AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images
Abstract
:1. Introduction
- We propose a novel kidney and renal mass diagnosis framework integrating 3D segmentation and renal mass subtype classification. It provides an easy-to-analyze 3D morphologic representation of the kidney and renal mass with the subtypes. The segmentation method adopts the basic 3D U-Net structure with residual blocks included for gaining the cross-layer connections. The postprocessing steps further improved the accuracy and reduced false positives by small region detection. The classification network applies the dual-path schema to combine the different fields of view for the prediction of subtypes.
- We propose a weakly supervised method to improve the robustness of the trained model on the datasets collected by various vendors with only a few slice-level annotations.
- The experimental results on the KiTs19 dataset demonstrate the state-of-the-art performance on kidney and renal mass segmentation and classification. Additionally, the results on three NIH datasets (the TCGA-KICH [15], TCGA-KIRP [16], and TCGA-KIRC [17] datasets) show that the proposed framework can be robust in different institutions with few annotations.
2. Related Work
2.1. 3D Semantic Segmentation
2.2. Weakly Supervised Learning
2.3. Kidney and Renal Mass Segmentation and Classification
3. Our Method
3.1. Kidney and Renal Mass Segmentation
3.2. Renal Mass Classification
3.3. Weakly Supervised Learning
4. Experimental Results and Discussions
4.1. Datasets
4.2. Experimental Settings
4.2.1. Pre-Processing
4.2.2. 3D Semantic Segmentation
4.2.3. Renal Mass Classification
4.3. Evaluation Metrics
4.4. Experimental Results and Analysis
4.4.1. Comparison with the State-of-the-Art Methods
4.4.2. Model Robustness
4.4.3. Renal Mass Subtype Classification
4.5. Ablation Study
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Motzer, R.J.; Bander, N.H.; Nanus, D.M. Renal-cell carcinoma. N. Engl. J. Med. 1996, 335, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 1–19. [Google Scholar] [CrossRef]
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of renal cell carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.; Sanford, T.; Parwani, A.V.; Pedrosa, I. Artificial Intelligence in Kidney Cancer. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, L.; Akin, O.; Tian, Y. 3DFPN-HS2: 3D Feature Pyramid Network Based High Sensitivity and Specificity Pulmonary Nodule Detection. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 513–521. [Google Scholar]
- Xi, I.L.; Zhao, Y.; Wang, R.; Chang, M.; Purkayastha, S.; Chang, K.; Huang, R.Y.; Silva, A.C.; Vallières, M.; Habibollahi, P.; et al. Deep Learning to Distinguish Benign from Malignant Renal Lesions Based on Routine MR ImagingDeep Learning for Characterization of Renal Lesions. Clin. Cancer Res. 2020, 26, 1944–1952. [Google Scholar] [CrossRef]
- Said, D.; Hectors, S.J.; Wilck, E.; Rosen, A.; Stocker, D.; Bane, O.; Beksaç, A.T.; Lewis, S.; Badani, K.; Taouli, B. Characterization of solid renal neoplasms using MRI-based quantitative radiomics features. Abdom. Radiol. 2020, 45, 2840–2850. [Google Scholar] [CrossRef]
- Nassiri, N.; Maas, M.; Cacciamani, G.; Varghese, B.; Hwang, D.; Lei, X.; Aron, M.; Desai, M.; Oberai, A.A.; Cen, S.Y.; et al. A radiomic-based machine learning algorithm to reliably differentiate benign renal masses from renal cell carcinoma. Eur. Urol. Focus 2022, 8, 988–994. [Google Scholar] [CrossRef]
- Liu, H.; Cao, H.; Chen, L.; Fang, L.; Liu, Y.; Zhan, J.; Diao, X.; Chen, Y. The quantitative evaluation of contrast-enhanced ultrasound in the differentiation of small renal cell carcinoma subtypes and angiomyolipoma. Quant. Imaging Med. Surg. 2022, 12, 106. [Google Scholar] [CrossRef]
- Yang, G.; Wang, C.; Yang, J.; Chen, Y.; Tang, L.; Shao, P.; Dillenseger, J.L.; Shu, H.; Luo, L. Weakly-supervised convolutional neural networks of renal tumor segmentation in abdominal CTA images. BMC Med. Imaging 2020, 20, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.; Kim, B.; Ham, B. Background-aware pooling and noise-aware loss for weakly-supervised semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6913–6922. [Google Scholar]
- Li, Y.; Kuang, Z.; Liu, L.; Chen, Y.; Zhang, W. Pseudo-mask matters in weakly-supervised semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 6964–6973. [Google Scholar]
- Bearman, A.; Russakovsky, O.; Ferrari, V.; Fei-Fei, L. What’s the point: Semantic segmentation with point supervision. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 549–565. [Google Scholar]
- Wei, Y.; Feng, J.; Liang, X.; Cheng, M.M.; Zhao, Y.; Yan, S. Object region mining with adversarial erasing: A simple classification to semantic segmentation approach. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1568–1576. [Google Scholar]
- Linehan, M.; Gautam, R.; Sadow, C.; Levine, S. Radiology data from the cancer genome atlas kidney chromophobe [TCGA-KICH] collection. Cancer Imaging Arch. 2016. [Google Scholar] [CrossRef]
- Linehan, M.; Gautam, R.; Kirk, S.; Lee, Y.; Roche, C.; Bonaccio, E.; Jarosz, R. Radiology data from the cancer genome atlas cervical kidney renal papillary cell carcinoma [KIRP] collection. Cancer Imaging Arch. 2016, 10, K9. [Google Scholar]
- Akin, O.; Elnajjar, P.; Heller, M.; Jarosz, R.; Erickson, B.; Kirk, S.; Filippini, J. Radiology data from the cancer genome atlas kidney renal clear cell carcinoma [TCGA-KIRC] collection. Cancer Imaging Arch. 2016. [Google Scholar] [CrossRef]
- He, T.; Guo, J.; Wang, J.; Xu, X.; Yi, Z. Multi-task Learning for the Segmentation of Thoracic Organs at Risk in CT images. In Proceedings of the SegTHOR@ ISBI, Venice, Italy, 8 April 2019; pp. 10–13. [Google Scholar]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. Automated Machine Learning: Methods, Systems, Challenges; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Jégou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The one hundred layers tiramisu: Fully convolutional densenets for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 11–19. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Zhang, M.; Zhou, Y.; Zhao, J.; Man, Y.; Liu, B.; Yao, R. A survey of semi-and weakly supervised semantic segmentation of images. Artif. Intell. Rev. 2020, 53, 4259–4288. [Google Scholar] [CrossRef]
- Dai, J.; He, K.; Sun, J. Boxsup: Exploiting bounding boxes to supervise convolutional networks for semantic segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1635–1643. [Google Scholar]
- Lin, D.; Dai, J.; Jia, J.; He, K.; Sun, J. Scribblesup: Scribble-supervised convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3159–3167. [Google Scholar]
- Papandreou, G.; Chen, L.C.; Murphy, K.P.; Yuille, A.L. Weakly-and semi-supervised learning of a deep convolutional network for semantic image segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1742–1750. [Google Scholar]
- Wei, Y.; Liang, X.; Chen, Y.; Shen, X.; Cheng, M.M.; Feng, J.; Zhao, Y.; Yan, S. Stc: A simple to complex framework for weakly-supervised semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 2314–2320. [Google Scholar] [CrossRef] [Green Version]
- Cholakkal, H.; Sun, G.; Khan, F.S.; Shao, L. Object counting and instance segmentation with image-level supervision. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 12397–12405. [Google Scholar]
- Sadeghi, M.H.; Zadeh, H.M.; Behroozi, H.; Royat, A. Accurate Kidney Tumor Segmentation Using Weakly-Supervised Kidney Volume Segmentation in CT images. In Proceedings of the 2021 28th National and 6th International Iranian Conference on Biomedical Engineering (ICBME), Tehran, Iran, 25–26 November 2021; pp. 38–43. [Google Scholar]
- Tanaka, T.; Huang, Y.; Marukawa, Y.; Tsuboi, Y.; Masaoka, Y.; Kojima, K.; Iguchi, T.; Hiraki, T.; Gobara, H.; Yanai, H.; et al. Differentiation of small (≤4 cm) renal masses on multiphase contrast-enhanced CT by deep learning. Am. J. Roentgenol. 2020, 214, 605–612. [Google Scholar] [CrossRef]
- Oberai, A.; Varghese, B.; Cen, S.; Angelini, T.; Hwang, D.; Gill, I.; Aron, M.; Lau, C.; Duddalwar, V. Deep learning based classification of solid lipid-poor contrast enhancing renal masses using contrast enhanced CT. Br. J. Radiol. 2020, 93, 20200002. [Google Scholar] [CrossRef]
- Zabihollahy, F.; Schieda, N.; Krishna, S.; Ukwatta, E. Automated classification of solid renal masses on contrast-enhanced computed tomography images using convolutional neural network with decision fusion. Eur. Radiol. 2020, 30, 5183–5190. [Google Scholar] [CrossRef]
- George, Y. A Coarse-to-Fine 3D U-Net Network for Semantic Segmentation of Kidney CT Scans. In Proceedings of the International Challenge on Kidney and Kidney Tumor Segmentation, Strasbourg, France, 27 September 2021; pp. 137–142. [Google Scholar]
- Li, D.; Xiao, C.; Liu, Y.; Chen, Z.; Hassan, H.; Su, L.; Liu, J.; Li, H.; Xie, W.; Zhong, W.; et al. Deep Segmentation Networks for Segmenting Kidneys and Detecting Kidney Stones in Unenhanced Abdominal CT Images. Diagnostics 2022, 12, 1788. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Li, G.; Pan, T.; Kong, Y.; Wu, J.; Shu, H.; Luo, L.; Dillenseger, J.L.; Coatrieux, J.L.; Tang, L.; et al. Automatic segmentation of kidney and renal tumor in ct images based on 3d fully convolutional neural network with pyramid pooling module. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3790–3795. [Google Scholar]
- Yu, Q.; Shi, Y.; Sun, J.; Gao, Y.; Zhu, J.; Dai, Y. Crossbar-net: A novel convolutional neural network for kidney tumor segmentation in CT images. IEEE Trans. Image Process. 2019, 28, 4060–4074. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Liu, C.; Bardis, M.; Martin, J.; Liu, H.; Ushinsky, A.; Glavis-Bloom, J.; Chantaduly, C.; Chow, D.S.; Houshyar, R.; et al. Deep learning segmentation of kidneys with renal cell carcinoma. J. Clin. Oncol. 2019, 37, e16098. [Google Scholar] [CrossRef]
- Xia, K.J.; Yin, H.S.; Zhang, Y.D. Deep semantic segmentation of kidney and space-occupying lesion area based on SCNN and ResNet models combined with SIFT-flow algorithm. J. Med. Syst. 2019, 43, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Li, D.; Marshall, H.; Miao, T.; Cossetto, T.; Chan, I.; Daher, O.; Accorsi, F.; Goela, A.; Li, S. MB-FSGAN: Joint segmentation and quantification of kidney tumor on CT by the multi-branch feature sharing generative adversarial network. Med. Image Anal. 2020, 64, 101721. [Google Scholar] [CrossRef]
- Han, S.; Hwang, S.I.; Lee, H.J. The classification of renal cancer in 3-phase CT images using a deep learning method. J. Digit. Imaging 2019, 32, 638–643. [Google Scholar] [CrossRef] [Green Version]
- Uhm, K.H.; Jung, S.W.; Choi, M.H.; Shin, H.K.; Yoo, J.I.; Oh, S.W.; Kim, J.Y.; Kim, H.G.; Lee, Y.J.; Youn, S.Y.; et al. Deep learning for end-to-end kidney cancer diagnosis on multi-phase abdominal computed tomography. NPJ Precis. Oncol. 2021, 5, 1–6. [Google Scholar] [CrossRef]
- Chen, Z.; Tian, Z.; Zhu, J.; Li, C.; Du, S. C-CAM: Causal CAM for Weakly Supervised Semantic Segmentation on Medical Image. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11676–11685. [Google Scholar]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The kits19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. arXiv 2019, arXiv:1904.00445. [Google Scholar]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.W.; Han, X.; Heng, P.A.; Hesser, J.; et al. The liver tumor segmentation benchmark (lits). arXiv 2019, arXiv:1901.04056. [Google Scholar] [CrossRef]

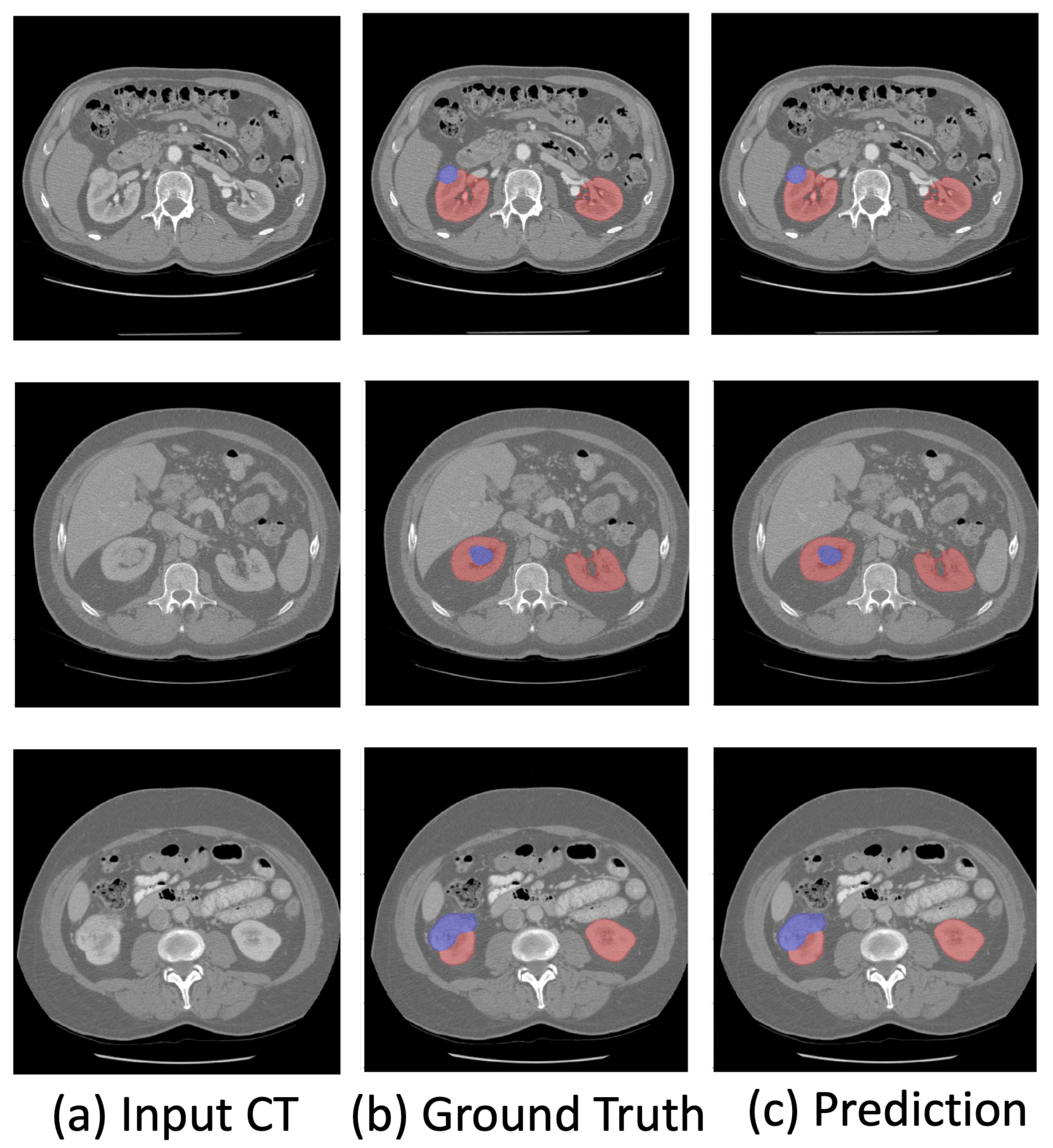
| Methods | Pixel Accuracy | Dice Coefficient | Specificity | Sensitivity |
|---|---|---|---|---|
| Xia et al. [45] | 92.1% | 79.6% | 86.7% | 83.4% |
| Yin et al. [44] | 89.4% | 83.8% | 81.1% | 85.4% |
| Yu et al. [43] | 87.7% | 80.4% | 83.4% | 82.2% |
| Ruan et al. [46] | 95.7% | 85.9% | 89.4% | 86.2% |
| Ours | 95.9% | 86.6% | 91.2% | 88.1% |
| Subtype | AUC | Specificity | Sensitivity |
|---|---|---|---|
| Clear Cell | 87.1% | 84.3 % | 84.1% |
| Papillary | 86.2% | 83.7% | 82.4 % |
| Chromophobe | 85.9% | 82.1% | 83.7 % |
| Oncocytoma | 79.9% | 77.7% | 76.9 % |
| Others | 85.8% | 81.6% | 80.4 % |
| 3D U-Net | ResNet | Preprocessing | Post-Processing | Pixel Accuracy | Dice Coefficient | Specificity | Sensitivity |
|---|---|---|---|---|---|---|---|
| √ | - | - | - | 86.2% | 80.9% | 84.1% | 78.8% |
| √ | √ | - | - | 89.1% | 82.4% | 86.8% | 83.2% |
| √ | √ | √ | - | 91.7% | 83.1% | 89.2% | 86.4% |
| √ | - | √ | √ | 91.4% | 82.7% | 87.3% | 83.7% |
| √ | √ | √ | √ | 95.9% | 86.6% | 91.2% | 88.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Yildirim, O.; Akin, O.; Tian, Y. AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images. Bioengineering 2023, 10, 116. https://doi.org/10.3390/bioengineering10010116
Liu J, Yildirim O, Akin O, Tian Y. AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images. Bioengineering. 2023; 10(1):116. https://doi.org/10.3390/bioengineering10010116
Chicago/Turabian StyleLiu, Jingya, Onur Yildirim, Oguz Akin, and Yingli Tian. 2023. "AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images" Bioengineering 10, no. 1: 116. https://doi.org/10.3390/bioengineering10010116
APA StyleLiu, J., Yildirim, O., Akin, O., & Tian, Y. (2023). AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images. Bioengineering, 10(1), 116. https://doi.org/10.3390/bioengineering10010116










