Does Constitutive Expression of Defense-Related Genes and Salicylic Acid Concentrations Correlate with Field Resistance of Potato to Black Scurf Disease?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiment
2.2. Preparing Plantlets for Field Experiment
2.3. In Vitro and Sand Culture Experiments
2.4. Expression Analysis of Defense-Related Genes in Potato Tissue
2.5. Determination of Salicylic Acid
2.6. Statistical Analyses
3. Results
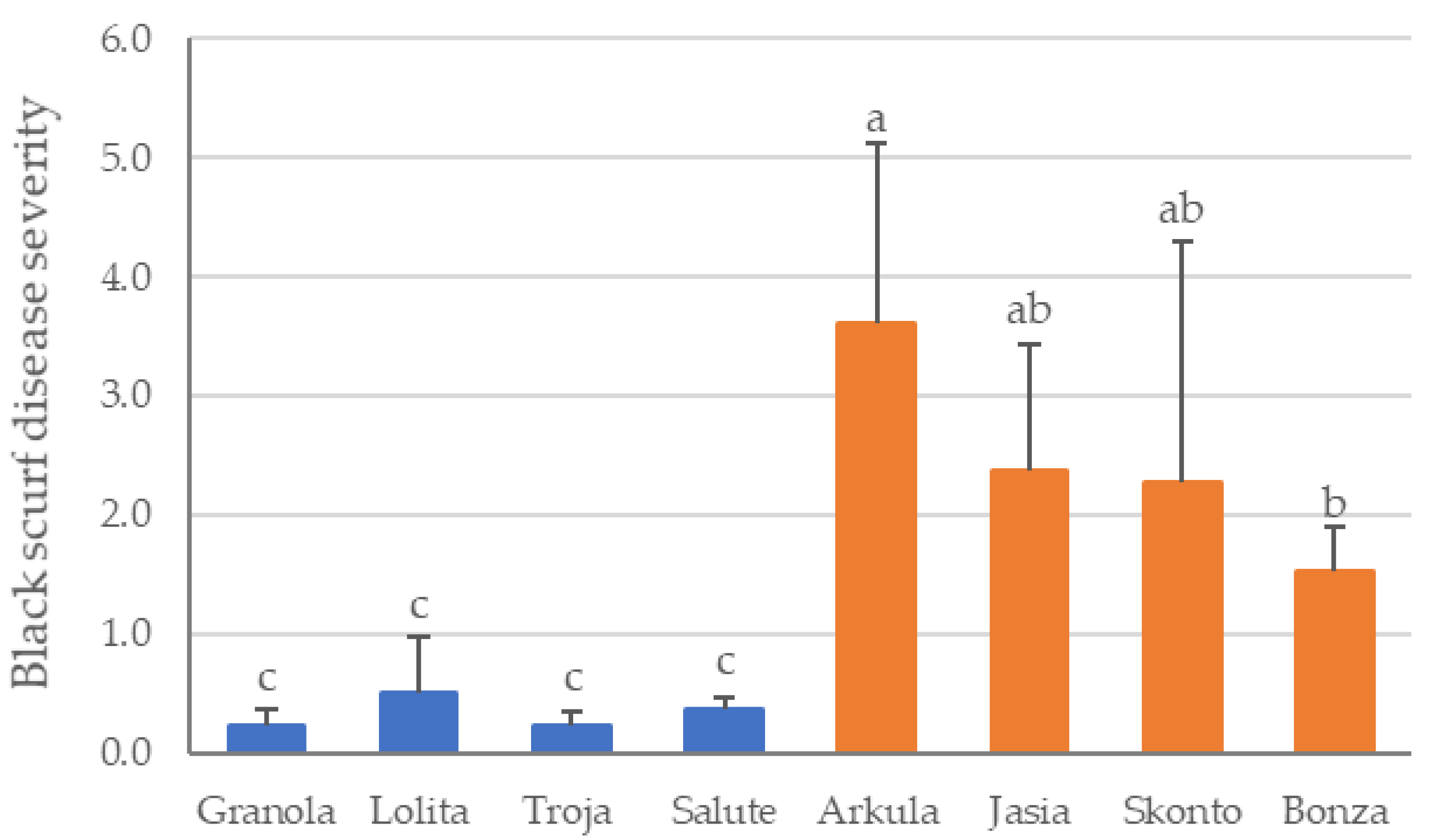
3.1. Field Experiment to Evaluate Disease Severity (DS) of Black Scurf on Potato Cultivars
3.2. Expression of Defense-Related Genes in Potato Cultivars
3.2.1. In Vitro Culture
3.2.2. Sand Culture
3.3. Salicylic Acid Concentration in Potato Cultivars
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, P.S.; Ketola, E.O.; Ahvenniemi, P.M.; Lehtonen, M.J.; Valkonen, J.P.T. Dynamics of soilborne Rhizoctonia solani in the presence of Trichoderma harzianum: Effects on stem canker, black scurf and progeny tubers of potato. Plant Pathol. 2008, 57, 152–161. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Yu, X.-X.; Yu, Z.; Xue, Y.-F.; Qi, L.-P. A simple method based on laboratory inoculum and field inoculum for evaluating potato resistance to black scurf caused by Rhizoctonia solani. Breed. Sci. 2014, 64, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Hide, G.A.; Horrocks, J.K. Influence of stem canker (Rhizoctonia solani Kühn) on tuber yield, tuber size, reducing sugars and crisp colour in cv. Record. Potato Res. 1994, 37, 43–49. [Google Scholar] [CrossRef]
- Keiser, A.; Häberli, M.; Stamp, P. Quality deficiencies on potato (Solanum tuberosum L.) tubers caused by Rhizoctonia solani, wireworms (Agriotes ssp.) and slugs (Deroceras reticulatum, Arion hortensis) in different farming systems. Field Crops Res. 2012, 128, 147–155. [Google Scholar] [CrossRef]
- Banville, G.J.; Carling, D.E.; Otrysko, B.E. Rhizoctonia Disease on Potato. In Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Sneh, B., Jabaji-Hare, S., Neate, S., Dijst, G., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 321–330. [Google Scholar] [CrossRef]
- Carling, D.E.; Leiner, R.H.; Westphale, P.C. Symptoms, signs and yield reduction associated with Rhizoctonia disease of potato induced by tuberborne inoculum of Rhizoctonia solani AG-3. Am. Potato J. 1989, 66, 693–701. [Google Scholar] [CrossRef]
- Bains, P.; Bennypaul, H.; Lynch, D.; Kawchuk, L.; Schaupmeyer, C.A. Rhizoctonia disease of potatoes (Rhizoctonia solani): Fungicidal efficacy and cultivar susceptibility. Am. J. Potato Res. 2002, 79, 99–106. [Google Scholar] [CrossRef]
- Lehtonen, M.J.; Wilson, P.S.; Ahvenniemi, P. Formation of canker lesions on stems and black scurf on tubers in experimentally inoculated potato plants by isolates of AG2-1, AG3 and AG5 of Rhizoctonia solani: A pilot study and literature review. Agric. Food Sci. 2009, 18, 223–233. [Google Scholar] [CrossRef]
- Woodhall, J.W.; Lees, A.K.; Edwards, S.G.; Jenkinson, P. Characterization of Rhizoctonia solani from potato in Great Britain. Plant Pathol. 2007, 56, 286–295. [Google Scholar] [CrossRef]
- Campion, C.; Chatot, C.; Perraton, B.; Andrivon, D. Anastomosis Groups, Pathogenicity and Sensitivity to Fungicides of Rhizoctonia solani Isolates Collected on Potato Crops in France. Eur. J. Plant Pathol. 2003, 109, 983–992. [Google Scholar] [CrossRef]
- Larkin, R.P.; Honeycutt, C.W. Effects of Different 3-Year Cropping Systems on Soil Microbial Communities and Rhizoctonia Diseases of Potato. Phytopathology 2006, 96, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Djébali, N.; Belhassen, T. Field study of the relative susceptibility of eleven potato (Solanum tuberosum L.) varieties and the efficacy of two fungicides against Rhizoctonia solani attack. Crop Prot. 2010, 29, 998–1002. [Google Scholar] [CrossRef]
- Olanya, O.M.; Lambert, D.H.; Reeves§, A.F.; Porter, G.A. Evaluation of potato clones for resistance to stem canker and tuber black scurf in field studies following artificial inoculation with Rhizoctonia solani AG-3 in Maine. Arch. Phytopathol. Plant Prot. 2009, 42, 409–418. [Google Scholar] [CrossRef]
- Okubara, P.A.; Dickman, M.B.; Blechl, A.E. Molecular and genetic aspects of controlling the soilborne necrotrophic pathogens Rhizoctonia and Pythium. Plant Sci. 2014, 228, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-J.; Wang, A.-R.; Shi, Y.-J.; Wang, L.-Q.; Liu, W.-D.; Wang, Z.-H.; Lu, G.-D. Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor. Appl. Genet. 2008, 116, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Foley, R.C.; Kidd, B.N.; Hane, J.K.; Anderson, J.P.; Singh, K.B. Reactive Oxygen Species Play a Role in the Infection of the Necrotrophic Fungi, Rhizoctonia solani in Wheat. PLoS ONE 2016, 11, e0152548. [Google Scholar] [CrossRef]
- Zhu, C.; Ai, L.; Wang, L.; Yin, P.; Liu, C.; Li, S.; Zeng, H. De novo Transcriptome Analysis of Rhizoctonia solani AG1 IA Strain Early Invasion in Zoysia japonica Root. Front. Microbiol. 2016, 7, 708. [Google Scholar] [CrossRef]
- Yadav, S.; Anuradha, G.; Kumar, R.R.; Vemireddy, L.R.; Sudhakar, R.; Donempudi, K.; Venkata, D.; Jabeen, F.; Narasimhan, Y.K.; Marathi, B.; et al. Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). SpringerPlus 2015, 4, 175. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Fu, C.; Wang, L.; Liu, H.; Cheng, Y.; Li, S.; Deng, Q.; Wang, S.; Zhu, J.; et al. Comparative Transcriptome Analyses of Gene Expression Changes Triggered by Rhizoctonia solani AG1 IA Infection in Resistant and Susceptible Rice Varieties. Front. Plant Sci. 2017, 8, 1422. [Google Scholar] [CrossRef]
- Sedláková, V.; Dejmalová, J.; Doležal, P.; Hausvater, E.; Sedlák, P.; Baštová, P. Characterization of forty-four potato varieties for resistance to common scab, black scurf and silver scurf. Crop Prot. 2013, 48, 82–87. [Google Scholar] [CrossRef]
- Hejazi, R.; Esfahani, M.N.; Maleki, M.; Sedaghatfar, E. Susceptibility assessment and genetic population structure associated with Rhizoctonia solani AG3-PT—Potato stem canker disease. Physiol. Mol. Plant Pathol. 2022, 119, 101835. [Google Scholar] [CrossRef]
- Lehtonen, M.J.; Somervuo, P.; Valkonen, J.P.T. Infection with Rhizoctonia solani Induces Defense Genes and Systemic Resistance in Potato Sprouts Grown without Light. Phytopathology 2008, 98, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Hejazi, R.; Esfahani, M.N.; Maleki, M.; Sedaghatfar, E. Comparative analysis of putative pathogenesis-related gene expression in potato associated with defense responses to Rhizoctonia solani. Sydowia 2022, 74, 251–262. [Google Scholar] [CrossRef]
- Soheili-Moghaddam, B.; Nasr-Esfahani, M.; Mousanejad, S.; Hassanzadeh-Khankahdani, H.; Karbalaie-Khiyavie, H. Biochemical defense mechanism associated with host-specific disease resistance pathways against Rhizoctonia solani AG3-PT potatoes canker disease. Planta 2023, 257, 13. [Google Scholar] [CrossRef]
- Zhao, B.; Liu, Q.; Wang, B.; Yuan, F. Roles of Phytohormones and Their Signaling Pathways in Leaf Development and Stress Responses. J. Agric. Food Chem. 2021, 69, 3566–3584. [Google Scholar] [CrossRef]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Kmiecik, P.; Willems, P.; Van Der Kelen, K.; Coll, N.S.; Teige, M.; Van Breusegem, F. Plant innate immunity—Sunny side up? Trends Plant Sci. 2015, 20, 3–11. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic Acquired Resistance and Salicylic Acid: Past, Present, and Future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Bauters, L.; Stojilković, B.; Gheysen, G. Pathogens pulling the strings: Effectors manipulating salicylic acid and phenylpropanoid biosynthesis in plants. Mol. Plant Pathol. 2021, 22, 1436–1448. [Google Scholar] [CrossRef]
- Nováková, M.; Šašek, V.; Dobrev, P.I.; Valentová, O.; Burketová, L. Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum—Reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 2014, 80, 308–317. [Google Scholar] [CrossRef]
- Jia, C.; Zhang, L.; Liu, L.; Wang, J.; Li, C.; Wang, Q. Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J. Exp. Bot. 2013, 64, 637–650. [Google Scholar] [CrossRef]
- An, C.; Mou, Z. Salicylic Acid and its Function in Plant Immunity. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Genzel, F.; Franken, P.; Witzel, K.; Grosch, R. Systemic induction of salicylic acid-related plant defences in potato in response to Rhizoctonia solani AG3PT. Plant Pathol. 2018, 67, 337–348. [Google Scholar] [CrossRef]
- Zrenner, R.; Verwaaijen, B.; Genzel, F.; Flemer, B.; Grosch, R. Transcriptional Changes in Potato Sprouts upon Interaction with Rhizoctonia solani Indicate Pathogen-Induced Interference in the Defence Pathways of Potato. Int. J. Mol. Sci. 2021, 22, 3094. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Van Buyten, E.; Satoh, K.; Balidion, J.; Mauleon, R.; Choi, I.-R.; Vera-Cruz, C.; Kikuchi, S.; Höfte, M. Brassinosteroids Antagonize Gibberellin- and Salicylate-Mediated Root Immunity in Rice. Plant Physiol. 2012, 158, 1833–1846. [Google Scholar] [CrossRef]
- Denancé, N.; Ranocha, P.; Oria, N.; Barlet, X.; Rivière, M.P.; Yadeta, K.A.; Hoffmann, L.; Perreau, F.; Clément, G.; Maia-Grondard, A.; et al. Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 2013, 73, 225–239. [Google Scholar] [CrossRef]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in Systemic Plant Immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Segarra, G.; Jáuregui, O.; Casanova, E.; Trillas, I. Simultaneous quantitative LC–ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 2006, 67, 395–401. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.A.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Meuwly, P.; Metraux, J.P. Ortho-Anisic Acid as Internal Standard for the Simultaneous Quantitation of Salicylic Acid and Its Putative Biosynthetic Precursors in Cucumber Leaves. Anal. Biochem. 1993, 214, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Métraux, J.-P. Salicylic Acid Induction–Deficient Mutants of Arabidopsis Express PR-2 and PR-5 and Accumulate High Levels of Camalexin after Pathogen Inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [CrossRef]
- Nietzsche, M.; Guerra, T.; Alseekh, S.; Wiermer, M.; Sonnewald, S.; Fernie, A.R.; Börnke, F. STOREKEEPER RELATED1/G-Element Binding Protein (STKR1) Interacts with Protein Kinase SnRK1. Plant Physiol. 2017, 176, 1773–1792. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Wanke, A.; Malisic, M.; Wawra, S.; Zuccaro, A. Unraveling the sugar code: The role of microbial extracellular glycans in plant-microbe interactions. J. Exp. Bot. 2021, 72, 15–35. [Google Scholar] [CrossRef]
- Mauch, F.; Mauch-Mani, B.; Boller, T. Antifungal Hydrolases in Pea Tissue: II. Inhibition of Fungal Growth by Combinations of Chitinase and beta-1,3-Glucanase. Plant Physiol. 1988, 88, 936–942. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Van Dooijeweert, W.; Govers, F.; Kamoun, S.; Colon, L.T. Does basal PR gene expression in Solanum species contribute to non-specific resistance to Phytophthora infestans? Physiol. Mol. Plant Pathol. 2000, 57, 35–42. [Google Scholar] [CrossRef]
- Vergne, E.; Grand, X.; Ballini, E.; Chalvon, V.; Saindrenan, P.; Tharreau, D.; Nottéghem, J.L.; Morel, J.B. Preformed expression of defense is a hallmark of partial resistance to rice blast fungal pathogen Magnaporthe oryzae. BMC Plant Biol. 2010, 10, 206. [Google Scholar] [CrossRef]
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 30. [Google Scholar] [CrossRef]
- Koley, P.; Brahmachari, S.; Saha, A.; Deb, C.; Mondal, M.; Das, N.; Das, A.; Lahiri, S.; Das, M.; Thakur, M.; et al. Phytohormone Priming of Tomato Plants Evoke Differential Behavior in Rhizoctonia solani During Infection, with Salicylate Priming Imparting Greater Tolerance than Jasmonate. Front. Plant Sci. 2022, 12, 766095. [Google Scholar] [CrossRef]
- Taheri, P.; Tarighi, S. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 2010, 167, 201–208. [Google Scholar] [CrossRef] [PubMed]
- El Oirdi, M.; El Rahman, T.A.; Rigano, L.; El Hadrami, A.; Rodriguez, M.C.; Daayf, F.; Vojnov, A.; Bouarab, K. Botrytis cinerea Manipulates the Antagonistic Effects between Immune Pathways to Promote Disease Development in Tomato. Plant Cell 2011, 23, 2405–2421. [Google Scholar] [CrossRef]
- Rahman, T.A.E.; Oirdi, M.E.; Gonzalez-Lamothe, R.; Bouarab, K. Necrotrophic Pathogens Use the Salicylic Acid Signaling Pathway to Promote Disease Development in Tomato. Mol. Plant-Microbe Interact. 2012, 25, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, S.M.; Odilbekov, F.; Burra, D.D.; Lenman, M.; Hedley, P.E.; Grenville-Briggs, L.; Alexandersson, E.; Liljeroth, E.; Andreasson, E. Intact salicylic acid signalling is required for potato defence against the necrotrophic fungus Alternaria solani. Plant Mol. Biol. 2020, 104, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Garcia, V.; Portal Onco, M.A.; Rubio Susan, V. Review. Biology and systematics of the form genus Rhizoctonia. Span. J. Agric. Res. 2006, 4, 55–79. [Google Scholar] [CrossRef]
- Liu, L.; Sonbol, F.-M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef]

| Cultivar | DS | PR1 | PR2 | PR3 | PR6 | PR10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 5.3 | abc | 1.63 | 2.3 | abc | 0.62 | 4.4 | a | 0.97 | 6.2 | abc | 1.35 | 5.9 | b | 0.29 |
| Lolita | low | 3.1 | bcd | 1.77 | −2.3 | d | 0.52 | 2.4 | a | 1.80 | 6.2 | abc | 1.26 | 5.4 | b | 0.25 |
| Troja | low | 2.2 | cd | 0.98 | −1.7 | d | 1.26 | 3.8 | a | 0.84 | 6.4 | ab | 1.01 | 5.6 | b | 0.68 |
| Salute | low | 0.3 | d | 0.29 | −0.4 | cd | 0.67 | 4.1 | a | 0.35 | 6.4 | abc | 1.32 | 5.8 | b | 0.20 |
| Arkula | high | 6.3 | ab | 3.68 | 0.5 | bcd | 1.76 | 4.6 | a | 2.68 | 6.8 | a | 1.16 | 5.1 | b | 0.73 |
| Jasia | high | 4.3 | a | 0.58 | 2.8 | a | 0.70 | 3.5 | a | 1.87 | 4.4 | d | 0.87 | 6.7 | a | 0.43 |
| Skonto | high | 4.4 | abc | 1.13 | 2.6 | ab | 2.50 | 4.0 | a | 1.66 | 4.1 | bcd | 1.03 | 6.5 | b | 1.06 |
| Bonza | high | 8.0 | abc | 2.11 | 4.6 | ab | 1.49 | 5.4 | a | 1.00 | 3.9 | cd | 0.70 | 9.0 | b | 1.70 |
| t-test | * | * | ns | * | * | |||||||||||
| Cultivar | DS | PR1 | PR2 | PR3 | PR6 | PR10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | S | SD | SD | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 1.7 | a | 0.71 | 0.90 | b | 2.54 | 3.4 | a | 0.50 | 3.4 | a | 0.90 | 0.4 | b | 1.42 |
| Lolita | low | −0.6 | a | 2.40 | 0.76 | a | 0.56 | 2.8 | a | 1.33 | 3.6 | a | 0.76 | 10.3 | a | 0.76 |
| Troja | low | −0.2 | a | 1.19 | 0.99 | a | 1.15 | 3.0 | a | 1.31 | 3.2 | a | 0.99 | 8.9 | a | 0.79 |
| Salute | low | −0.2 | a | 1.42 | 0.84 | a | 0.29 | 2.9 | a | 0.66 | 3.1 | a | 0.84 | 9.5 | a | 0.84 |
| Arkula | high | 1.2 | a | 0.56 | 0.98 | b | 2.20 | 4.2 | a | 1.67 | 3.4 | a | 0.98 | 0.1 | b | 1.02 |
| Jasia | high | 0.7 | a | 2.08 | 0.71 | a | 0.70 | 3.1 | a | 3.88 | 2.5 | a | 0.71 | 9.4 | a | 1.46 |
| Skonto | high | −1.0 | a | 1.12 | 1.30 | a | 0.33 | 3.5 | a | 0.40 | 2.4 | a | 1.30 | 8.8 | a | 0.56 |
| Bonza | high | 0.3 | a | 1.88 | 0.62 | a | 0.23 | 3.2 | a | 1.36 | 1.9 | a | 0.62 | 11.0 | a | 1.96 |
| t-test | ns | ns | ns | * | ns | |||||||||||
| Shoots | Roots | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cultivar | DS | PAL | ICS | PAL | ICS | ||||||||
| MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 6.3 | a | 1.67 | 2.0 | bc | 0.38 | 8.2 | a | 0.70 | 1.4 | bc | 0.72 |
| Lolita | low | 1.7 | b | 0.73 | 1.1 | c | 0.18 | 1.7 | b | 0.74 | 1.0 | bc | 0.36 |
| Troja | low | 2.1 | b | 0.76 | 1.4 | c | 0.73 | 1.6 | b | 0.55 | 2.2 | ab | 0.48 |
| Salute | low | 5.8 | a | 0.53 | 1.1 | c | 0.37 | 7.7 | a | 0.99 | 1.4 | bc | 0.19 |
| Arkula | high | 6.2 | a | 0.95 | 1.6 | c | 0.95 | 8.1 | a | 0.96 | 1.1 | bc | 0.88 |
| Jasia | high | 2.3 | b | 0.93 | 3.1 | a | 0.68 | 2.4 | b | 0.69 | 1.4 | a | 0.85 |
| Skonto | high | 1.8 | b | 0.85 | 1.5 | ab | 0.45 | 1.2 | b | 0.84 | 0.1 | bc | 0.25 |
| Bonza | high | 1.3 | b | 0.41 | 4.4 | c | 0.88 | 1.8 | b | 0.96 | 3.5 | c | 0.96 |
| t-test | ns | * | ns | ns | |||||||||
| Cultivar | DS | PR1 | PR2 | PR3 | PR6 | PR10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 5.1 | bc | 1.14 | 3.2 | a | 1.31 | 7.8 | a | 1.74 | 6.7 | b | 1.05 | 8.3 | a | 1.42 |
| Lolita | low | 3.9 | bc | 1.00 | 1.6 | a | 0.89 | 7.0 | a | 1.23 | 6.1 | b | 0.80 | 8.1 | a | 1.03 |
| Troja | low | 5.9 | abc | 2.34 | 2.5 | a | 1.94 | 7.7 | a | 2.24 | 7.2 | ab | 2.13 | 8.5 | a | 1.78 |
| Salute | low | 3.4 | c | 0.87 | 2.9 | a | 1.02 | 7.7 | a | 1.40 | 6.6 | b | 1.63 | 8.0 | a | 0.84 |
| Arkula | high | 7.4 | abc | 0.40 | 3.3 | a | 1.04 | 7.6 | a | 1.43 | 6.9 | b | 1.76 | 8.1 | a | 1.32 |
| Jasia | high | 4.8 | ab | 0.63 | 2.4 | a | 1.01 | 8.1 | a | 0.75 | 8.1 | ab | 0.40 | 7.6 | a | 0.20 |
| Skonto | high | 9.3 | bc | 2.34 | 5.5 | a | 2.24 | 10.2 | a | 2.45 | 11.1 | ab | 2.02 | 10.0 | a | 2.14 |
| Bonza | high | 7.9 | a | 1.35 | 5.3 | a | 1.88 | 9.0 | a | 1.62 | 8.0 | a | 0.86 | 9.3 | a | 1.19 |
| t-test | * | * | ns | * | ns | |||||||||||
| Cultivar | DS | PR1 | PR2 | PR3 | PR6 | PR10 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 2.4 | bc | 0.30 | 6.1 | ab | 1.27 | 6.5 | a | 1.46 | 3.8 | abc | 1.04 | 9.4 | ab | 0.95 |
| Lolita | low | 1.7 | bc | 0.94 | 5.6 | ab | 1.76 | 4.1 | a | 0.78 | 3.9 | abc | 1.25 | 9.2 | ab | 0.57 |
| Troja | low | 2.1 | bc | 0.65 | 4.6 | ab | 1.62 | 4.5 | a | 0.25 | 2.1 | c | 0.91 | 8.4 | b | 0.44 |
| Salute | low | 1.3 | c | 1.55 | 3.8 | b | 2.07 | 3.4 | a | 0.30 | 2.7 | bc | 0.63 | 8.6 | b | 0.56 |
| Arkula | high | 1.9 | bc | 0.56 | 4.5 | ab | 0.86 | 4.6 | a | 1.40 | 2.9 | abc | 0.09 | 9.8 | ab | 1.37 |
| Jasia | high | 3.5 | a | 0.79 | 6.1 | a | 0.40 | 5.4 | a | 1.18 | 3.4 | a | 0.97 | 9.2 | ab | 0.88 |
| Skonto | high | 2.7 | ab | 1.28 | 5.9 | ab | 1.18 | 6.6 | a | 2.50 | 4.3 | abc | 0.88 | 11.4 | ab | 1.80 |
| Bonza | high | 4.9 | bc | 0.26 | 7.2 | ab | 0.37 | 5.3 | a | 0.30 | 4.9 | ab | 0.47 | 9.1 | a | 0.20 |
| t-test | * | ns | ns | ns | * | |||||||||||
| Shoots | Roots | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cultivar | DS | PAL | ICS | PAL | ICS | ||||||||
| MV | S | SD | MV | S | SD | MV | S | SD | MV | S | SD | ||
| Granola | low | 5.5 | a | 0.74 | 1.7 | a | 0.67 | 6.7 | a | 0.89 | −1.7 | a | 1.45 |
| Lolita | low | −2.6 | b | 0.45 | 1.2 | a | 0.63 | −0.7 | b | 0.76 | −2.2 | a | 0.35 |
| Troja | low | −2.5 | b | 1.32 | 1.5 | a | 0.87 | −1.3 | b | 1.38 | −2.5 | a | 0.63 |
| Salute | low | 5.4 | a | 0.59 | 1.6 | a | 0.14 | 6.8 | a | 1.11 | −1.4 | a | 0.60 |
| Arkula | high | 4.5 | a | 1.57 | 1.5 | a | 0.99 | 6.5 | a | 0.39 | −0.6 | a | 1.49 |
| Jasia | high | 6.6 | b | 0.48 | 1.8 | a | 0.36 | 7.9 | b | 0.79 | −2.3 | a | 1.25 |
| Skonto | high | −1.1 | a | 1.21 | 1.8 | a | 0.66 | −0.7 | a | 0.92 | −0.6 | a | 1.01 |
| Bonza | high | −2.0 | b | 1.01 | 1.9 | a | 0.82 | −1.4 | b | 1.07 | −2.0 | a | 0.40 |
| t-test | ns | ns | ns | ns | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zrenner, R.; Genzel, F.; Baldermann, S.; Guerra, T.; Grosch, R. Does Constitutive Expression of Defense-Related Genes and Salicylic Acid Concentrations Correlate with Field Resistance of Potato to Black Scurf Disease? Bioengineering 2023, 10, 1244. https://doi.org/10.3390/bioengineering10111244
Zrenner R, Genzel F, Baldermann S, Guerra T, Grosch R. Does Constitutive Expression of Defense-Related Genes and Salicylic Acid Concentrations Correlate with Field Resistance of Potato to Black Scurf Disease? Bioengineering. 2023; 10(11):1244. https://doi.org/10.3390/bioengineering10111244
Chicago/Turabian StyleZrenner, Rita, Franziska Genzel, Susanne Baldermann, Tiziana Guerra, and Rita Grosch. 2023. "Does Constitutive Expression of Defense-Related Genes and Salicylic Acid Concentrations Correlate with Field Resistance of Potato to Black Scurf Disease?" Bioengineering 10, no. 11: 1244. https://doi.org/10.3390/bioengineering10111244
APA StyleZrenner, R., Genzel, F., Baldermann, S., Guerra, T., & Grosch, R. (2023). Does Constitutive Expression of Defense-Related Genes and Salicylic Acid Concentrations Correlate with Field Resistance of Potato to Black Scurf Disease? Bioengineering, 10(11), 1244. https://doi.org/10.3390/bioengineering10111244






