Liu C-W, Ru H. Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions. Toxics. 2019; 7(3):41. https://doi.org/10.3390/toxics7030041
Chicago/Turabian StyleXue, Jingchuan, Yunjia Lai, Chih-Wei Liu, and Hongyu Ru. 2019. "Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions" Toxics 7, no. 3: 41. https://doi.org/10.3390/toxics7030041
APA StyleXue, J., Lai, Y., Liu, C.-W., & Ru, H. (2019). Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions. Toxics, 7(3), 41. https://doi.org/10.3390/toxics7030041




