Fibroblasts Impair Migration and Antitumor Activity of NK-92 Lymphocytes in a Melanoma-on-Chip Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Microfluidic Device Features
2.2. Cell Lines
2.3. Three-Dimensional Spheroid Experimental Setup
2.4. Viability Assay and Confocal Microscopy
3. Results
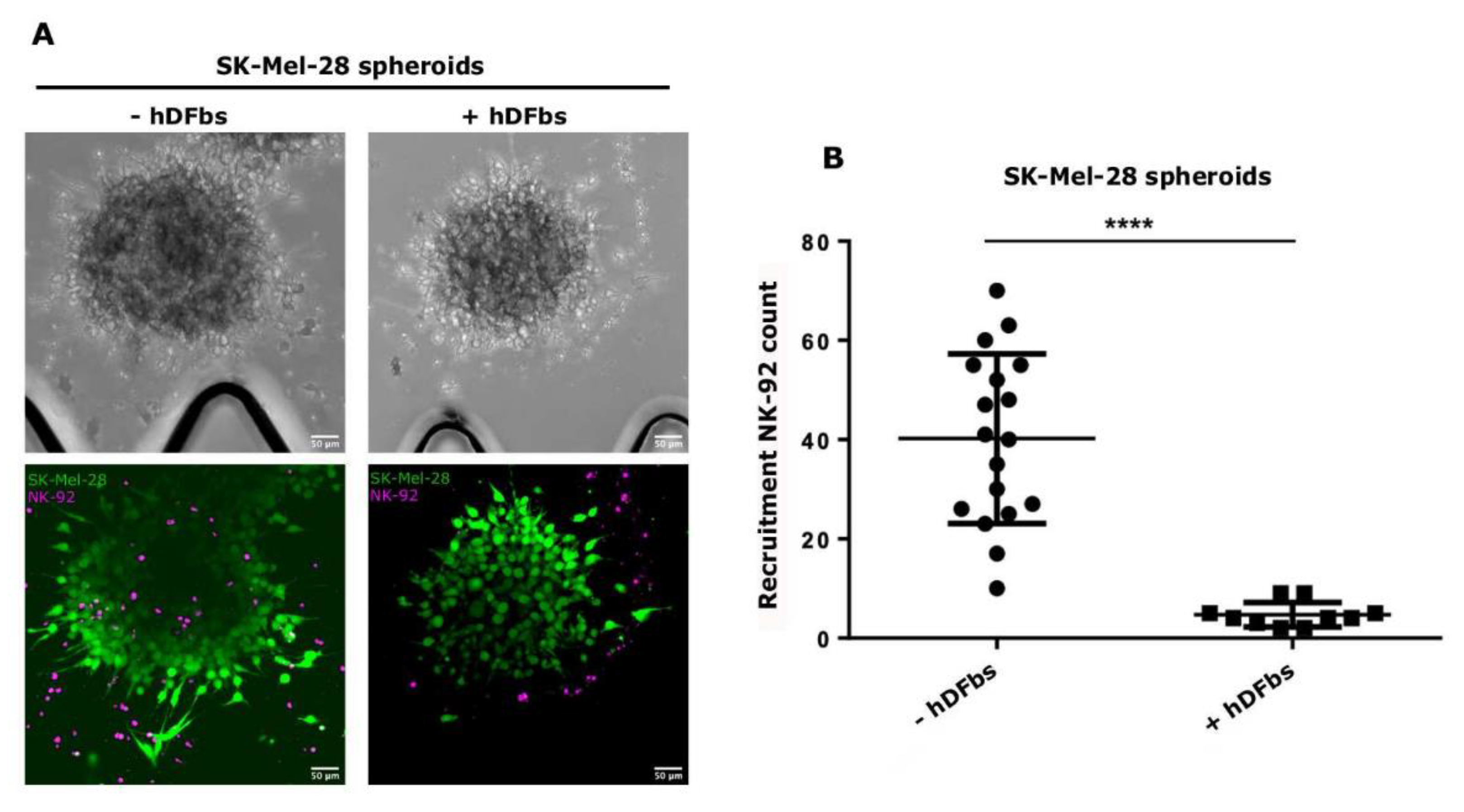
3.1. Recruitment Inhibition of NK-92 by Fibroblasts
3.2. Inhibition of NK-92 Anti-Melanoma Cytotoxicity by Fibroblasts
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aranda, F.; Buqué, A.; Bloy, N.; Castoldi, F.; Eggermont, A.; Cremer, I.; Fridman, W.H.; Fucikova, J.; Galon, J.; Spisek, R.; et al. Trial Watch: Adoptive cell transfer for oncological indications. Oncoimmunology 2015, 4, e1046673. [Google Scholar] [CrossRef]
- Fournier, C.; Martin, F.; Zitvogel, L.; Kroemer, G.; Galluzzi, L.; Apetoh, L. Trial Watch: Adoptively transferred cells for anticancer immunotherapy. Oncoimmunology 2017, 6, e1363139. [Google Scholar] [CrossRef]
- Rohaan, M.W.; Wilgenhof, S.; Haanen, J.B.A.G. Adoptive cellular therapies: The current landscape. Virchows Arch. 2019, 474, 449–461. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Restifo, N.P.; Yang, J.C.; Morgan, R.A.; Dudley, M.E. Adoptive cell transfer: A clinical path to effective cancer immunotherapy. Nat. Rev. Cancer 2008, 8, 299–308. [Google Scholar] [CrossRef]
- Holstein, S.A.; Lunning, M.A. CAR T-Cell Therapy in Hematologic Malignancies: A Voyage in Progress. Clin. Pharmacol. Ther. 2020, 107, 112–122. [Google Scholar] [CrossRef]
- Miliotou, A.N.; Papadopoulou, L.C. CAR T-cell Therapy: A New Era in Cancer Immunotherapy. Curr. Pharm. Biotechnol. 2018, 19, 5–18. [Google Scholar] [CrossRef]
- Marofi, F.; Motavalli, R.; Safonov, V.A.; Thangavelu, L.; Yumashev, A.V.; Alexander, M.; Shomali, N.; Chartrand, M.S.; Pathak, Y.; Jarahian, M.; et al. CAR T cells in solid tumors: Challenges and opportunities. Stem Cell Res. Ther. 2021, 12, 81. [Google Scholar] [CrossRef]
- Gammaitoni, L.; Giraudo, L.; Leuci, V.; Todorovic, M.; Mesiano, G.; Picciotto, F.; Pisacane, A.; Zaccagna, A.; Volpe, M.G.; Gallo, S.; et al. Effective activity of cytokine-induced killer cells against autologous metastatic melanoma including cells with stemness features. Clin. Cancer Res. 2013, 19, 4347–4358. [Google Scholar] [CrossRef]
- Ando, Y.; Siegler, E.; Ta, H.P.; Cinay, G.E.; Zhou, H.; Gorrell, K.A.; Au, H.; Jarvis, B.M.; Wang, P.; Shen, K. Evaluating CAR-T Cell Therapy in a Hypoxic 3-D Tumor Model. Adv. Heal. Mater. 2019, 8, e1900001. [Google Scholar] [CrossRef]
- Becker, J.C.; Andersen, M.H.; Schrama, D.; Thor Straten, P. Immune-suppressive properties of the tumor microenvironment. Cancer Immunol. Immunother. 2013, 62, 1137–1148. [Google Scholar] [CrossRef]
- Franchi-Mendes, T.; Eduardo, R.; Domenici, G.; Brito, C. 3D Cancer Models: Depicting Cellular Crosstalk within the Tumour Microenvironment. Cancers 2021, 13, 4610. [Google Scholar] [CrossRef]
- Iaia, I.; Gammaitoni, L.; Cattaneo, G.; Giraudo, L.; Donini, C.; Fiorino, E.; Primo, L.; Carnevale-Schianca, F.; Aglietta, M.; Puliafito, A.; et al. Recruitment, Infiltration, and Cytotoxicity of HLA-Independent Killer Lymphocytes in Three-Dimensional Melanoma Models. Cancers 2021, 13, 2302. [Google Scholar] [CrossRef]
- Pietras, K.; Östman, A. Hallmarks of cancer: Interactions with the tumor stroma. Exp. Cell Res. 2010, 316, 1324–1331. [Google Scholar] [CrossRef]
- Ziani, L.; Chouaib, S.; Thiery, J. Alteration of the Antitumor Immune Response by Cancer-Associated Fibroblasts. Front. Immunol. 2018, 9, 414. [Google Scholar] [CrossRef]
- An, Y.; Liu, F.; Chen, Y.; Yang, Q. Crosstalk between cancer-associated fibroblasts and immune cells in cancer. J. Cell. Mol. Med. 2020, 24, 13–24. [Google Scholar] [CrossRef]
- Gieniec, K.A.; Butler, L.M.; Worthley, D.L.; Woods, S.L. Cancer-associated fibroblasts—Heroes or villains? Br. J. Cancer 2019, 121, 293–302. [Google Scholar] [CrossRef]
- Salih, H.R.; Rammensee, H.-G.; Steinle, A. Cutting Edge: Down-Regulation of MICA on Human Tumors by Proteolytic Shedding. J. Immunol. 2002, 169, 4098–4102. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L. The emerging world of TCR-T cell trials against cancer: A systematic review. Technol. Cancer Res. Treat. 2019, 18, 1533033819831068. [Google Scholar] [CrossRef]
- Brancato, V.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Could 3D models of cancer enhance drug screening? Biomaterials 2020, 232, 119744. [Google Scholar] [CrossRef]
- Di Modugno, F.; Colosi, C.; Trono, P.; Antonacci, G.; Ruocco, G.; Nisticò, P. 3D models in the new era of immune oncology: Focus on T cells, CAF and ECM. J. Exp. Clin. Cancer Res. 2019, 38, 117. [Google Scholar] [CrossRef]
- Liu, X.; Fang, J.; Huang, S.; Wu, X.; Xie, X.; Wang, J.; Liu, F.; Zhang, M.; Peng, Z.; Hu, N. Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 2021, 7, 50. [Google Scholar] [CrossRef]
- Poggi, A.; Villa, F.; Fernadez, J.L.C.; Costa, D.; Zocchi, M.R.; Benelli, R. Three-Dimensional Culture Models to Study Innate Anti-Tumor Immune Response: Advantages and Disadvantages. Cancers 2021, 13, 3417. [Google Scholar] [CrossRef]
- Adriani, G.; Pavesi, A.; Kamm, R.D. Studying TCR T cell anti-tumor activity in a microfluidic intrahepatic tumor model. Methods Cell Biol 2018, 146, 199–214. [Google Scholar]
- Marasso, S.L.; Puliafito, A.; Mombello, D.; Benetto, S.; Primo, L.; Bussolino, F.; Pirri, C.F.; Cocuzza, M. Optimized design and fabrication of a microfluidic platform to study single cells and multicellular aggregates in 3D. Microfluid. Nanofluidics 2017, 21, 29. [Google Scholar] [CrossRef]
- Parlato, S.; De Ninno, A.; Molfetta, R.; Toschi, E.; Salerno, D.; Mencattini, A.; Romagnoli, G.; Fragale, A.; Roccazzello, L.; Buoncervello, M.; et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep. 2017, 7, 1093. [Google Scholar] [CrossRef]
- Maio, M. Melanoma as a model tumour for immuno-oncology. Ann. Oncol. 2012, 23, viii10–viii14. [Google Scholar] [CrossRef]
- Merhavi-Shoham, E.; Itzhaki, O.; Markel, G.; Schachter, J.; Besser, M.J. Adoptive Cell Therapy for Metastatic Melanoma. Cancer J. 2017, 23, 48–53. [Google Scholar] [CrossRef]
- Fabian, K.P.; Hodge, J.W. The emerging role of off-the-shelf engineered natural killer cells in targeted cancer immunotherapy. Mol. Ther. Oncolytics 2021, 23, 266–276. [Google Scholar] [CrossRef]
- Klingemann, H.; Boissel, L.; Toneguzzo, F. Natural killer cells for immunotherapy—Advantages of the NK-92 cell line over blood NK cells. Front. Immunol. 2016, 7, 91. [Google Scholar] [CrossRef]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011, 51, e2720. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iaia, I.; Brancato, V.; Caballero, D.; Reis, R.L.; Aglietta, M.; Sangiolo, D.; Kundu, S.C. Fibroblasts Impair Migration and Antitumor Activity of NK-92 Lymphocytes in a Melanoma-on-Chip Model. Bioengineering 2023, 10, 52. https://doi.org/10.3390/bioengineering10010052
Iaia I, Brancato V, Caballero D, Reis RL, Aglietta M, Sangiolo D, Kundu SC. Fibroblasts Impair Migration and Antitumor Activity of NK-92 Lymphocytes in a Melanoma-on-Chip Model. Bioengineering. 2023; 10(1):52. https://doi.org/10.3390/bioengineering10010052
Chicago/Turabian StyleIaia, Ilenia, Virginia Brancato, David Caballero, Rui L. Reis, Massimo Aglietta, Dario Sangiolo, and Subhas C. Kundu. 2023. "Fibroblasts Impair Migration and Antitumor Activity of NK-92 Lymphocytes in a Melanoma-on-Chip Model" Bioengineering 10, no. 1: 52. https://doi.org/10.3390/bioengineering10010052
APA StyleIaia, I., Brancato, V., Caballero, D., Reis, R. L., Aglietta, M., Sangiolo, D., & Kundu, S. C. (2023). Fibroblasts Impair Migration and Antitumor Activity of NK-92 Lymphocytes in a Melanoma-on-Chip Model. Bioengineering, 10(1), 52. https://doi.org/10.3390/bioengineering10010052










